Abstract
Survival for patients with acute myeloid leukemia (AML) is limited by treatment-related mortality (TRM) and relapse after unrelated donor (URD) hematopoietic cell transplantation (HCT). Natural killer (NK)–cell alloreactivity, determined by donor killer-cell immunoglobulin-like receptors (KIRs) and recipient HLA, correlates with successful HCT for AML. Hypothesizing that donor KIR genotype (A/A: 2 A KIR haplotypes; B/x: at least 1 B haplotype) would affect outcomes, we genotyped donors and recipients from 209 HLA-matched and 239 mismatched T-replete URD transplantations for AML. Three-year overall survival was significantly higher after transplantation from a KIR B/x donor (31% [95% CI: 26-36] vs 20% [95% CI: 13-27]; P = .007). Multivariate analysis demonstrated a 30% improvement in the relative risk of relapse-free survival with B/x donors compared with A/A donors (RR: 0.70 [95% CI: 0.55-0.88]; P = .002). B/x donors were associated with a higher incidence of chronic graft-versus-host disease (GVHD; RR: 1.51 [95% CI: 1.01-2.18]; P = .03), but not of acute GVHD, relapse, or TRM. This analysis demonstrates that unrelated donors with KIR B haplotypes confer significant survival benefit to patients undergoing T-replete HCT for AML. KIR genotyping of prospective donors, in addition to HLA typing, should be performed to identify HLA-matched donors with B KIR haplotypes.
Introduction
Allogeneic hematopoietic cell transplantation (HCT) using related or unrelated donors (URDs) is standard treatment for many patients with hematologic malignancies who are unlikely to be cured by chemotherapy alone. Although the most important variable for donor selection is the match for HLA class I and class II, other relevant factors include donor sex, parity, cytomegalovirus (CMV) serostatus, and age.1 Alloreactive donor-derived natural killer (NK) cells have been correlated with improved survival after HCT for acute myelogenous leukemia, and are thought to promote engraftment, reduce graft-versus-host disease (GVHD), and decrease leukemic relapse.2,3 NK-cell function is determined by the net effect of signaling through several receptor families, including activating and inhibitory killer-cell immunoglobulin-like receptors (KIRs). Although the ligands for activating KIRs remain elusive, the interactions between inhibitory KIRs on the donor-derived NK cells and HLA class I molecules on the recipient's healthy and leukemic cells determine NK-cell alloreactivity. All HLA-C allotypes have either the C1 epitope, the ligand for KIR2DL2/3, or the C2 epitope, the ligand for KIR2DL1. Analogously, all HLA-B allotypes have either the Bw4 or Bw6 epitope, but only the Bw4 epitope is a ligand for KIRs: its cognate inhibitory receptor being KIR3DL1.4 In healthy individuals, interactions of inhibitory KIRs with cognate HLA class I prevent NK cells from attacking healthy cells.5 In the setting of allogeneic transplantation, donor NK cells attack the allogeneic cells if the recipient HLA class I ligands do not sufficiently engage their inhibitory receptors.
The KIR genes on chromosome 19 segregate independently of HLA, with the important consequence for unrelated allogeneic transplantation that matching for HLA does not match for KIRs. Diverse KIR haplotypes can be simplified into 2 biologically distinct groups, A and B. Group A haplotypes have a fixed number of genes that encode inhibitory receptors with the exception of 2DS4, whereas group B haplotypes have variable gene content including additional activating receptor genes. These KIR haplotypes have been associated with reproductive success, responses to viral infections such as HIV and hepatitis C virus (HCV), susceptibility to autoimmune disease, and outcomes of HCT.6-8 All individuals can be categorized as having 1 of 2 KIR genotypes: A/A, which is homozygous for group A KIR haplotypes, or B/x, which contains either 1 (A/B heterozygotes) or 2 (B/B homozygotes) group B haplotypes. Previous HCT studies have reported varied and sometimes inconsistent associations between KIR haplotype and clinical outcome.9-16 The results were confounded by the small size and heterogeneity of the cohorts studied. In aggregate, these results suggested the hypothesis that combining KIR and HLA genotyping could help selection of transplant donors and improve the outcome of transplantation. To test this hypothesis, we analyzed the effects of KIR genotype in a homogeneous cohort of 448 AML patients who received transplants from unrelated donors.
Methods
Population
Collection of DNA samples from 448 donor/recipient pairs from URD HCT performed to treat AML was facilitated by the National Marrow Donor Program (NMDP, Minneapolis, MN) Research Sample Repository. Outcome data were obtained from the Center for International Blood and Marrow Transplant Research (CIBMTR, Milwaukee, WI). Samples and clinical data were collected after informed consent and approval from the NMDP and University of Minnesota Institutional Review Boards were obtained in accordance with the Declaration of Helsinki. To decrease the cohort's clinical heterogeneity, samples from 3 groups (HLA matched/KIR-ligand matched, HLA mismatched/KIR-ligand matched, and HLA-mismatched/KIR-ligand mismatched) were selected using an algorithm that matched on important demographic and clinical variables (Table 1). KIR-ligand matching status was determined using high-resolution HLA-B and HLA-C genotypes,17 following the algorithm available on the KIR-ligand calculator tool maintained by the HLA Informatics Group of the Anthony Nolan Research Institute (http://www.ebi.ac.uk/ipd/kir/).18
Patient demographics
| . | Donor KIR genotype . | P* . | |
|---|---|---|---|
| Genotype A/A, n = 131 (29%) . | Genotype B/x, n = 317 (71%) . | ||
| Median age, y (range) | 34.1 (1.8-58.4) | 33.3 (0.8-60.8) | .57 |
| Disease status (%) | .53 | ||
| Early | 25 (19.1) | 61 (19.2) | |
| Intermediate | 42 (32.1) | 118 (37.2) | |
| Advanced | 64 (48.9) | 138 (43.5) | |
| Donor: recipient HLA-allele matching at A, B, C, DRB1, and DQB1 (%) | .01 | ||
| 10/10 allele matched | 51 (38.9) | 158 (49.8) | |
| 9/10 | 28 (21.4) | 67 (21.1) | |
| 8/10 | 39 (29.8) | 51 (16.1) | |
| 7/10 | 13 (9.9) | 41 (12.9) | |
| HLA match group (%) | .10 | ||
| HLA 10/10 allele matched | 51 (38.9) | 158 (49.8) | |
| HLA mismatched KIR-ligand matched | 58 (44.3) | 111 (35.0) | |
| HLA mismatched KIR-ligand mismatched | 22 (16.8) | 48 (15.1) | |
| Sex match, donor/recipient (%) | .33 | ||
| Male/male | 40 (30.5) | 117 (36.9) | |
| Male/female | 32 (24.4) | 85 (26.8) | |
| Female/male | 24 (18.3) | 52 (16.4) | |
| Female/female | 35 (26.7) | 63 (19.9) | |
| CMV serostatus (%) | .62 | ||
| Donor −/recipient − | 40 (30.5) | 106 (33.4) | |
| Donor +/recipient − | 17 (13.0) | 47 (14.8) | |
| Donor any/recipient + | 73 (55.7) | 160 (50.5) | |
| Missing | 1 (0.8) | 4 (1.3) | |
| Graft type (%) | .89 | ||
| Bone marrow | 116 (88.5) | 281 (88.6) | |
| Peripheral blood progenitor cells | 15 (11.5) | 36 (11.4) | |
| Karnofsky score (%) | .08 | ||
| 90 to 100 | 77 (58.8) | 196 (61.8) | |
| 10 to 80 | 52 (39.7) | 103 (32.5) | |
| Unknown | 2 (1.5) | 18 (5.7) | |
| Median time from diagnosis to transplantation y (range) | 1.1 (0.0-4.0) | 1.2 (0.0-8.2) | .46 |
| Race (%) | .91 | ||
| White | 117 (89.3) | 286 (90.2) | |
| Nonwhite | 14 (10.7) | 31 (10.8) | |
| . | Donor KIR genotype . | P* . | |
|---|---|---|---|
| Genotype A/A, n = 131 (29%) . | Genotype B/x, n = 317 (71%) . | ||
| Median age, y (range) | 34.1 (1.8-58.4) | 33.3 (0.8-60.8) | .57 |
| Disease status (%) | .53 | ||
| Early | 25 (19.1) | 61 (19.2) | |
| Intermediate | 42 (32.1) | 118 (37.2) | |
| Advanced | 64 (48.9) | 138 (43.5) | |
| Donor: recipient HLA-allele matching at A, B, C, DRB1, and DQB1 (%) | .01 | ||
| 10/10 allele matched | 51 (38.9) | 158 (49.8) | |
| 9/10 | 28 (21.4) | 67 (21.1) | |
| 8/10 | 39 (29.8) | 51 (16.1) | |
| 7/10 | 13 (9.9) | 41 (12.9) | |
| HLA match group (%) | .10 | ||
| HLA 10/10 allele matched | 51 (38.9) | 158 (49.8) | |
| HLA mismatched KIR-ligand matched | 58 (44.3) | 111 (35.0) | |
| HLA mismatched KIR-ligand mismatched | 22 (16.8) | 48 (15.1) | |
| Sex match, donor/recipient (%) | .33 | ||
| Male/male | 40 (30.5) | 117 (36.9) | |
| Male/female | 32 (24.4) | 85 (26.8) | |
| Female/male | 24 (18.3) | 52 (16.4) | |
| Female/female | 35 (26.7) | 63 (19.9) | |
| CMV serostatus (%) | .62 | ||
| Donor −/recipient − | 40 (30.5) | 106 (33.4) | |
| Donor +/recipient − | 17 (13.0) | 47 (14.8) | |
| Donor any/recipient + | 73 (55.7) | 160 (50.5) | |
| Missing | 1 (0.8) | 4 (1.3) | |
| Graft type (%) | .89 | ||
| Bone marrow | 116 (88.5) | 281 (88.6) | |
| Peripheral blood progenitor cells | 15 (11.5) | 36 (11.4) | |
| Karnofsky score (%) | .08 | ||
| 90 to 100 | 77 (58.8) | 196 (61.8) | |
| 10 to 80 | 52 (39.7) | 103 (32.5) | |
| Unknown | 2 (1.5) | 18 (5.7) | |
| Median time from diagnosis to transplantation y (range) | 1.1 (0.0-4.0) | 1.2 (0.0-8.2) | .46 |
| Race (%) | .91 | ||
| White | 117 (89.3) | 286 (90.2) | |
| Nonwhite | 14 (10.7) | 31 (10.8) | |
P values from Welch t test or a χ2 test.
Procedures
KIR genotyping.
The presence or absence of 16 KIR genes (KIR2DL5A and KIR2DL5B were considered together) was determined using a high-throughput SNP-based Sequenom MassARRAY system (Sequenom, San Diego, CA) and the matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) platform for the large scale KIR genotyping of DNA samples as previously validated.19
Haplotype assignment.
Detection of at least 1 of the KIR B haplotype-defining loci (KIR2DL5, 2DS1, 2DS2, 2DS3, 2DS5, or 3DS1) in a sample dictated that the genotype contains at least 1 B haplotype. Such samples were assigned the genotype designation B/x. Samples lacking all KIR B loci were assigned the genotype A/A.
Statistical analysis
We considered 6 outcomes in the analysis: (survival, relapse-free survival [RFS], relapse, treatment-related mortality [TRM], and grades II-IV acute and chronic GVHD). Overall survival (OS) and RFS were summarized by Kaplan-Meier curves, whereas the other 4 events were summarized by cumulative incidence functions. OS and RFS were compared between KIR genotype groups using the log-rank test. Other outcomes were compared using Gray test.20 The completeness of follow-up was more than 90% at 3 years after follow-up and 95% of events occurred by 3 years after HCT.
For OS and RFS, a Cox proportional hazards model was used to adjust for important clinical factors. All variables were tested for proportional hazards using a time-dependent covariate approach. The multivariate model accounted for HLA matching, and other covariates listed in Table 1 were examined using backward stepwise selection. Variables were removed when the multiple degree of freedom test P value exceeded .10. A second model that stratified on HLA type gave similar results.
For the other events (relapse, TRM, acute and chronic GVHD), the Cox model was replaced by the model of Fine and Gray for the subdistributional hazard based on the cumulative incidence function.21 Relapse and death were considered as competing risks.
Results
The 448 AML patients underwent URD myeloablative, T cell–replete transplantations facilitated by the National Marrow Donor Program between 1988 and 2003. The cohort consisted of 3 HLA-matched groups (Table 1). Approximately half (47%) were 10/10 HLA-allele matched (and thus KIR-ligand matched) at HLA-A, -B, -C, -DRB1, and -DQB1. The HLA-mismatched group (53%) was further divided into KIR-ligand–mismatched (29%) and KIR ligand–matched (71%) groups. KIR genotyping for the presence of 16 KIR genes shows that the donors and recipients in this predominantly white population had similar and typical KIR frequencies. In this cohort, 29.2% of donors and 34.8% of recipients had the A/A KIR genotype; the remainder had the B/x genotype. There were no significant differences in KIR gene or haplotype frequencies between the 3 HLA match groups; nor did they vary for transplantations performed at different times during the period 1988 to 2002: in any given year approximately 66% of donors were B/x.
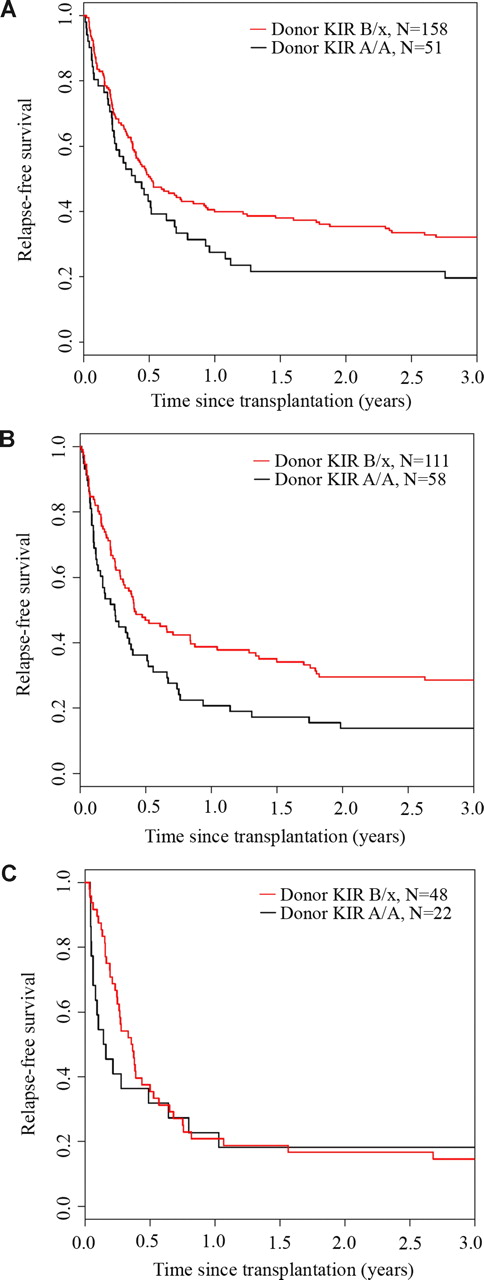
We analyzed the effect of donor and recipient KIR genotype on outcomes after URD HCT for AML. Whereas the recipient KIR haplotype content had no effect on relapse-free survival (P = .75), KIR B/x donors were associated with significantly better 3-year relapse-free survival (donor KIR B/x: 28% [95% CI: 23-33] vs donor KIR A/A: 17% [95% CI: 11-24]; P = .003) (Figure 1). In univariate analysis, 3-year overall survival was also significantly higher after transplantation using a B/x donor (31% [95% CI: 26-36] vs 20% [95% CI: 13-27]; P = .007) (Figure 2). Stratifying patients by HLA matching, we found that the 3-year relapse-free survival was higher with KIR B/x versus KIR A/A donors after 10/10 allele HLA-matched/KIR ligand–matched (32% vs 20%) and HLA-mismatched but KIR ligand–matched (29% vs 14%) transplantations, but not in the cohort that received HLA-mismatched/KIR ligand–mismatched transplants (15% vs 18%) (Figure 3).
Transplantation using donors with a KIR B haplotype improves relapse-free survival (RFS) irrespective of the recipient KIR haplotype status. Donor (D) and recipient (R) DNA samples from 448 URD transplants were genotyped for 16 KIR loci. KIR gene content was used to identify haplotypes (A or B), from which KIR genotypes were assigned (KIR A/A or KIR B/x). The Kaplan-Meier curves demonstrate RFS for each donor/recipient genotype pairing (A/A into A/A, A/A into B/x, B/x into A/A, and B/x into B/x). RFS was significantly better after transplantation using KIR B/x donors (28% [95% CI: 23-33]; n = 317) compared with A/A donors (17% [11%-24%]; n = 131; P = .003).The recipient KIR genotype had no effect (P = .75).
Transplantation using donors with a KIR B haplotype improves relapse-free survival (RFS) irrespective of the recipient KIR haplotype status. Donor (D) and recipient (R) DNA samples from 448 URD transplants were genotyped for 16 KIR loci. KIR gene content was used to identify haplotypes (A or B), from which KIR genotypes were assigned (KIR A/A or KIR B/x). The Kaplan-Meier curves demonstrate RFS for each donor/recipient genotype pairing (A/A into A/A, A/A into B/x, B/x into A/A, and B/x into B/x). RFS was significantly better after transplantation using KIR B/x donors (28% [95% CI: 23-33]; n = 317) compared with A/A donors (17% [11%-24%]; n = 131; P = .003).The recipient KIR genotype had no effect (P = .75).
Transplantation using donors with KIR B haplotypes improves overall survival. The univariate Kaplan-Meier curves for overall survival (OS) are shown for patients receiving transplants from donors with (KIR B/x; n = 317) or without (KIR A/A; n = 131) KIR B haplotypes. Survival is significantly better if the donor had a KIR B haplotype (31% [95% CI: 26-36] vs 20% [95% CI: 13-27]; P = .007).
Transplantation using donors with KIR B haplotypes improves overall survival. The univariate Kaplan-Meier curves for overall survival (OS) are shown for patients receiving transplants from donors with (KIR B/x; n = 317) or without (KIR A/A; n = 131) KIR B haplotypes. Survival is significantly better if the donor had a KIR B haplotype (31% [95% CI: 26-36] vs 20% [95% CI: 13-27]; P = .007).
Relapse-free survival benefit from KIR B haplotype donors after HLA-matched and HLA-matched/KIR ligand–matched transplantation. The univariate Kaplan-Meier curves demonstrate the relapse-free survival (RFS) for patients receiving transplants from donors with or without KIR B haplotypes who were (A) 10/10 HLA matched, (B) HLA mismatched and KIR-ligand matched, or (C) HLA mismatched and KIR-ligand mismatched.
Relapse-free survival benefit from KIR B haplotype donors after HLA-matched and HLA-matched/KIR ligand–matched transplantation. The univariate Kaplan-Meier curves demonstrate the relapse-free survival (RFS) for patients receiving transplants from donors with or without KIR B haplotypes who were (A) 10/10 HLA matched, (B) HLA mismatched and KIR-ligand matched, or (C) HLA mismatched and KIR-ligand mismatched.
To explore further the influence of donor KIR haplotypes, a multivariate Cox proportional hazards model was constructed adjusting for statistically significant (P < .10) clinical and demographic factors including HLA match group, disease status (first or greater complete remission [CR] vs primary induction failure [PIF] or relapse), time from diagnosis to transplantation, age, race, and Karnofsky performance status (KPS). After adjustment, the benefit of transplantation using KIR B/x donors (compared with A/A) remained highly significant, with a favorable RR of 3-year relapse-free survival of 0.70 (95% CI: 0.55-0.88; P = .002). Graft type (peripheral blood progenitor cell [PBPC] vs marrow) was not a significant covariate and was not included in the multivariate model. There was no significant interaction between graft type and the donor KIR haplotype effect and the conclusions were unchanged in a subset analysis that excluded those patients (11% of total) who received PBPCs; RR with B/x donors of 3-year relapse-free survival of 0.71 (95% CI: 0.56-0.92; P = .008).
As expected, older and less fit patients being treated for intermediate and advanced leukemia or receiving grafts from HLA-mismatched donors had poorer relapse-free survival (Table 2). The multivariate analysis confirmed that donor KIR B/x genotype conferred a significant relapse-free survival benefit (RR: 0.69 [95% CI: 0.54-0.90), P = .005) for all KIR ligand–matched transplantations (HLA matched and HLA mismatched). We also analyzed the group with 1 or 2 HLA allele mismatches only (n = 185), excluding those with greater degrees of HLA mismatch. In this partially HLA-matched group, the 3-year estimate of relapse-free survival using KIR B/x genotype donors (26% [95% CI: 19-34]) was comparable with the 3-year relapse-free survival estimate for all (donor KIR A/A or B/x) 10/10 HLA-allele–matched transplantations (29% [95% CI: 23-35]). A multivariate Cox model comparing these groups showed no difference (RR: 1.07 [95% CI: 0.76-1.50]), suggesting that favorable donor KIR genotype can, at least in part, overcome the adverse effect of partial HLA mismatch.
Relapse-free survival after URD HCT: multivariate analysis
| All samples . | RR of relapse or death . | 95% CI . | P . |
|---|---|---|---|
| KIR genotype | |||
| B/x (compared with A/A) | 0.70 | 0.55-0.88 | .002 |
| Transplant group | |||
| HLA mismatched/KIR-ligand match (compared with 10/10 HLA matched) | 1.21 | 0.95-1.54 | .13 |
| HLA mismatched/KIR-ligand mismatch (compared with10/10 HLA matched) | 1.65 | 1.22-2.23 | .001 |
| Disease status | |||
| Intermediate (compared with early) | 1.41 | 0.99-2.02 | .06 |
| Advanced (compared with early) | 2.17 | 1.58-2.99 | <.001 |
| Age (categoric) | .03 | ||
| Karnofsky performance status | .01 | ||
| Time from diagnosis to transplantation, per year | 0.88 | 0.77-1.02 | .08 |
| Nonwhite | 1.34 | 0.95-1.88 | .10 |
| All samples . | RR of relapse or death . | 95% CI . | P . |
|---|---|---|---|
| KIR genotype | |||
| B/x (compared with A/A) | 0.70 | 0.55-0.88 | .002 |
| Transplant group | |||
| HLA mismatched/KIR-ligand match (compared with 10/10 HLA matched) | 1.21 | 0.95-1.54 | .13 |
| HLA mismatched/KIR-ligand mismatch (compared with10/10 HLA matched) | 1.65 | 1.22-2.23 | .001 |
| Disease status | |||
| Intermediate (compared with early) | 1.41 | 0.99-2.02 | .06 |
| Advanced (compared with early) | 2.17 | 1.58-2.99 | <.001 |
| Age (categoric) | .03 | ||
| Karnofsky performance status | .01 | ||
| Time from diagnosis to transplantation, per year | 0.88 | 0.77-1.02 | .08 |
| Nonwhite | 1.34 | 0.95-1.88 | .10 |
After establishing that B/x haplotype donors conferred a survival advantage over A/A donors, we performed further analyses to explore the underlying mechanism. The influence of individual donor KIR genes on survival outcomes was examined (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The 2 KIRs showing independent effects on overall survival were KIR2DL2 (P = .07) and KIR2DS2 (P = .02), which are in strong linkage disequilibrium with each other. Of the 448 donors, 246 had both KIR2DL2 and KIR2DS2, 195 had neither, and only 7 had just 1 of them (6 2DS2+/2DL2− and 1 2DS2−/2DL2+). To assess the quantitative contribution of KIR2DS2/L2 to the favorable relapse-free survival benefit using B/x donors, we constructed a multivariate Cox model adjusting for the same covariates and comparing A/A donors to B/x donors who either have or lack KIR2DS2. Transplantation using either KIR B/x genotype group (KIR2DS2+ or KIR2DS2−) was associated with similarly significant improvements in relapse-free survival (2DS2+ vs A/A: RR: 0.70 [95% CI: 0.55-0.89], P = .003; 2DS2− vs A/A: RR: 0.69 [95% CI: 0.49-0.97], P = .03). Thus presence of KIR2DS2 is not uniquely responsible for the survival benefit associated with B KIR haplotypes (Figure 4). Using regression modeling to assess the impact of increasing numbers of activating or inhibitory KIR genes, we found no additive effect. Only the presence of at least 1 (vs none) KIR B haplotype-defining gene was associated with significant improvement in outcome. Nor did we observe any interaction between the donor KIR genotype (A/A or B/x) and the KIR-ligand status of the recipient. The protective effect of B/x donors was similar in recipients having different combinations of the HLA-C (C1/C1, C2/C2, and C1/C2) and HLA-B (Bw4/x vs Bw6/Bw6) epitopes that bind to KIRs. In these recipient subsets, we detected no significant interaction between donor KIR and recipient HLA KIR ligands using the same model for relapse-free survival (HLA-C: P = .8; HLA-B: P = .5). Similarly, no differences were observed in acute GVHD, chronic GVHD, or overall survival based on KIR-ligand status (all P = NS). In summary, transplantation using a KIR B/x donor was independently associated with improved relapse-free survival.
The survival benefit of a donor KIR B haplotype is not dependent on the presence of KIR2DS2. Donors with KIR B haplotypes were divided into 2 groups based on the presence (2DS2+, n = 252) or absence (2DS2−, n = 65) of KIR2DS2. Survival was significantly better for patients who received transplants from donors with either KIR B haplotype (2DS2+: 31% [95% CI: 25-37] or 2DS2−: 31% [95% CI: 20-42]) versus donors homozygous for the A haplotype (A/A: 20% [95% CI: 13-27]; P = .02).
The survival benefit of a donor KIR B haplotype is not dependent on the presence of KIR2DS2. Donors with KIR B haplotypes were divided into 2 groups based on the presence (2DS2+, n = 252) or absence (2DS2−, n = 65) of KIR2DS2. Survival was significantly better for patients who received transplants from donors with either KIR B haplotype (2DS2+: 31% [95% CI: 25-37] or 2DS2−: 31% [95% CI: 20-42]) versus donors homozygous for the A haplotype (A/A: 20% [95% CI: 13-27]; P = .02).
To assess how KIR B/x donors improve transplantation outcome, we analyzed the cumulative incidence estimates and multivariate competing risks models for differences in risk of relapse, TRM, and acute and chronic GVHD in transplantations using donors with KIR A/A versus B/x genotypes (Figure 5). All multivariate competing risks models were adjusted for HLA match group, disease status, age, KPS, and time from diagnosis to transplantation. Transplantations using KIR B/x donors were associated with modest though not significantly decreased risks for both relapse and TRM. No interaction was detected between CMV serostatus and the effect of a donor B haplotype on any outcomes. A donor KIR B haplotype did not alter the incidence of grades II to IV acute GVHD (adjusted RR: 1.10 [95% CI: 0.76-1.59], P = .61), but was associated with increased 3-year cumulative incidence of chronic GVHD (B/x: 39% [95% CI: 34-45] vs A/A: 28% [95% CI: 20-35], P = .02). Multivariate models confirmed the significant association of donor B/x with increased RR of chronic GVHD (B/x vs A/A RR: 1.51 [95% CI: 1.01-2.18], P = .03).
Transplantation using donors with KIR B haplotypes leads to less relapse and TRM, and equivalent acute GVHD, but increased chronic GVHD. The cumulative incidences of relapse, TRM, acute GVHD, and chronic GVHD are shown for transplantation using donors with (KIR B/x) or without KIR B (KIR A/A) haplotypes. The incidence of chronic GVHD was higher using KIR B/x donors (39% [95% CI: 34-45]) than when using KIR A/A donors (28% [95% CI: 20-35], P = .02). Acute GHVD, relapse, and TRM were not significantly different between donor KIR haplotype groups (P > .15).
Transplantation using donors with KIR B haplotypes leads to less relapse and TRM, and equivalent acute GVHD, but increased chronic GVHD. The cumulative incidences of relapse, TRM, acute GVHD, and chronic GVHD are shown for transplantation using donors with (KIR B/x) or without KIR B (KIR A/A) haplotypes. The incidence of chronic GVHD was higher using KIR B/x donors (39% [95% CI: 34-45]) than when using KIR A/A donors (28% [95% CI: 20-35], P = .02). Acute GHVD, relapse, and TRM were not significantly different between donor KIR haplotype groups (P > .15).
Discussion
Previous studies on the effects of KIR genotype in HCT involved small cohorts of patients treated for different diseases and reported conflicting results. Here we report our analysis of a large homogeneous cohort of AML patients receiving myeloablative conditioning followed by a T cell–replete unrelated donor grafts. Moreover, we used careful matching criteria to select clinically comparable cohorts of donor-recipient pairs, and we controlled for important clinical covariates using multivariate models.
In this multicenter study, we found that transplantation using a donor with 1 or 2 KIR B haplotypes was associated with significant improvements in overall and relapse-free survival, with more than 30% better relative risk in both these end points for the KIR B/x donor group. The clinical benefit associated with donor KIR B haplotypes was evident across the entire KIR ligand–matched cohort, and was not influenced by the recipients' KIR genotype, the extent of HLA match, or the recipients' disease status at the time of transplantation. In contrast, the only group showing no benefit from a B/x donor was the small subset of transplants in which there was both HLA and KIR-ligand mismatch. The HLA-C group mismatches that impart KIR-ligand mismatches are known to adversely affect clinical outcomes.17 Consequently, it is possible that the clinical advantage conferred by a KIR B/x donor is insufficient to overcome the disadvantage arising from a KIR-ligand (HLA-C) mismatch.
Although our results suggest that donor 2DL2 and 2DS2 may independently be advantageous, the analysis (not adjusted for multiple comparisons) was not definitive because of its limited power. Importantly, the clinical benefit of a B/x donor did not depend on the presence of either KIR2DL2 or KIR2DS2, or any other particular B haplotype–defining KIR gene; nor was there an association with increasing number of activating KIR genes. Ongoing analyses of a larger cohort will address the effect of individual KIR genes. In conclusion, our results indicate that all donors with 1 or 2 KIR B haplotypes have the potential to provide an improved clinical benefit in HCT for AML and should be selected over an otherwise equivalent donor who lacks a KIR B haplotype.
The benefit of donor KIR B haplotypes has also been observed for T cell–replete transplantations using HLA-matched sibling donors. There the improved survival and reduced transplantation-related mortality were attributed to lower rates of cytomegalovirus reactivation,22 an effect reported in other settings23 and paralleled in the mouse model of cytomegalovirus infection.24,25 Independent evidence for human activating KIRs in resisting infection is the reduced progression to AIDS in HIV-infected patients having KIR3DS1 and a cognate HLA Bw4 ligand.26 Beneficial and detrimental clinical effects on outcomes after HCT have been associated with activating KIRs.9-11,27 Increased acute GVHD was associated with higher numbers of donor activating KIRs in HLA-matched URD transplantations. KIR2DS3, in particular, was strongly associated with GVHD. In our study, donor KIR B/x genotype (but not individual KIR genes) was associated with higher rates of chronic, but not acute, GVHD. Activating KIRs are similarly associated with the chronic inflammation that characterizes autoimmune diseases.7,28
In haploidentical, T cell–depleted (TCD) HCT, donors mismatched for KIR ligands were associated with NK-cell alloreactivity and improved survival.3,29 In URD HCT for myeloid malignancies, mismatching for KIR ligands can confer clinical benefit but only for T cell–depleted grafts,2 not T cell–replete grafts.1,30 This difference is attributed to T cells in the graft that interfere with NK-cell development and KIR reconstitution after URD HCT.31 Here we observed no benefit associated with mismatch of KIR ligands, consistent with the previous studies of T cell–replete URD HCT.32,33
Although the underlying mechanism remains uncertain, this multicenter analysis demonstrates a significant and substantial (> 10%) survival benefit for AML patients receiving grafts from unrelated donors having 1 or 2 KIR B haplotypes. In our cohort, approximately two-thirds of the donors had at least 1 KIR B haplotype, indicating that KIR genotyping of donors to preferentially select for the presence of a B haplotype is a feasible proposition. If implemented, such typing could yield a KIR B haplotype donor option for a majority of the patients who have several high-resolution HLA-matched donors. Although a sophisticated and high-throughput method based upon mass spectrometry was used in this analytic study, simpler and less expensive methods could be used in clinical immunogenetics laboratories to reliably distinguish A/A and B/x donors.34 The significant survival benefit observed in this multicenter study of HCT for AML provides compelling evidence that KIR B haplotype donors should be used preferentially in HLA-matched or HLA-mismatched unrelated donor transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Stephen Spellman, Martin Maiers, and Michael Haagenson and the Immunobiology committee at the National Marrow Donor Program/Center for International Blood and Marrow Transplant Research for assistance in providing transplant samples and outcome data for this analysis and for assisting in KIR-ligand coding.
This research was funded by National Institutes of Health grant NIH/NCI P01 111412. The CIBMTR is supported by Public Health Service Grant U24-CA76518 from the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Heart, Lung and Blood Institute; Office of Naval Research (grant to the NMDP N00014-06-1-0704); Health Resources and Services Administration (DHHS); and others.
Any opinions, findings, and conclusions or recommendations expressed in this article are those of the author(s) and do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, National Marrow Donor Program, or any other agency of the US government. Only the authors participated in study design, the collection or interpretation of the data, and preparation of the paper.
National Institutes of Health
Authorship
Contribution: S.C. participated in the design and interpretation of the analysis and the writing of the paper; D.J.W., P.P., S.G.E.M., and J.S.M. planned, directed, and coordinated the research and revised the paper; E.T. participated in the design and interpretation of the analysis, led the team who performed genotyping of the DNA samples, and revised the paper; K.S. performed all of the genotyping, interpreted the data, and revised the paper; L.A.G. provided quality control data, helped interpret the genotyping data, and revised the paper; and T.L.B. performed the statistical analyses and helped write the paper with input from J.K. and C.T.L.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sarah Cooley, Assistant Professor of Medicine, University of Minnesota Cancer Center, MMC 480, Division of Hematology, Oncology, and Transplantation, 420 Delaware Street SE, Minneapolis, MN 55455; e-mail: cool0023@umn.edu.

![Figure 1. Transplantation using donors with a KIR B haplotype improves relapse-free survival (RFS) irrespective of the recipient KIR haplotype status. Donor (D) and recipient (R) DNA samples from 448 URD transplants were genotyped for 16 KIR loci. KIR gene content was used to identify haplotypes (A or B), from which KIR genotypes were assigned (KIR A/A or KIR B/x). The Kaplan-Meier curves demonstrate RFS for each donor/recipient genotype pairing (A/A into A/A, A/A into B/x, B/x into A/A, and B/x into B/x). RFS was significantly better after transplantation using KIR B/x donors (28% [95% CI: 23-33]; n = 317) compared with A/A donors (17% [11%-24%]; n = 131; P = .003).The recipient KIR genotype had no effect (P = .75).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2008-07-171926/5/m_zh80020929410001.jpeg?Expires=1767739550&Signature=5BmTq-aKjendUM9kMCBDAO1iX3DSxsnKRvz-TPS-gQygm-qL7GnJj31kmdfG0v7J2eqUjplTRPKJyi6p~69zPGXOH~GMAe9jGOKlpCoITraQ3aRsJy4xEDVaLoqLlTloXhYB1iWLPMYhz-I0aSd72AbjAp33bIylzV3olxnm3uUsvC9040hbjYbjilDlS0rfvSLMWst3uChLLLZ92kRc8WDVXh-2-6WxIHTv7nvGsdsAsWzi6Cl84AqrU0CIZt~ccUI1sEaNLcj5XKM-koX8uBQ0vTNv78EVEIvkWRCOA45SZWvhIwcDL6g4SWaqEERpbdv0i7SIfsASj5Ys8Rbguw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Transplantation using donors with KIR B haplotypes improves overall survival. The univariate Kaplan-Meier curves for overall survival (OS) are shown for patients receiving transplants from donors with (KIR B/x; n = 317) or without (KIR A/A; n = 131) KIR B haplotypes. Survival is significantly better if the donor had a KIR B haplotype (31% [95% CI: 26-36] vs 20% [95% CI: 13-27]; P = .007).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2008-07-171926/5/m_zh80020929410002.jpeg?Expires=1767739550&Signature=Lwy~~kPbiuIhZuvyCVIqfdWLDwGiryOVEPAFFRiyYSGjROjwyJPYnHXJDvHFKbH4YoF5Dq8jDUFSmQsmRcauiMrSi2aJihLB-sjN2qGO6zejTcVxKRylzcAHulKOQ1L5VgACevuDojMtKLHY71KQYf5Z9YEYZAAkV0j9giQj0mFoMB8kd2G5Aq88mD-e~Cvq~J8oiBa9ha7hPk4X8pw9F6SMZb0eBhVRtQIWj-YpiUmAhoBOJ1JwZC4ugiUvHrJK8il6HmviDojzvKDd7sZwnM7oNiCIldRCQQr8JEWKYMt00diulINS81wthZJY5Z9ymtUZwRJpv8Dax8~fDI5xrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. The survival benefit of a donor KIR B haplotype is not dependent on the presence of KIR2DS2. Donors with KIR B haplotypes were divided into 2 groups based on the presence (2DS2+, n = 252) or absence (2DS2−, n = 65) of KIR2DS2. Survival was significantly better for patients who received transplants from donors with either KIR B haplotype (2DS2+: 31% [95% CI: 25-37] or 2DS2−: 31% [95% CI: 20-42]) versus donors homozygous for the A haplotype (A/A: 20% [95% CI: 13-27]; P = .02).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2008-07-171926/5/m_zh80020929410004.jpeg?Expires=1767739550&Signature=yy6EDq6GfTEzAuuwpe6n94NnRcQWwL1jw9sioWfzMX9vAimzRcvHOLMhXG2LiHyAp6rPnozD6dKABQVf6LGBYXy33NGBe~jb-Fwez21mb19rSiwPeHAWSgXGbegQha2GytydsdL~Whms4fyq~DzguAgEqFAJXO0hnKKYL4XI2Mp~81pH8oLezfIvBgnXtO7s81P4HDyFlurC5g0~P-ElvPpAbe9Iu2MY8Z2i8svnycll-wSATb13HbL7m1emMVdy0SfHUOlHUeRZ9DMSDpZJ9Q8wODiFiQgu8OufmMv~qa20ayQLV5JBg84xCBWpf~3TbzwnBiXqOpMgLawDqzQXQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Transplantation using donors with KIR B haplotypes leads to less relapse and TRM, and equivalent acute GVHD, but increased chronic GVHD. The cumulative incidences of relapse, TRM, acute GVHD, and chronic GVHD are shown for transplantation using donors with (KIR B/x) or without KIR B (KIR A/A) haplotypes. The incidence of chronic GVHD was higher using KIR B/x donors (39% [95% CI: 34-45]) than when using KIR A/A donors (28% [95% CI: 20-35], P = .02). Acute GHVD, relapse, and TRM were not significantly different between donor KIR haplotype groups (P > .15).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/3/10.1182_blood-2008-07-171926/5/m_zh80020929410005.jpeg?Expires=1767739550&Signature=pslx5htTae0NcVzhavHKFhn2Kt2CA4IuKIhWTrwwina6nvKmnfEY31GZDasUvHyUQOvkfP8~vAxHf2DDS7uI0OYcIaABZX74eKTjYGQyna287ntLPJlg~B~D5jcq9wdtc3ifA97LVyTEt52vXjHS96J4e1FBysqrJBaBQWyijmyCcgnGkcFw7PqWEt6AVp5UVlN4S1Rs6NVXNAlSsIbyox14S~a1ijGPDtNEJrpdX9utA9BkWtwEVlJDN532a4zAaO~Azo0ACyzo2QX5h7POaQuc3BWSqqfFBVpWP5KnY3FYo6Ai-kxxYgE4MLLv0uf-IlJLL33cFw6iev-pEQKkFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal