Abstract
STAT5 is a critical mediator of a variety of cytokine signaling whose transcriptional activity is regulated by associating with various proteins. During a search for STAT5-interacting proteins, we identified SHD1, a mammalian homologue of yeast gene Sac3, as a potential interacter. SHD1 was localized in the nucleus, and induced by cytokines that activate STAT5, such as erythropoietin, interleukin-2 (IL-2), or IL-3. SHD1 interacted specifically with STAT5A and STAT5B, and interestingly, it specifically repressed STAT5-dependent transcription in vitro without affecting the stability or phosphorylation of STAT5 protein. Gene disruption study revealed that T, B, or bone marrow cells from mice lacking SHD1 were hyperresponsive to T-cell–receptor engagement, or stimulation with various STAT5-activating cytokines. These results suggest that SHD1 is a novel cytokine-inducible negative feedback regulator of STAT5.
Introduction
Signal transducer and activator of transcription (STAT) is one of the central mediators of cytokine signaling.1 Of the 7 known members of mammalian STATs, 2 highly related STAT5 molecules, namely STAT5A and STAT5B, are considered to be of particular interest since they are activated by a wide array of cytokines including interleukin-2 (IL-2), IL-3, IL-5, IL-7, erythropoietin (Epo), and granulocyte macrophage–colony-stimulating factor (GM-CSF).1 STAT5 is implicated in the self-renewal of hematopoietic stem cells2 and is constitutively activated in a variety of leukemias,3 indicating that STAT5 plays a critical role in normal and malignant hematopoiesis.4
The activities of STATs are regulated by various protein modifications, such as tyrosine phosphorylation/dephosphorylation, serine phosphorylation, ubiquitination, and arginine methylation.5 The phosphorylation and dephosphorylation of tyrosine residue is the most important posttranslational modification common to all STAT proteins, directly affecting dimerization, nuclear translocation and export, and DNA binding.1,6,7 The phosphorylation of serine residue in the C-terminal activation domain is essential for the maximal transcriptional activity of STAT1.8,9 Arginine methylation also affects the DNA-binding activity of STAT1.10 Ubiquitination is particularly important for terminating the STAT activities by targeting the protein to the ubiquitin-proteasome degradation pathway.11 These regulations are, in part, achieved by association of STATs with other regulatory molecules, which include SLIM and PIAS family of proteins.12-16 In addition to protein modifications, STATs are also regulated at the level of transcription by associating with transcriptional coactivators and corepressors. STATs associate with CREB-binding protein (CBP)/p300, universal coactivators for many transcription factors, through the C-terminal activation domain.17-19 We have previously shown that a nuclear receptor corepressor, SMRT, associated with STAT5 through its coiled-coil domain and repressed the STAT5-dependent transcription.20 However, it remains unclear precisely how such transcriptional repression plays a role in the STAT5-dependent transcription in a physiological context.
As is commonly seen with other signaling pathways, the Jak-STAT pathway also succumbs to the negative feedback regulation of cytokines.21-23 The SOCS family, which comprises 8 family members (Cis and SOCS1-SOCS7), is a major negative feedback regulator induced by a variety of cytokines.24 Cytokines rapidly induce SOCS proteins, some of which are the direct targets of STATs, and they down-regulate Jak-STAT signaling by binding through their SH2 domain to the phosphorylated tyrosine residues of activated Jaks or cytokine receptors. Other negative regulators of cytokine signaling include SHP-1,25 LNK,26 and APS27 proteins. All these proteins affect signaling cascade in the cytoplasm, and negative feedback regulator directly affecting STAT5 transcription has not yet been reported.
To reveal the molecular mechanism of STAT5 signaling, we sought to identify the proteins that regulate the function of STAT5 by yeast 2-hybrid screening. We herein report a Sac3 domain-containing protein, SHD1, as a novel cytokine-inducible negative feedback regulator of STAT5, which represses STAT5-dependent transcription.
Methods
Yeast 2-hybrid screening
Yeast 2-hybrid screening was performed as previously described using carboxyl-terminal truncated STAT5B as a bait.20 A quantitative analysis of the protein-protein interaction by yeast 2-hybrid system was performed according to the manufacturer's protocol (Matchmaker 2 yeast 2-hybrid system; BD Clontech, Palo Alto, CA).
Cells
BaF3 and 32Dcl3 cells were cultured in an RPMI1640 medium (Life Technologies, Bethesda, MD) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin G, 100 μg/mL streptomycin, 2 mM l-glutamine, and 2.5 U/mL recombinant murine IL-3. 293T and Hela cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, and 2 mM l-glutamine. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2. Mouse embryonic fibroblasts (MEFs) were generated from E14.5 embryo as described previously.28
Northern blot and Southern blot
Total RNA was extracted from 107 cells by Trizol (Invitrogen) according to the manufacturer's protocol. The RNA samples (20 μg/lane) were separated on formaldehyde-denaturing 1.0% agarose gel and transferred onto Hybond N+ membrane (GE Healthcare, Arlington Heights, IL). Mouse multiple tissue Northern blot and mouse embryo Northern blot were purchased from BD Clontech. Genomic DNA was extracted by a standard phenol/chloroform method, run on gel, and transferred onto Hybond N+ membrane by the alkaline transfer method. The probe was labeled using Rediprime kit (GE Healthcare). Hybridizations with 32P-labeled probes were carried out in ExpressHyb buffer (BD Clontech) according to the manufacturer's protocol.
RT-PCR
Synthesis of complementary DNA and standard reverse-transcription–polymerase chain reaction (RT-PCR) were performed as previously described.29 Quantitative RT-PCR was performed as described previously29 using Light Cycler (Roche Diagnostics, Indianapolis, IN). cDNA quantity was normalized by 18S rRNA using Light Cycler Fast Start DNA SYBR Green I kit (Roche Diagnostics). Primer sequences are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Immunoprecipitation and Western blot
Cell extracts were made by lysing cells in 0.5% NP-40 buffer (0.5% NP-40, 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF). Nuclear extracts were prepared as previously described.30 Nuclear extracts were directly used for SDS–polyacrylamide gel electrophoresis (PAGE), or were diluted in 0.5% NP-40 buffer for coimmunoprecipitation studies. Immunoprecipitation and Western blot were performed as previously described.20
Transfection and luciferase assays
293T cells were transfected using FuGENE 6 (Roche Diagnostics) according to the manufacturer's protocol. Transfection of BaF3 cells was performed by DEAE-dextran method as previously described.31 Transfected cells were stimulated with 25 U/mL Epo or 20 ng/mL IL-3 at 24 hours after transfection. The cells were harvested and lysed at 24 hours after cytokine stimulation, and luciferase assay was performed using Luciferase Assay Systems (Promega, Madison, WI) according to the manufacturer's protocol. Transfections were performed in duplicate and the data were averaged from 3 independent experiments.
Antibodies
Antiphosphotyrosine antibody (clone 4G10) was purchased from Upstate Biotechnologies (Lake Placid, NY). Anti-STAT5 (C-17) was from Santa Cruz Biotechnology (Santa Cruz, CA) and anti–α-tubulin monoclonal antibody was from Sigma-Aldrich (St Louis, MO). Anti-SHD1 polyclonal antibody was raised against a peptide corresponding to the C-terminus of SHD1 coupled to KLH. The peptide was used to immunize rabbits, and specific antibodies were purified by protein A column.
Plasmids
A variety of deletion mutants of STAT5 and SHD1 were generated by PCR using Pfu polymerase (Stratagene, La Jolla, CA). The integrity of the amplified sequence was verified by DNA sequencing. A mutation of the amino acids was introduced using a QuikChange site-directed mutagenesis kit (Stratagene). pGEX-4T-1/SHD1-S was generated by subcloning SHD1-S cDNA in frame with the preceding GST sequence.
Immunostaining
Hela cells transfected with SHD1-HA vector were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.1% NP-40/PBS, and stained sequentially with anti-HA mouse monoclonal antibody (12CA5) and fluorescein isothiocyanate (FITC)–conjugated goat anti–mouse Ig antibody (Wako, Osaka, Japan). Nuclei were counterstained with DAPI (4′, 6-diamino-2-phenylindole), and the images were captured by confocal laser microscopy (Olympus, Melville, NY).
In vitro protein-binding assay
GST-SHD1 was produced in E coli (DH5α) by transforming pGEX-4T-1/SHD1-S. The proteins were purified by glutathione-sepharose beads (Pharmacia, Piscataway, NJ) according to the manufacturer's protocol. 35S-methionine labeling and in vitro protein-binding assays were performed as previously described.20
Generation of SHD1-deficient mice
Murine SHD1 genomic DNA was obtained from W9.5 embryonic stem cell BAC library. SpeI-NotI fragment containing the proximal promoter and the 5′-portion of exon 1 was replaced with neomycin resistance gene. Diphtheria toxin A gene driven by the thymidine kinase promoter was used for negative selection.32 The targeting vector was electroporated into E14 embryonic stem (ES) cells, and G418-resistant clones were screened by Southern blot to identify homologous recombinants. Three correctly targeted clones were injected into blastocysts of C57BL/6 mice to obtain chimeric mice, which were then crossed to get germ-line transmission. The animals were housed under specific pathogen-free conditions under institutional guidelines. All animal experiments were reviewed and approved by the institutional review board of the Institute of Medical Science, University of Tokyo.
T-cell proliferation and stimulation assays
T cells were purified from spleens of 8- to 12-week old wild-type or mutant mice using Imag CD4+ T-cell enrichment system according to the manufacturer's protocol (BD Pharmingen, San Diego, CA). The purity of T cells was more than 90% in all experiments. For the proliferation assays, 105 purified T cells were placed in a flat-bottom 96-well plate precoated with anti-CD3 (145-2C11; BD Pharmingen) antibody at the indicated concentrations, and cultured in RPMI1640 medium containing 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, and 1 μg/mL anti-CD28 (37.51; BD Pharmingen). The cells were cultured for 24 to 72 hours, labeled with 1 μCi (0.037 MBq)/well 3H-thymidine (GE Healthcare) for 6 hours, and then were harvested for the analysis using a Matrix 9600 beta counter (Packard, San Jose, CA). The culture supernatants harvested at 48 hours were examined for IL-2 production from T cells by enzyme-linked immunosorbent assay (ELISA).
B-cell assay
B cells were purified from spleens of 8- to 12-week old wild-type or mutant mice by negative selection using anti-CD43 microbeads and magnetic cell sorting (MACS) system (Miltenyi Biotec, Auburn, CA). The purity of B cells was more than 95% in all experiments. Purified B cells were cultured at 5 × 105/well in flat-bottom 96-well plates in RPMI1640 medium containing 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol. The cells were stimulated with IL-5 at the indicated concentrations in the presence of anti-CD40 (1 μg/mL) for 7 days, and the culture supernatants were harvested for the examination of IgM concentration by ELISA.
Proliferation assay for bone marrow cells and mast cells
Whole BM cells were obtained by flushing out the femurs with phosphate-buffered saline (PBS), and BM mononuclear cells (BMMNCs) were separated by density gradient centrifugation using Lymphoprep (Nycomed, Munich, Germany). Mast cells were prepared from the bone marrow cells as previously described.33 BMMNCs were cultured in RPMI1640 medium supplemented with 10% FBS, 100 U/mL penicillin G, 100 μg/mL streptomycin, 2 mM l-glutamine, plus 10 ng/mL IL-3 and 10 ng/mL GM-CSF. Mast cells were cultured in RPMI1640 medium supplemented with 10% FBS, 100 μM nonessential amino acids, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 100 U/mL penicillin G, 100 μg/mL streptomycin, and 10 ng/mL IL-3. The cells were counted every day and diluted as necessary to keep the concentration between 2 and 8 × 105/mL.
Results
Identification of SHD1 as a STAT5-interacting protein
To obtain further insight into the regulation of STAT5 signaling, we sought to identify STAT5-interacting proteins by yeast 2-hybrid screening using carboxyl-terminal truncated STAT5B as bait.20 Of the 70 positive clones isolated by screening mouse bone marrow library, one clone showed a specific association with STAT5A and STAT5B, but not with other STATs (STAT1, STAT3, STAT4, and STAT6; Figure 1A). Sequence analysis revealed that this clone contained almost the entire coding sequence of Sac3 domain containing protein, SHD1.34
Cloning and characterization of SHD1. (A) Interaction of SHD1 and STATs by 2-hybrid assay. C-terminal truncated forms of various STATs were subcloned into pAS2-1 vector and examined for the interaction with SHD1. The interaction was verified by the β-gal assay. (B) Schematic structure of SHD1. Asterisks denote potential start codons. Dark gray areas indicate 2 LXXLL motifs. An area with oblique lines is SAC3-homology domain. (C,D) Expression of SHD1 in a variety of mouse tissues (C) and embryos (D). Sk muscle indicates skeletal muscle. (E) SHD1-L and SHD1-S proteins expressed in 293T cells. Cell lysates were resolved by SDS-PAGE and Western blot was performed with anti-HA antibody. (F) Intracellular localization of SHD1 by immunostaining. A vector expressing SHD1-HA or a mock-HA vector was transfected into Hela cells and immunostained with anti-HA antibody. Original magnification: ×600.
Cloning and characterization of SHD1. (A) Interaction of SHD1 and STATs by 2-hybrid assay. C-terminal truncated forms of various STATs were subcloned into pAS2-1 vector and examined for the interaction with SHD1. The interaction was verified by the β-gal assay. (B) Schematic structure of SHD1. Asterisks denote potential start codons. Dark gray areas indicate 2 LXXLL motifs. An area with oblique lines is SAC3-homology domain. (C,D) Expression of SHD1 in a variety of mouse tissues (C) and embryos (D). Sk muscle indicates skeletal muscle. (E) SHD1-L and SHD1-S proteins expressed in 293T cells. Cell lysates were resolved by SDS-PAGE and Western blot was performed with anti-HA antibody. (F) Intracellular localization of SHD1 by immunostaining. A vector expressing SHD1-HA or a mock-HA vector was transfected into Hela cells and immunostained with anti-HA antibody. Original magnification: ×600.
SHD1 has a region homologous to a yeast gene Sac3 in the middle, and 2 LXXLL motifs, a signature motif for transcriptional coactivators (Figure 1B). It has 5 translational start sites clustered into 2 small regions in the N-terminus (as shown in asterisks in Figure 1B). SHD1 was ubiquitously expressed in various tissues in the adult mice (Figure 1C), and its expression was up-regulated from E11.0 in the murine embryonic development (Figure 1D). It was also weakly expressed in the hematopoietic cell lines of various lineages including myeloid, B, T, and erythroid (data not shown). We subcloned cDNAs starting from the first ATG and the third ATG into the expression vector, thus representing the long and the short form of SHD1 (SHD1-L and SHD1-S, respectively). The expressed proteins in mammalian cells were about 50 kDa and 40 kDa in size, respectively (Figure 1E), and they localized mainly in the nucleus by immunofluorescence (Figure 1F). The following in vitro experiments were performed with SHD1-S and SHD1-L, both of which gave the same results. Therefore, only the data of SHD1-S are shown hereafter.
Association of SHD1 and STAT5
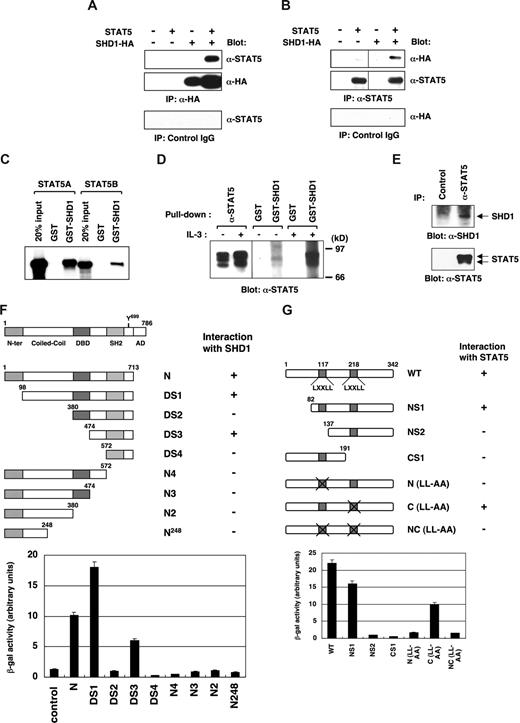
To verify the interaction of STAT5 and SHD1, we coexpressed STAT5 and HA-tagged SHD1 in 293T cells and examined their association (Figure 2A,B). The immunoprecipitation of either SHD1 or STAT5 proteins resulted in the coprecipitation of STAT5 or SHD1, respectively. In addition, the purified SHD1 protein associated with in vitro translated STAT5A or STAT5B proteins in vitro (Figure 2C). Notably, SHD1 interacted more strongly with phosphorylated STAT5 in comparison with nonphosphorylated STAT5, thus suggesting that STAT5-SHD1 interaction is induced by cytokine stimulation (Figure 2D). Lastly, physiological interaction of STAT5 and SHD1 was demonstrated by the coimmunoprecipitation of endogenous STAT5 and SHD1 proteins from the nuclear extracts of BaF3 cells (Figure 2E). Taken together, these results indicate that STAT5 interacts with SHD1 both in vitro and in vivo.
Interaction of SHD1 and STAT5. (A,B) In vivo binding of SHD1 and STAT5. Plasmids expressing HA-tagged SHD1 and STAT5A were transfected into 293T cells, and the cell lysates were immunoprecipitated with either anti-HA (A) or anti-STAT5 (B) antibodies. The precipitated proteins were detected by Western blotting using the same sets of antibodies. (C) The in vitro binding of purified SHD1 and in vitro–translated STAT5. In vitro–translated STAT5A or STAT5B labeled with 35S-methionine was mixed with GST-SHD1 in binding buffer, and pulled down with glutathione sepharose beads. The pulled down proteins were separated on SDS-PAGE gel, which was dried and exposed on x-ray film. (D) GST or GST-SHD1 protein was mixed with the lysates of starved BaF3 cells or cells stimulated with IL-3 (25 U/mL) for 10 minutes, and pulled down with glutathione sepharose beads. Immunoprecipitation using anti-STAT5 antibody was performed as a control. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-STAT5 antibody. (E) Association of endogenous SHD1 and STAT5. STAT5 was immunoprecipitated from the nuclear extracts of BaF3 cells using anti-STAT5 antibody or control IgG. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-SHD1 or anti-STAT5 antibody. (F) Analysis of SHD1-interacting domain in STAT5B. A variety of deletion mutants of STAT5B were assessed for their ability to interact with SHD1 by yeast 2-hybrid assay. The numbers denote the position of the amino acids of STAT5B. The interaction is presented as the β-gal activity. N-ter indicates N-terminus; DBD, DNA-binding domain; SH2, Src-homology 2 domain; and AD, activation domain. (G) An analysis of STAT5 interacting domain in SHD1. Various mutants of SHD1-S were assessed for their ability to interact with STAT5B by yeast 2-hybrid assay similarly as in panel F. Numbers denote the position of amino acids of SHD1-S. In (LL-AA) mutants, LXXLL motif was mutated to LXXAA.
Interaction of SHD1 and STAT5. (A,B) In vivo binding of SHD1 and STAT5. Plasmids expressing HA-tagged SHD1 and STAT5A were transfected into 293T cells, and the cell lysates were immunoprecipitated with either anti-HA (A) or anti-STAT5 (B) antibodies. The precipitated proteins were detected by Western blotting using the same sets of antibodies. (C) The in vitro binding of purified SHD1 and in vitro–translated STAT5. In vitro–translated STAT5A or STAT5B labeled with 35S-methionine was mixed with GST-SHD1 in binding buffer, and pulled down with glutathione sepharose beads. The pulled down proteins were separated on SDS-PAGE gel, which was dried and exposed on x-ray film. (D) GST or GST-SHD1 protein was mixed with the lysates of starved BaF3 cells or cells stimulated with IL-3 (25 U/mL) for 10 minutes, and pulled down with glutathione sepharose beads. Immunoprecipitation using anti-STAT5 antibody was performed as a control. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-STAT5 antibody. (E) Association of endogenous SHD1 and STAT5. STAT5 was immunoprecipitated from the nuclear extracts of BaF3 cells using anti-STAT5 antibody or control IgG. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-SHD1 or anti-STAT5 antibody. (F) Analysis of SHD1-interacting domain in STAT5B. A variety of deletion mutants of STAT5B were assessed for their ability to interact with SHD1 by yeast 2-hybrid assay. The numbers denote the position of the amino acids of STAT5B. The interaction is presented as the β-gal activity. N-ter indicates N-terminus; DBD, DNA-binding domain; SH2, Src-homology 2 domain; and AD, activation domain. (G) An analysis of STAT5 interacting domain in SHD1. Various mutants of SHD1-S were assessed for their ability to interact with STAT5B by yeast 2-hybrid assay similarly as in panel F. Numbers denote the position of amino acids of SHD1-S. In (LL-AA) mutants, LXXLL motif was mutated to LXXAA.
To determine the region of STAT5 required for the interaction with SHD1, we generated a series of truncation mutants of STAT5B and examined the interaction in yeast-2 hybrid system. As shown in Figure 2F, N-terminal 98 amino acids were not absolutely required for the interaction. However, a further deletion of the coiled-coil domain or the SH2 domain disrupted the binding with SHD1. These results indicate that almost the entire STAT5B protein, excluding the N-terminal region and the C-terminal activation domain, was essential for strong binding with SHD1. Interestingly, the DS3 mutant that contained the SH2 domain and the surrounding sequences showed a weak binding with SHD1. This indicates that amino acids 474 to 713 of STAT5 are a minimal requirement for the interaction with SHD1. In contrast, DS2 mutant that has a DNA-binding domain added to DS3 mutant did not interact with SHD1, suggesting that the DNA-binding domain of STAT5 works negatively on the interaction.
We also determined the STAT5-interacting domain in SHD1. As shown in Figure 2G, amino acids 82 to 137 and 191 to 342 of SHD1-S were essential for the binding with STAT5. The LXXLL motif is known to be a signature motif for coactivators serving as a binding surface for transcriptional activator complex.35-38 SHD1 possesses 2 LXXLL motifs and we speculated that these motifs might be critical for STAT5 binding. As expected, the first LXXLL motif was critical for the binding with STAT5 as revealed by the 2-hybrid assay. In contrast, the second LXXLL motif was dispensable for the interaction. These results indicate that the C-terminal 151 amino acids together with amino acids 82 to 137 including the first LXXLL motif of SHD1-S are essential for the binding with STAT5.
Induction of SHD1 by cytokines that activate STAT5
To investigate the possible involvement of SHD1 in STAT5 function, we first examined whether SHD1 was regulated by cytokines. Interestingly, SHD1 mRNA was induced by Epo as early as 2 hours after stimulation in BaF3 cells expressing Epo receptor (BaF3/EpoR; Figure 3A). In addition, IL-2 induced the expression of SHD1 in murine primary T cells after 2 hours of stimulation, with the maximal expression observed after 12 hours (Figure 3B). The expression level returned to the baseline after 24 hours, and asynchronously growing cells expressed low levels of SHD1. In contrast, granulocyte colony-stimulating factor (G-CSF) could not maintain the expression of SHD1 in 32D cells when they were transferred from IL-3– to G-CSF–containing media, indicating that G-CSF does not support SHD1 expression, although IL-3 did (Figure 3C).
SHD1 is induced by STAT5-activating cytokines. (A) BaF3/EpoR cells were starved overnight, and then stimulated with 10 U/mL Epo for the indicated times. The expression of SHD1 was analyzed by Northern blotting. (B) Murine splenic T cells were expanded as described in “T-cell proliferation and stimulation assays.” The cells were starved, and then stimulated with IL-2 (100 U/mL) for the indicated times. A indicates asynchronously growing cells. (C) 32D cells growing in the media containing IL-3 were washed with PBS, and immediately transferred into the media containing 10 ng/mL IL-3 or 50 ng/mL G-CSF. The cells were harvested at the indicated time points, and the expression of SHD1 was analyzed by Northern blotting.
SHD1 is induced by STAT5-activating cytokines. (A) BaF3/EpoR cells were starved overnight, and then stimulated with 10 U/mL Epo for the indicated times. The expression of SHD1 was analyzed by Northern blotting. (B) Murine splenic T cells were expanded as described in “T-cell proliferation and stimulation assays.” The cells were starved, and then stimulated with IL-2 (100 U/mL) for the indicated times. A indicates asynchronously growing cells. (C) 32D cells growing in the media containing IL-3 were washed with PBS, and immediately transferred into the media containing 10 ng/mL IL-3 or 50 ng/mL G-CSF. The cells were harvested at the indicated time points, and the expression of SHD1 was analyzed by Northern blotting.
These data demonstrate that SHD1 is induced by STAT5-activating cytokines, such as Epo, IL-2, and IL-3, but not by G-CSF that activates mainly STAT3. This raises a possibility that SHD1 could be involved in the physiological function of STAT5.
SHD1 represses STAT5 transcription in vitro
To elucidate the role of SHD1 in STAT5 signaling, we investigated the effect of SHD1 on STAT5-mediated transcription. 293T cells were transfected with a luciferase-reporter construct for STAT5, together with Epo receptor (EpoR) and various amounts of SHD1 expression vector. As shown in Figure 4A, SHD1 clearly repressed the STAT5-mediated transcription in a dose-dependent manner. In addition, a similar experiment using BaF3 cells with a different reporter gene showed the same result (Figure 4B). The levels of endogenous (Figure 4A) or expressed (Figure 4B) STAT5 protein were not affected by SHD1 overexpression either in a stimulated or an unstimulated condition. Moreover, the phosphorylation of STAT5 was not affected by the SHD1 overexpression (data not shown). Taken together, these results indicate that the SHD1 represses STAT5 transcription without modulating either the protein levels or tyrosine phosphorylation of STAT5.
Repression of STAT5-dependent transcription by SHD1. (A) 293T cells were transfected with plasmids expressing Epo receptor, SHD1, and the luciferase reporter construct containing 4 tandem repeats of optimal STAT5-binding site. One microgram of each plasmid was used for transfection unless otherwise indicated. The cells were stimulated with recombinant human Epo (25 U/mL) after 24 hours of transfection, and harvested for luciferase assay after 24 hours of stimulation. The data are the mean plus or minus SD (n = 3). The lysates were analyzed by Western blotting to determine the expression of STAT5 and α-tubulin as a loading control. (B) The indicated amounts of SHD1 or STAT5A expression vector together with luciferase reporter construct containing β-casein promoter were transfected into BaF3 cells by DEAE-dextran method. The cells were stimulated with recombinant murine IL-3 after 24 hours of transfection and harvested for the analysis after 24 hours of stimulation. The data are the mean plus or minus SD (n = 3). The expression level of STAT5 was analyzed by immunoprecipitation and Western blotting. (C) A vector expressing full-length STAT5B fused to GAL4 DNA-binding domain (pM/STAT5, 1 μg) was transfected into 293T cells together with indicated amounts of SHD1 expression vector and the luciferase reporter plasmid bearing 5 copies of GAL4-binding sites (GAL4-luc). The cells were lysed for the analysis after 48 hours of transfection. The data are the mean plus or minus SD (n = 3).
Repression of STAT5-dependent transcription by SHD1. (A) 293T cells were transfected with plasmids expressing Epo receptor, SHD1, and the luciferase reporter construct containing 4 tandem repeats of optimal STAT5-binding site. One microgram of each plasmid was used for transfection unless otherwise indicated. The cells were stimulated with recombinant human Epo (25 U/mL) after 24 hours of transfection, and harvested for luciferase assay after 24 hours of stimulation. The data are the mean plus or minus SD (n = 3). The lysates were analyzed by Western blotting to determine the expression of STAT5 and α-tubulin as a loading control. (B) The indicated amounts of SHD1 or STAT5A expression vector together with luciferase reporter construct containing β-casein promoter were transfected into BaF3 cells by DEAE-dextran method. The cells were stimulated with recombinant murine IL-3 after 24 hours of transfection and harvested for the analysis after 24 hours of stimulation. The data are the mean plus or minus SD (n = 3). The expression level of STAT5 was analyzed by immunoprecipitation and Western blotting. (C) A vector expressing full-length STAT5B fused to GAL4 DNA-binding domain (pM/STAT5, 1 μg) was transfected into 293T cells together with indicated amounts of SHD1 expression vector and the luciferase reporter plasmid bearing 5 copies of GAL4-binding sites (GAL4-luc). The cells were lysed for the analysis after 48 hours of transfection. The data are the mean plus or minus SD (n = 3).
We next examined whether the repression of STAT5 transcription by SHD1 required the dimerization or DNA-binding capacity of STAT5. We fused full-length STAT5 monomer to GAL4-DNA–binding domain (pM/STAT5), and examined its activity against the reporter containing GAL4-binding site (Figure 4C). Interestingly, SHD1 did not repress the activity of pM/STAT5, although pM/STAT5 demonstrated strong transactivating capacity. This result indicates that (1) SHD1-mediated STAT5 repression requires dimerization or DNA-binding activity of STAT5, and (2) SHD1 does not disrupt the interaction of STAT5 and the transcriptional coactivators.
It is of note that transcriptional repression by SHD1 was specific to STAT5, since non-STAT5 promoters such as STAT1-, STAT3-, NFκB-, and SMAD3-responsive promoter were not sensitive to SHD1 repression (Figure S1).
A disruption of the SHD1 gene results in the hyperresponsiveness of T, B, and myeloid cells
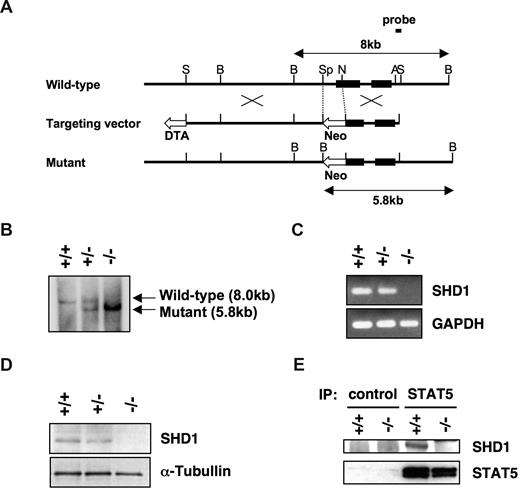
To investigate the physiological role of SHD1 in vivo, we generated SHD1-deficient mice. A genomic region containing the proximal promoter and 5′-portion of exon 1 was replaced with a neo gene cassette in E14 ES cells (Figure 5A). Three correctly targeted ES cell clones were used to generate chimeric mice, which were then crossed to obtain germline transmission. The transmission of the mutated allele was confirmed by a Southern blot analysis (Figure 5B), and the absence of SHD1 mRNA and protein was verified by RT-PCR and Western blotting (Figure 5C,D). A slight reduction in the levels of mRNA and the protein was noted in heterozygous mice, indicating that the disruption of one allele slightly affected the expression of SHD1. We have also confirmed the association of SHD1 and STAT5 using wild-type and mutant mast cells (Figure 5E).
Generation of SHD1-deficient mice. (A) Structure of the targeting vector and the targeting strategy. The filled boxes represent exons. The targeting vector was designed to replace SpeI-NotI fragment containing the proximal promoter and the 5′-portion of exon 1 with neomycin resistance cassette. This removes all 5 potential translational start sites in exon 1. Neo indicates neomycin resistance cassette; DTA, diphtheria toxin A; S, SalI; B, BamHI; Sp, SpeI; N, NotI; and A, ApaI. (B) Southern blot analysis of BamHI-digested genomic DNA from wild-type (+/+), heterozygous (+/−), or homozygous (−/−) mice. The wild-type (8.0 kb) and the mutant (5.8 kb) bands are indicated by arrows. (C) The expression of SHD1 in mutant mice. Total RNA was extracted from splenocytes of mutant mice, and the expression of SHD1 was analyzed by RT-PCR. GAPDH is shown as a control. (D) The absence of SHD1 protein in SHD1-deficient mice. The nuclear extracts of thymocytes from the mutant mice were subjected to a Western blot analysis using anti-SHD1 antibody. α-Tubulin was probed to indicate equal loading of the protein extracts. (E) Association of SHD1 and STAT5 in wild-type (+/+) and mutant (−/−) mast cells. STAT5 was immunoprecipitated from the nuclear extracts with anti-STAT5 antibody or control IgG. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-SHD1 or anti-STAT5 antibody.
Generation of SHD1-deficient mice. (A) Structure of the targeting vector and the targeting strategy. The filled boxes represent exons. The targeting vector was designed to replace SpeI-NotI fragment containing the proximal promoter and the 5′-portion of exon 1 with neomycin resistance cassette. This removes all 5 potential translational start sites in exon 1. Neo indicates neomycin resistance cassette; DTA, diphtheria toxin A; S, SalI; B, BamHI; Sp, SpeI; N, NotI; and A, ApaI. (B) Southern blot analysis of BamHI-digested genomic DNA from wild-type (+/+), heterozygous (+/−), or homozygous (−/−) mice. The wild-type (8.0 kb) and the mutant (5.8 kb) bands are indicated by arrows. (C) The expression of SHD1 in mutant mice. Total RNA was extracted from splenocytes of mutant mice, and the expression of SHD1 was analyzed by RT-PCR. GAPDH is shown as a control. (D) The absence of SHD1 protein in SHD1-deficient mice. The nuclear extracts of thymocytes from the mutant mice were subjected to a Western blot analysis using anti-SHD1 antibody. α-Tubulin was probed to indicate equal loading of the protein extracts. (E) Association of SHD1 and STAT5 in wild-type (+/+) and mutant (−/−) mast cells. STAT5 was immunoprecipitated from the nuclear extracts with anti-STAT5 antibody or control IgG. The precipitated proteins were analyzed by SDS-PAGE and Western blotting using anti-SHD1 or anti-STAT5 antibody.
Gross analyses of peripheral blood and differentiation profiles of BM, spleen, and thymus revealed no substantial difference between wild-type, heterozygous, and homozygous mice (Figures S2Figure S3. Flow cytometric analysis of spleen cells (JPG, 119 KB)–S4). However, closer examination revealed that the total number of splenocytes and thymocytes slightly increased in the homozygous mice, which were 1.4 and 1.9 times higher than those in wild-type mice, respectively (Table S2). In addition, the number of various hematopoietic progenitors in the bone marrow (BM) slightly increased in the heterozygous and homozygous mice by colony assays (Table S3).
In T cells, STAT5 is essential for their proliferation in response to T-cell receptor (TCR) ligation and/or IL-2 stimulation.39 Since SHD1 repressed STAT5-dependent transcription in vitro, it is speculated that SHD1-deficient T cells are hyperresponsive. To test this hypothesis, we purified CD4+ T cells from SHD1 wild-type, heterozygous, and -deficient mice, and examined their mitogenic response to TCR stimulation. As expected, SHD1-deficient T cells showed a greater proliferative response, approximately 1.5- to 11-fold compared with that of wild type (Figure 6A). This phenotype was evident in the low concentration range of anti-CD3 (0.01-0.1 μg/mL), and there was little difference in higher concentrations (more than 1.0 μg/mL) where the response was saturated (data not shown). Importantly, the hyperproliferation of SHD1-deficient T cells was not the result of either enhanced autocrine production of IL-2 (Figure 6B), increased phosphorylation of STAT5 in response to IL-2 (Figure 6C), or an enhanced induction of IL-2 receptor α chain (Figure S5). Of note, there was no alteration of Th1 and Th2 differentiation from naive T cells in response to anti-CD3 stimulation in SHD1 mutant mice (Figure S6).
Hyperresponsive phenotype of SHD1−/− T, B, and bone marrow cells. (A) Splenic T cells from wild-type, SHD1+/−, or SHD1−/− mice were stimulated with the indicated concentrations of anti-CD3 in the presence of anti-CD28 (1 μg/mL). Proliferation was assessed by the 3H-thymidine uptake after 72 hours of stimulation. Each assay was conducted in triplicate and the data are representative of 4 experiments. The data are the mean plus or minus SD. (B) The IL-2 production by splenic T cells from wild-type, SHD1+/−, or SHD1−/− mice. The culture supernatants from panel A were analyzed by ELISA for IL-2 concentration. The data are the mean plus or minus SD (n = 3). (C) Cultured splenic T cells from wild-type or SHD1−/− mice were starved, and then stimulated with IL-2 (10 ng/mL) for 10 or 30 minutes. The cells were lysed and subjected to immunoprecipitation and a Western blot analysis for the phosphorylation of STAT5. (D) IgM production by B cells. Purified splenic B cells from wild-type, SHD1+/−, or SHD1−/− mice were stimulated with the indicated concentrations of IL-5 plus anti-CD40 (1 μg/mL) for 7 days. The concentration of IgM in the culture supernatants was analyzed by ELISA. The data are the mean plus or minus SD (n = 3). (E) BM cell proliferation. BM cells from wild-type, SHD1+/−, or SHD1−/− mice were cultured in the media containing IL-3 and GM-CSF. Cell numbers were enumerated every day as described in “Proliferation assays for bone marrow cells and mast cells.” The data are the mean plus or minus SD (n = 3). (F) Proliferation of mast cells. Mast cells derived from SHD1-mutant mice were cultured in the presence of 1 or 10 ng/mL IL-3, and the cell numbers were enumerated at the indicated time points (mean ± SD, n = 3). (G) Induction of STAT5 target genes in wild-type, SHD1+/−, or SHD1−/− mice. Total RNA was extracted from cultured T cells stimulated with the indicated concentrations of IL-2 for the indicated times. Quantitative RT-PCR analysis was performed as described in “RT-PCR.” Data are shown as a ratio compared with the average value of prestimulated (0 minutes) wild-type in each graph.
Hyperresponsive phenotype of SHD1−/− T, B, and bone marrow cells. (A) Splenic T cells from wild-type, SHD1+/−, or SHD1−/− mice were stimulated with the indicated concentrations of anti-CD3 in the presence of anti-CD28 (1 μg/mL). Proliferation was assessed by the 3H-thymidine uptake after 72 hours of stimulation. Each assay was conducted in triplicate and the data are representative of 4 experiments. The data are the mean plus or minus SD. (B) The IL-2 production by splenic T cells from wild-type, SHD1+/−, or SHD1−/− mice. The culture supernatants from panel A were analyzed by ELISA for IL-2 concentration. The data are the mean plus or minus SD (n = 3). (C) Cultured splenic T cells from wild-type or SHD1−/− mice were starved, and then stimulated with IL-2 (10 ng/mL) for 10 or 30 minutes. The cells were lysed and subjected to immunoprecipitation and a Western blot analysis for the phosphorylation of STAT5. (D) IgM production by B cells. Purified splenic B cells from wild-type, SHD1+/−, or SHD1−/− mice were stimulated with the indicated concentrations of IL-5 plus anti-CD40 (1 μg/mL) for 7 days. The concentration of IgM in the culture supernatants was analyzed by ELISA. The data are the mean plus or minus SD (n = 3). (E) BM cell proliferation. BM cells from wild-type, SHD1+/−, or SHD1−/− mice were cultured in the media containing IL-3 and GM-CSF. Cell numbers were enumerated every day as described in “Proliferation assays for bone marrow cells and mast cells.” The data are the mean plus or minus SD (n = 3). (F) Proliferation of mast cells. Mast cells derived from SHD1-mutant mice were cultured in the presence of 1 or 10 ng/mL IL-3, and the cell numbers were enumerated at the indicated time points (mean ± SD, n = 3). (G) Induction of STAT5 target genes in wild-type, SHD1+/−, or SHD1−/− mice. Total RNA was extracted from cultured T cells stimulated with the indicated concentrations of IL-2 for the indicated times. Quantitative RT-PCR analysis was performed as described in “RT-PCR.” Data are shown as a ratio compared with the average value of prestimulated (0 minutes) wild-type in each graph.
STAT5 also plays a critical role in the IgM production in response to IL-5 in B cells.40 We purified the splenic B cells from SHD1-deficient mice, and examined their response to IL-5. As expected, they presented approximately a 3 times greater IgM production in response to IL-5 compared with that of wild type (Figure 6D). It is of note that there was no difference in the production of IgG1. In addition, the proliferation of B cells in response to IL-4 or anti-CD40 was not different between wild-type and SHD1 mutant mice (data not shown).
Other cytokines that activate STAT5 include IL-3 and GM-CSF. To test the response to these cytokines, we cultured BM cells from the mutant animals in the presence of IL-3 and GM-CSF, and examined their growth response. As shown in Figure 6E, SHD1-deficient cells showed approximately a 3 to 6 times faster proliferation rate in comparison with that of wild-type cells, and the heterozygous cells showed an intermediate response over a 14-day period. Furthermore, SHD1-deficient mast cells proliferated faster than wild-type cells in response to both low (1 ng/mL) and high (10 ng/mL) concentrations of IL-3 (Figure 6F).
Taken together, these results demonstrate that the disruption of SHD1 results in a hyperresponsive phenotype of T, B, and myeloid cells in response to STAT5-activating cytokines such as IL-2, IL-3, IL-5, and GM-CSF.
Induction of STAT5 target genes in SHD1-deficient cells
We next examined whether the expression of STAT5 target genes was enhanced in SHD1-deficient cells. The purified T cells of mutant animals were expanded in culture in vitro, starved, and stimulated with various concentrations of IL-2 for the times indicated (Figure 6G). Real-time reverse-transcription–polymerase chain reaction (real-time RT-PCR) analysis revealed that STAT5 target genes,1 Cis, oncostatin M, bcl-x,41 and cyclin D2,42 were strongly induced in SHD1-deficient T cells in comparison with wild type, and the heterozygous cells showed a moderate response. These results indicate that SHD1 negatively regulates the induction of STAT5 target genes in vivo. It is of note that the inductions of STAT1-responsive gene, IRF1, and STAT3-responsive gene, SOCS-3, were not affected in SHD1 mutant mice (Figure S7), again showing that SHD1-mediated repression was specific to STAT5.
Discussion
In this study, we identified a novel negative feedback regulator of STAT5 that represses STAT5 transcription. Although several mechanisms including protein modifications and protein-protein interactions have been implicated in the STAT5 regulation, cytokine-inducible negative feedback regulator directly affecting STAT5 transcription has not yet been reported. Therefore, SHD1 establishes a novel class of negative feedback loop evoked by cytokine stimulation.
We showed that STAT5 repression by SHD1 does not involve either tyrosine phosphorylation or protein degradation of STAT5. A finding that the transcriptional activity of STAT5 monomer cannot be repressed by SHD1 (Figure 4C) suggests that SHD1 does not disrupt STAT5-coactivator (ie, CBP/p300) complex, and the repression requires dimerization and/or DNA binding of STAT5. The former notion is also supported by the fact that an overexpression of CBP/p300 did not reverse SHD1-mediated STAT5 repression (H.N., unpublished observation, October 2006). In addition, SHD1 does not disrupt the STAT5 DNA-binding complex, since adding a purified SHD1 protein to STAT5 gel-shift reaction did not affect the formation of either the dimers or the tetramers in vitro (H.N., unpublished data, April 2007). With all these, we speculate that SHD1 could repress STAT5 transcription by tethering transcriptional repressor complex to STAT5. In support of this notion, mutant SHD1 lacking STAT5-binding capacity (such as NS2, CS1, and N(LL-AA) mutants) enhanced the STAT5-dependent transcription in reporter assays (data not shown), probably by titrating out repressors available to STAT5. Furthermore, we observed association of SHD1 and transcriptional corepressor, N-CoR, in vitro (H.N., unpublished data, May 2007).
SHD1 contains 2 LXXLL motifs that are known for signature motifs of transcriptional coactivators. It serves as a binding surface for nuclear receptors.35-38 In the case of SHD1, the first LXXLL motif was critical both for the interaction with (Figure 2G) and the repression of (H.N., unpublished data, May 2007) STAT5. PIASy, a transcriptional repressor for STAT1, also contains an LXXLL motif that is essential for STAT1 repression.16 Therefore, the LXXLL motif is not only critical for nuclear receptor-coactivator interaction, but also for the function of STAT repressors.
SHD1 shares homology with a yeast gene, Sac3, which is a suppressor of actin formation and plays a critical role in the G2/M transition of the cell cycle. SHD1 was also reported to be involved in the mitotic progression of mammalian cells, as the suppression of SHD1 mRNA using siRNA resulted in abnormal centromere duplication and spindle assembly.34 However, we did not observe any proliferative defects in SHD1−/− T, B, and BM cells, and the SHD1-deficient mice were grossly normal with no growth retardation. The reason for these conflicting results is not clear at present. It is possible that the siRNA for SHD1 used in an in vitro study could have exerted a nonspecific effect on other genes critical for mitosis. Alternatively, other proteins with similar functions could compensate for the loss of SHD1 in the mutant mice to secure proper mitosis. In this regard, it is noteworthy that another Sac3-homology domain containing protein, GANP has been reported as a gene expressed in the germinal center of lymph nodes, thus being a possible candidate for such compensation.43 In view of the cell cycle progression, it is interesting that SHD1 is induced only by mitogenic cytokines, such as IL-2, IL-3, and Epo. This observation seems to suggest that the expression of SHD1 is cell cycle dependent. Regardless, a detailed analysis of SHD1 mutant mice addressing a defect in the cell cycle, if any, is thus required to reveal the precise role of SHD1 in vivo.
We and others have shown that nuclear receptor corepressor SMRT or ligand-activated peroxisome proliferator-activated receptor (PPAR) α and PPARγ were negative regulators of STAT5-dependent transcription.20,44 In addition, PIASy has been shown to be a corepressor for STAT1.16 In this study, we demonstrated that SHD1 was another example of a negative transcriptional regulator for STAT5. Particularly, SHD1 is distinct from other negative regulators in that it can be induced by cytokine stimulation and it forms a negative feedback loop. The well-known negative feedback regulators in cytokine signaling are SOCS family proteins.21,23,45 However, the SOCS family and SHD1 are different in a few points. First, SOCS proteins block the most upstream step of cytokine signaling by binding to the phosphorylated tyrosine residues of Jaks or the receptors.46,47 In contrast, SHD1 specifically down-regulates STAT5 signaling at the level of transcription. Second, SOCS proteins are immediately (within 30 minutes) induced by cytokine stimulation, whereas SHD1 is induced at least 2 hours after stimulation, a process that requires new protein synthesis (data not shown). Third, a negative regulation by SHD1 is not as strong as that by SOCS proteins. These data suggest that SHD1 is fine-tuning the strength of STAT5 signaling particularly at low cytokine concentrations, whereas SOCS proteins counterbalance positive signals to ensure their prompt regulation. Taken together, SHD1 is a novel class negative feedback regulator specific for STAT5, which is critical for the fine-tuned transcription of STAT5 at a weak signaling strength.
In summary, we herein identified a novel negative feedback transcriptional regulator of STAT5. These findings not only introduce a new mechanism for STAT5 regulation, but they may also help to uncover the still unrecognized regulatory mechanisms of other STAT molecules.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr James N. Ihle and Evan Parganas for their continuous support.
This work was supported in part by a grant from the Ministry of Education, Science, and Culture of Japan (Tokyo, Japan).
Authorship
Contribution: H.N. designed and performed research, analyzed the data, and wrote the paper; T.T., M.I., F.S., K.K., Y.F., and N.W. performed research; T.K. contributed vital new reagents; and Y.I. and M.H. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hideaki Nakajima, Division of Hematology, Department of Internal Medicine, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan; e-mail: hnakajim@sc.itc.keio.ac.jp.
References
Author notes
*T.T., M.I., and F.S. contributed equally to this work.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal