Abstract
The 8;21 translocation, which involves the gene encoding the RUNX family DNA-binding transcription factor AML1 (RUNX1) on chromosome 21 and the ETO (MTG8) gene on chromosome 8, generates AML1-ETO fusion proteins. Previous analyses have demonstrated that full-length AML1-ETO blocks AML1 function and requires additional mutagenic events to promote leukemia. More recently, we have identified an alternatively spliced form of AML1-ETO, AML1-ETO9a, from t(8;21) acute myeloid leukemia (AML) patient samples. AML1-ETO9a lacks the C-terminal NHR3 and NHR4 domains of AML1-ETO and is highly leukemogenic in the mouse model. Here, we report that the AML1 DNA-binding domain and the ETO NHR2-dimerization domain, but not the ETO NHR1 domain, are critical for the induction of AML by AML1-ETO9a. A region between NHR1 and NHR2 affects latency of leukemogenesis. These results provide valuable insight into further analysis of the molecular mechanism of t(8;21) in leukemogenesis.
Introduction
The AML1-ETO (AE) protein from a common chromosomal translocation t(8;21) contains 5 known functional domains (Figure 1A), the Drosophila runt homology domain (RHD) from AML1 and 4 Drosophila Nervy homology regions (NHRs) from ETO. The Runt domain is responsible for binding to DNA, interacting with its heterodimerization partner core-binding factor–β (CBF-β), and binding to other transcription regulators.1,2 NHR1 shares sequence similarity with TAF110. NHR2 is critical for ETO oligomerization; NHR3 contains a predicted coiled-coil structure; NHR4 is a myeloid-Nervy-DEAF1 homology domain with zinc-chelating motifs
To understand the molecular mechanism of AE in leukemogenesis, several mouse models have been established. Studies using these models revealed that AE expression was not sufficient to induce leukemia.3-7 However, with additional mutation(s), AE is necessary for causing acute myeloid leukemia (AML).8,9 Recently, we identified C-terminal–truncated AE proteins (AEtr and AE9a) as potent inducers of leukemia development in mice.10,11 Interestingly, the naturally occurring spliced isoform AE9a was detected in t(8;21) patients.11 In this report, we investigated the importance of different known AE domains in leukemogenesis using the AE9a mouse leukemia model.
Methods
Animals
Retroviral construction
The different ETO deletion constructs were as previously described.12 The cDNA fragments with deletions were inserted into the MigR1-AE9a construct. MigR1-AE9aR174Q and MigR1-AEtrL148D were generated from pCDNA6-HA-AE-R174Q and pCMV5-AE-L148D, respectively. All deletion and point mutation constructs were confirmed by sequencing.
Fetal liver cell isolation, retroviral infection, transplantation, and flow cytometry
Mouse survival and statistic analyses
The Kaplan-Meier survival curve and statistic analyses were performed using GraphPad Prism4 software (GraphPad Software, San Diego, CA).
Results and discussion
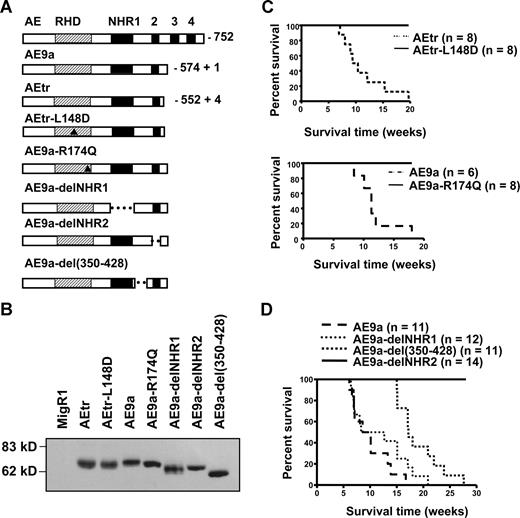
To investigate the important domains of AE in leukemogenesis, we used a series of point mutation and internal deletion mutants of C-terminal–truncated AE proteins to perform hematopoietic cell retroviral transduction and transplantation assays (Figure 1A). The expression of these fusion proteins was confirmed by Western blotting (Figure 1B). Based on previous reports, the L148D mutation should disrupt AE binding to DNA and CBF-β,13 and the R174Q mutation should prevent AE DNA binding but not significantly interfere with the interaction between AE and CBF-β.14 Furthermore, AE9a-delNHR1 will not interact with E proteins,15 AE9a-delNHR2 cannot form oligomers with itself (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) or with any MTG family members,12,16 and AE9a-del(350-428) lacks one of the NCoR-interacting regions of AE.12
Characterization of AE9a in leukemogenesis. (A) The structure of the AE, AEtr, AEtr-L148D, AE9a, AE9a-R174Q, AE9a-delNHR1, AE9a-delNHR2, and AE9a-del(350-428). Triangles point to the location of 2 mutations in the RHD domain. (B) The expression of different AE mutant proteins. (C) The Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing AEtr and AEtr-L148D (top panel) or AE9a and AE9a-R174Q (bottom panel). (D) Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing AE9a or its mutant forms. At least 2 independent sets of experiments were performed with each group of transplantations.
Characterization of AE9a in leukemogenesis. (A) The structure of the AE, AEtr, AEtr-L148D, AE9a, AE9a-R174Q, AE9a-delNHR1, AE9a-delNHR2, and AE9a-del(350-428). Triangles point to the location of 2 mutations in the RHD domain. (B) The expression of different AE mutant proteins. (C) The Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing AEtr and AEtr-L148D (top panel) or AE9a and AE9a-R174Q (bottom panel). (D) Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing AE9a or its mutant forms. At least 2 independent sets of experiments were performed with each group of transplantations.
All of the mice containing AE9a- or AEtr-transduced hematopoietic cells developed AML as expected.10,11 In contrast, all AEtr-L148D and AE9a-R174Q mice were leukemia free over 1 year after transplantation (Figure 1C), suggesting that DNA binding is required for promoting leukemia development. However, because many important transcription factors have been reported to interact with this domain,1,2 it is still possible that the binding of certain factor(s) to the RHD contributes to the oncogenic function of this fusion protein.
The median survival time of AE9a mice and AE9a-delNHR1 mice was 10.1 and 10.5 weeks, respectively (Figure 1D). In contrast, a significant delay in leukemia development was observed in AE9a-del(350-428) mice. Their median survival time was 17.2 weeks (P = .005). None of the mice transplanted with AE9a-delNHR2-expressing cells developed leukemia. Expression of AE9a-delNHR1 and AE9a-del(350-428) proteins was confirmed in the spleens of leukemic mice (data not shown). Thus, the NHR2 domain is required for AE9a-induced leukemogenesis. The NHR2 domain was reported to mediate oligomerization, and interaction with mSin3A16-19 and structural analysis revealed that this domain forms a homo-tetramer. Disruption of NHR2 impaired the ability of AE to increase self-renewal of hematopoietic progenitors.20,21 Using DNA-binding site selection coupled with polymerase chain reaction amplification, we discovered that AE and AE9a preferentially bind to DNA fragments containing duplicated AML1-binding sites, suggesting selective regulation of AML1 target genes by t(8;21) fusion proteins. This preferential binding also depended on the presence of the intact NHR2 domain.22
NHR1 is the contact site for binding of AE to E proteins and for the inhibitory effect of AE on an E-box containing promoter.15 Moreover, AE blocked the recruitment of p300/CBP to HEB to inhibit the transactivation. Because E proteins are known to play critical roles in hematopoietic cell proliferation and differentiation, it has been proposed that E proteins are major targets of AE in t(8;21)-related leukemogenesis. However, deletion of NHR1 from AE9a did not change the leukemogenic potential or the leukemia phenotypes seen in AE9a leukemic mice (Figures 1D, 2).11 Therefore, protein interactions via NHR1 domain do not contribute to AE9a-induced leukemogenesis.
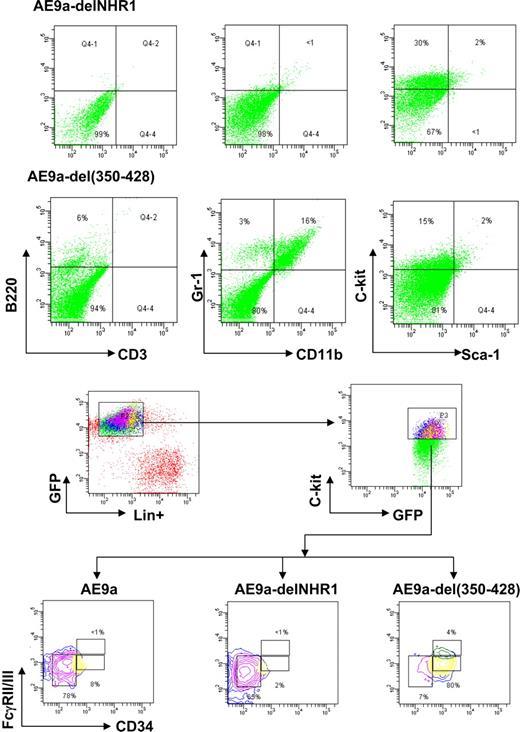
Flow cytometric analyses of various leukemic cells. (A) Hematopoietic lineage marker expression was analyzed in leukemic cells expressing AE9a-delNHR1 or AE9a-del(350-428) (EGFP+). (B) Analyses of surface myeloid progenitor marker expression in AE9a, AE9a-delNHR1, or AE9a-del(350-428) leukemic cells. Data represent the results from analyses of 4 samples of each leukemogenic construct.
Flow cytometric analyses of various leukemic cells. (A) Hematopoietic lineage marker expression was analyzed in leukemic cells expressing AE9a-delNHR1 or AE9a-del(350-428) (EGFP+). (B) Analyses of surface myeloid progenitor marker expression in AE9a, AE9a-delNHR1, or AE9a-del(350-428) leukemic cells. Data represent the results from analyses of 4 samples of each leukemogenic construct.
Amino acids 350 to 428, NHR2, and NHR4 of AE are important for AE-induced activation of the Wnt-signaling pathway by enhancing γ-catenin promoter activity.23 Furthermore, amino acids 350 to 428 are one of the NCoR-binding sites in AE protein.17 Deletion of this region of AE caused a significant delay in leukemia development. Secondary bone marrow transplantation experiments demonstrated that both AE9a-del(350-428) and AE9a-delNHR1 leukemic cells were transplantable (data not shown). We also performed Southern blot analysis to examine the clonality of these leukemia cells. Both AE9a-delNHR1 and AE9a-del(350-428) leukemia cells displayed oligo retroviral integration sites, as we have reported previously about AE9a (data not shown).
Although hematopoietic cell morphology and differential counts observed in AE9a, AE9a-delNHR1, and AE9a-del(350-428) leukemic mice were similar to those we reported previously (Figure S2),11 flow cytometry revealed differences in the expression of certain cell surface markers of leukemic cells derived from these mice. AE9a-delNHR1 leukemic cells are negative for the differentiation markers CD3, B220, CD11b, and Gr-1 but show an increased population of myeloid progenitors as we reported previously for AE9a mice11 (c-kit+/sca-1−/Lin−; Figure 2A). AE9a-del(350-428) leukemic mice had increased numbers of CD11b+/Gr-1+ cells (15.9% ± 3.3 vs < 1%) and decreased numbers of c-kit+/sca-1−/Lin− cells (14.6% ± 2.9 vs 31% ± 5.7). Moreover, all cells with AE9a, AE9a-delNHR1, or AE9a-del(350-428) expression lost the typical distribution of the 3 myeloid progenitor populations (Figure 2B).10,11 Both AE9a- and AE9a-delNHR1-induced leukemic mice displayed homogeneous accumulation of FcγRII/IIImedCD34− cells as was reported previously for AEtr- and AE9a-related leukemia.10,11 However, the leukemic cells from the AE9a-del(350-428) mice showed that most of their c-kit+/sca-1−/Lin− cells are FcγRII/IIImedCD34+. Together, these results indicate that AE9a-del(350-428) may have a relatively weaker leukemogenic potential than AE9a and AE9a-delNHR1.
In conclusion, as a first step to elucidate the molecular mechanism of AE9a in promoting leukemia development, we investigated the regions of the AE9a protein that are critical for its oncogenic potential. The results indicate that the AML1 DNA-binding domain and ETO oligomerization domain are required for AE9a-involved leukemogenesis; the NCoR-binding domain between NHR1 and NHR2 is not required but contributes in terms of the latency of leukemogenesis. The E protein–interacting NHR1 domain is not required in this AE9a model. Further studies should be conducted in the future to elucidate the function of these important regions and their binding factors in t(8;21)–related leukemogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Joseph Biggs for editing the manuscript and all other members of the DEZ laboratory for valuable discussions.
This work was supported by National Institutes of Health grants CA104509 (D.E.Z.) and CA64140 (S.W.H.).
National Institutes of Health
Authorship
Contribution: M.Y. designed and performed the research and wrote the paper; E.-Y.A. performed the research; S.W.H. provided critical reagents; and D.E.Z. supervised experimental design, data analysis, and paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, Mail Drop 0815, Room5328, Moores UCSD Cancer Center, University of California San Diego, 3855 Health Sciences Drive, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal