Abstract
Hematopoiesis consists of a series of lineage decisions controlled by specific gene expression that is regulated by transcription factors and intracellular signaling events in response to environmental cues. Here, we demonstrate that the balance between E-protein transcription factors and their inhibitors, Id proteins, is important for the myeloid-versus-lymphoid fate choice. Using Id1-GFP knockin mice, we show that transcription of the Id1 gene begins to be up-regulated at the granulocyte-macrophage progenitor stage and continues throughout myelopoiesis. Id1 expression is also stimulated by cytokines favoring myeloid differentiation. Forced expression of Id1 in multipotent progenitors promotes myeloid development and suppresses B-cell formation. Conversely, enhancing E-protein activity by expressing a variant of E47 resistant to Id-mediated inhibition prevents the myeloid cell fate while driving B-cell differentiation from lymphoid-primed multipotent progenitors. Together, these results suggest a crucial function for E proteins in the myeloid-versus-lymphoid lineage decision.
Introduction
During hematopoiesis, stem cells lose lineage plasticity as they become more differentiated. In general, long-term hematopoietic stem cells (LT-HSCs) give rise to short-term HSCs (ST-HSCs), which have a limited ability to self-renew and can further differentiate into the earliest progenitors of specific hematopoietic lineages.1-3 Early work on the hierarchy of progenitor populations suggested that ST-HSCs or multipotent progenitors could differentiate into either common myeloid progenitors (CMPs) or common lymphoid progenitors (CLPs), and these exclusively formed myeloid and lymphoid lineage cells, respectively.4,5 More recent studies have shown that other intermediary progenitors and alternative pathways exist.6,7 For instance, lymphoid-primed multipotent progenitors (LMPPs) have been identified as a subset of the heterogeneous lin−c-kit+Sca-1+ (LSK) progenitor pool by their high levels of Flt3 expression.6,8 LMPPs have a limited ability to produce megakaryocyte and erythroid lineage cells but can generate both myeloid and lymphoid lineage cells.6,9 Therefore, LMPPs have been suggested to represent a possible branch point in myeloid-versus-lymphoid lineage specification and that regulation of specific gene programs at this stage may instruct lineage fate.
Transcription factors play crucial roles in lineage decisions through the regulation of specific gene expression.10-13 For example, ablation of the transcription factor PU.1 eliminates lineage differentiation of primitive progenitors, whereas partial or full reconstitution of PU.1 in fetal liver progenitors results in biased production of either pro-B cells or macrophages, respectively.13 Moreover, lineage-specific genes are affected by the levels of PU.1 in early hematopoietic progenitors, suggesting a mechanism for the determination of lineage cell fate.13,14 Transcription factors also play crucial roles in maintaining the balance between differentiation and proliferation, which is best demonstrated by the deregulated expression of these proteins in leukemic processes.15,16 Therefore, it is important to determine the role of transcription factors in physiologic processes, such as lineage decisions, and to pinpoint the stage at which they control cell fate.
Basic helix-loop-helix (bHLH) proteins, such as E2A, E2-2, and HEB (collectively named E proteins), are transcription factors known to modulate lymphopoiesis.17,18 E proteins form homodimers or heterodimers, bind to specific DNA sequences called E boxes, and activate transcription of lymphoid-specific target genes.19,20 E proteins are essential for B lymphopoiesis because E2A-deficient mice fail to produce B lymphocytes.21,22 Because E proteins are ubiquitously expressed, their regulation is largely dependent on the tissue- and stage-specific expression of their natural inhibitors, Id proteins.23,24 Id proteins are HLH proteins that can heterodimerize with E proteins but lack a basic DNA-binding region.23 Therefore, these heterodimers are unable to bind DNA, thus inhibiting E-protein function.23 Ectopic Id expression inhibits early stages of B lymphopoiesis by blocking E protein–dependent transcription of critical mediators of lymphoid development.25 Although it is commonly accepted that Id proteins inhibit lymphopoiesis through the negative regulation of E-protein activity, it has been suggested that Id proteins may modulate cellular processes in an E protein–independent manner.26 Therefore, it is important to determine whether physiologic processes, such as the myeloid-versus-lymphoid lineage decision, are regulated by Id proteins through the inhibition of E-protein activities.
Compared with lymphopoiesis, the role of E proteins in myelopoiesis is less well delineated. However, previous work does suggest that Id proteins promote myeloid differentiation. Studies using cell lines indicate that both Id1 and Id2 are highly expressed in myeloid lineage cells, and Id1 expression can be induced by interleukin-3 (IL-3) or granulocyte-macrophage colony-stimulatingfactor (GM-CSF).27-29 Consistent with its expression profile, it has been demonstrated that overexpression of Id1 in 5-fluorouracil (5-FU)–enriched bone marrow promotes commitment toward the myeloid lineage.28 However, these studies have not addressed important questions, including whether Id proteins play a physiologic role in lineage decision, at what stage in hematopoiesis Id1 exerts its effect, whether Id isoforms play redundant roles, and whether the myeloid-versus-lymphoid lineage choice is E protein–dependent.
To obtain high-resolution information about Id1 expression in hematopoietic progenitors and myeloid lineage cells, we have generated Id1 knockin mice, designated as Id1G/G, where green fluorescent protein (GFP) is inserted into the Id1 gene downstream of its transcriptional start site. This model allowed us to examine Id1 expression in the bone marrow on an individual cell basis. We demonstrate that Id1 as indicated by GFP fluorescence is not expressed in CMP or megakaryocyte and erythroid progenitors (MEPs) but is up-regulated in granulocyte and macrophage progenitors (GMPs) as well as downstream myeloid lineage cells. Although ectopic expression of Id1 in LSK and 5-FU–enriched progenitors promotes myeloid and inhibits lymphoid differentiation, no defect in myelo- or lymphopoiesis in Id1-deficient (Id1G/G) mice was detected,30 suggesting that different Id isoforms play redundant roles in regulating lineage fate. Indeed, we show that Id-mediated inhibition of E proteins is involved in myeloid lineage commitment because enforcing E-protein activity at the lymphoid-primed progenitor (Flt3HiLSK) but not committed myeloid progenitor stages prevents myeloid differentiation. Together, these data suggest that the balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions.
Methods
Mice
C57BL/6 (CD45.2) and B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Id1/EGFP knockin mice were described previously.31 ROSA26-CreERT2 mice were generous gifts from Dr Thoma Ludwig (Columbia University, New York, NY).32
To generate ROSA26-ET2 mice, the construct was created by inserting a SalI-SacII fragment containing ET2 and GFP sequences into the pBigT vector,33 from which a PacI-AscI (partial digest) fragment was isolated and inserted into the ROSA26PA vector.33,34 The construct was linearized and electroporated into 129 × 1/SvJ embryonic stem cells. Embryonic stem cells containing properly targeted ROSA26-ET2 alleles were identified by Southern blotting and polymerase chain reaction (PCR) assays and then injected into C57BL/6 blastocysts. ROSA26-ET2 progenies were backcrossed to C57BL/6 mice for 8 generations.
Mice were bred and maintained in the Laboratory Animal Resource Center of the Oklahoma Medical Research Foundation in a specific pathogen–free environment and handled according to protocols preapproved by the Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee.
Isolation of cell populations and flow cytometry
Collection and staining of cells were performed in Hank balanced salt solution (Invitrogen, Carlsbad, CA) containing 5% fetal bovine serum and 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer. In addition, all bone marrow cells were treated with a hypotonic solution to lyse red blood cells. Lineage-negative bone marrow cells were obtained by a 30-minute incubation at 4°C with antibodies against Gr1 (Ly-6C/G; RB6-8C5), Mac-1 (M1/70), Ter-119, CD2 (RM2-5), CD3 (17A2), CD5 (53-7.3), CD8 (53-6.7), CD19 (1D3), and B220 (RA3/6B2). Labeled cells were then incubated for 30 minutes at 4°C with sheep anti–rat Ig-coupled magnetic beads (DynaTech, Oslo, Norway) to remove lineage-positive cells. Lineage-negative cells were stained first with biotin-conjugated antibodies against CD8a (RM2215), Gr-1, Mac-1, NK1.1 (NKR-P1B and NKR-P1C; PK136), Ter-119, and B220; and then with a streptavidin phycoerythrin–Texas Red antibody to gate out any remaining positive cells. Subsequently, cells were stained with antibodies against Sca-1 (D7; eBioscience, San Diego, CA) and c-kit (2B8) to isolate LSK progenitors. Myeloid progenitor compartments were resolved with antibodies against Sca-1 and c-kit, FcγR, and CD34. LMPPs were isolated by gating on 25% of LSKs with the highest levels of Flt3 expression. Cell sorting was performed using a MoFlo cell sorter (Dako Colorado, Fort Collins, CO), and fluorescence-activated cell sorter (FACS) analyses were carried out using an LSRII (BD Biosciences, San Jose, CA). All antibodies described in this section were purchased from BD Biosciences PharMingen (San Diego, CA) unless otherwise stated.
Cytokine stimulation of LSK and WBM
LSK progenitors were isolated from Id1+/G mice and cultured for 48 hours in X-VIVO15 media (Lonza BioWhittaker, Walkersville, MD), containing 1% detoxified bovine serum albumin (StemCell Technologies, Vancouver, BC), 100 ng/mL stem cell factor (SCF) plus 10 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL GM-CSF, or 20 ng/mL IL-7 (R&D Systems, Minneapolis, MN). Whole bone marrow cells from Id1G/G mice were stimulated with the same set of cytokines for 24 hours and stained with antibodies against CD19 and Mac-1. Recombinant cytokines were purchased from PeproTech (Rocky Hill, NJ) unless indicated.
Retroviral infection and bone marrow transplant
LSK progenitors were isolated and placed into a 6-well tissue-culture dish at a density of 80 000 cells/well. For transplantation studies, whole bone marrow cells were obtained from mice treated with 2 μg 5-FU for 4 days and used for transduction. These cells were resuspended in X-VIVO15 media containing 1% detoxified bovine serum albumin and prestimulated for 12 hours with 100 ng/mL SCF, 20 ng/mL of thrombopoietin, and 100 ng/mL Flt3-L. Retroviral supernatants generated by transient transfection of the Phoenix-E packaging cell line were mixed at a 1:1 ratio with the cell suspension for spin-infection at 1800g for 1.5 hours at 26°C followed by incubation at 37°C for 4 hours. A second spin-infection with fresh media and retroviral supernatants was performed. Cells were cultured for 36 hours in media containing fresh cytokines. GFP-positive cells were sorted into 96-well plates at 2500 cells/well or used for intravenous transplantation into lethally irradiated hosts.
Cell culture
For cultures in myeloid-promoting cytokines, cells were sorted into 96-well round-bottom plates in X-VIVO15 media containing 1% detoxified bovine serum albumin, 100 ng/mL SCF, 10 ng/mL IL-3, 20 ng/mL IL-6, and 20 ng/mL granulocyte colony-stimulating factor (G-CSF) and cultured for 4 days. For cultures in lymphoid-promoting conditions, cells were plates in the same media supplemented with 50 ng/mL SCF, 100 ng/mL Flt3-L, and 1 ng/mL IL-7. Cells were split at 1:3 on days 7, 9, and 11, and harvested on day 14 for FACS analyses.
For colony assays, LSK or myeloid progenitors (1000 cells/plate) were mixed with methylcellulose media for myeloid cells (MC3434; StemCell Technologies). Cells were dispensed with an 18-gauge needle into 35-mm plates and incubated for 6 days. Colonies were enumerated with an inverted microscope.
RT-PCR analysis of gene expression in early hematopoietic progenitors
Total RNA was isolated from sorted progenitor populations. Real-time PCR was performed with the following pairs of primers: M-CSFR, CCGGCCCACTCTTGGAATTT and AGACCGTTTTGCGTAAGACCT; Ig-Iμ GGGAATGTATGGTTGTGGC and CCAGGTGAA GGAAATGG. Primers for p21 (Cdkn1a) were purchased from QIAGEN (Valencia, CA).
Results
Id1 is expressed in the myeloid lineage
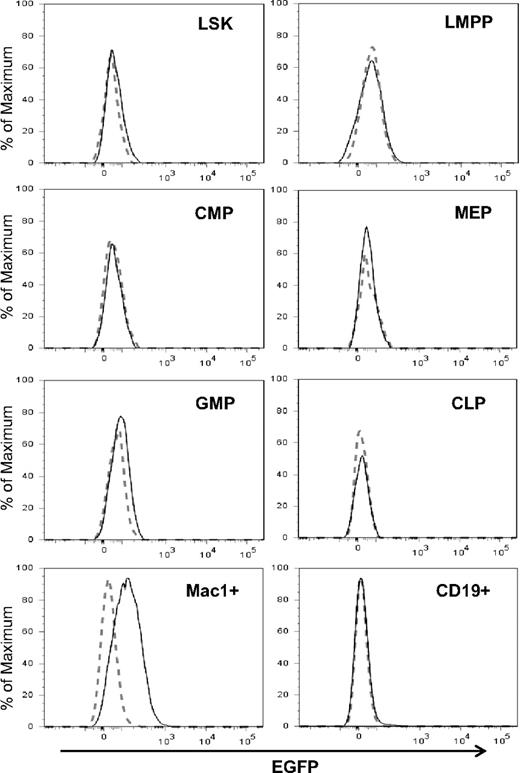
To determine levels of Id1 expression in hematopoietic progenitor populations, Id1/GFP knockin mice were created by inserting the GFP coding sequence downstream of the Id1 transcription start site as described previously.31 In these knockin mice, GFP levels have been shown to correlate with levels of Id1 mRNA.31 Insertion of GFP prevents the synthesis of Id1 protein and homozygous mice; Id1G/G are therefore deficient of Id1. Because steady-state hematopoiesis in Id1-deficient mice was not perturbed,30 we were able to determine Id1 expression using Id1G/G mice, which showed higher intensity of GFP fluorescence and more consistent results than heterozygous mice. To determine Id1/GFP expression in various phenotypic subsets of the bone marrow, we stained cells for markers that define each of these populations (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and compared GFP fluorescence between wild-type and Id1G/G cells by overlaying the histograms (Figure 1). Consistent with our previous findings,31 LSK from Id1 knockin mice exhibited a slight shift in GFP fluorescence, which at least in part, reflects Id1/GFP expression in long-term stem cells, which represent a small fraction of LSK. In contrast, Flt3HiLSK (LMPP) did not show any detectable levels of expression. In the Lin−c-kit+Sca-1− compartment, only the GMP but not MEP and CMP subsets expressed GFP. Both CLP and CD19+ B-lineage cells were negative for GFP. However, Mac-1+ myeloid cells produced high levels of Id1/GFP (Figure 1). Robust Id1/GFP expression was also found in myeloid lineage cells in the blood and peritoneum (data not shown). Therefore, it appears that Id1 expression is closely associated with myeloid lineage cells and begins to increase at the transition from LMPP to GMP in the hierarchy of hematopoiesis.
Id1 expression in myeloid lineage cells. Indicated bone marrow subsets were gated as shown in Figure S1. The definitions of the subsets are as follows: LSK, Lin−Sca-1+c-kit+; LMPP, Flt3HiLSK; CMP, Lin−Sca-1−c-kit+CD34+FcγR−; MEP, Lin−Sca-1−c-kit+CD34−FcγR+; GMP, Lin−Sca-1−c-kit+CD34+FcγR+; CLP, Lin−Sca-1+c-kitloIL7R+. Histograms of GFP fluorescence in Id1G/G cells (solid line) were superimposed on those in wild-type cells (dotted line). Data are representative of at least 3 independent experiments.
Id1 expression in myeloid lineage cells. Indicated bone marrow subsets were gated as shown in Figure S1. The definitions of the subsets are as follows: LSK, Lin−Sca-1+c-kit+; LMPP, Flt3HiLSK; CMP, Lin−Sca-1−c-kit+CD34+FcγR−; MEP, Lin−Sca-1−c-kit+CD34−FcγR+; GMP, Lin−Sca-1−c-kit+CD34+FcγR+; CLP, Lin−Sca-1+c-kitloIL7R+. Histograms of GFP fluorescence in Id1G/G cells (solid line) were superimposed on those in wild-type cells (dotted line). Data are representative of at least 3 independent experiments.
Myeloid-promoting cytokines stimulate Id1 expression
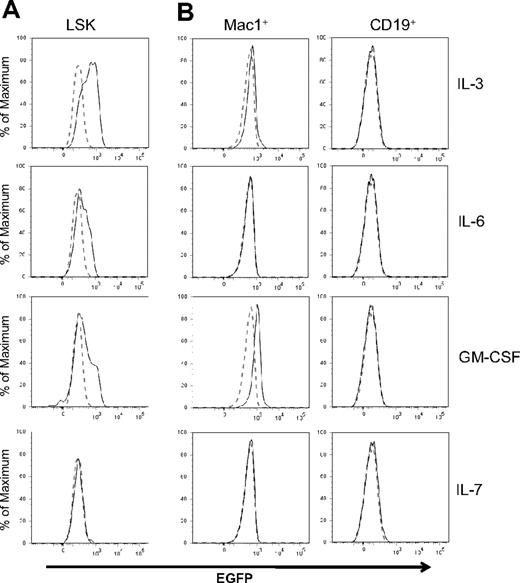
Id1 expression depends on STAT5-mediated signal transduction in IL-3–treated cell lines and bulk cultures.29,35 Using Id/GFP knockin mice, we determined levels of Id1 expression in LSK progenitors in response to various cytokines important in hematopoiesis. SCF was included in all cultures because it facilitates the survival of progenitors but it by itself did not stimulate Id1 expression (data not shown). However, Id1/GFP expression was greatly enhanced in LSK progenitors by a 48-hour treatment with IL-3, GM-CSF, or IL-6 (Figure 2A). An increase in GFP expression was observed as early as 6 hours after IL-3 stimulation of LSK progenitors (data not shown). In contrast, these progenitors did not express GFP after stimulation with IL-7 (Figure 2A), IL-4, IL-5, Flt3-L, G-CSF, macrophage colony-stimulating factor (M-CSF), erythropoietin, or thrombopoietin (data not shown). In lineage-positive bone marrow cells, we detected a significant stimulation of GFP expression by GM-CSF and a less robust increase by IL-3 in Mac-1+ cells (Figure 2B). IL-6 did not appear to have any effect on these cells. In contrast to myeloid cells, CD19+ B lineage cells did not respond to any of these cytokines. Together, these results suggest that cytokines promoting myeloid differentiation are capable of up-regulating Id1 expression in multipotent progenitors and myeloid lineage cells.
Regulation of Id1 expression by cytokines. (A) LSK progenitors from Id1/GFP knockin mice were treated with SCF plus indicated cytokines for 48 hours. (B) Mac-1+ or CD19+ whole bone marrow cells of Id1/GFP knockin mice were treated with the same cytokines for 24 hours. Histograms of GFP fluorescence in cells treated with indicated cytokines are indicated by solid lines, whereas those in cells treated with SCF alone are indicated by dotted lines. Data are representative of at least 3 independent experiments.
Regulation of Id1 expression by cytokines. (A) LSK progenitors from Id1/GFP knockin mice were treated with SCF plus indicated cytokines for 48 hours. (B) Mac-1+ or CD19+ whole bone marrow cells of Id1/GFP knockin mice were treated with the same cytokines for 24 hours. Histograms of GFP fluorescence in cells treated with indicated cytokines are indicated by solid lines, whereas those in cells treated with SCF alone are indicated by dotted lines. Data are representative of at least 3 independent experiments.
Ectopic expression of Id1 in LSK progenitors enhances myeloid differentiation but suppresses B lymphopoiesis
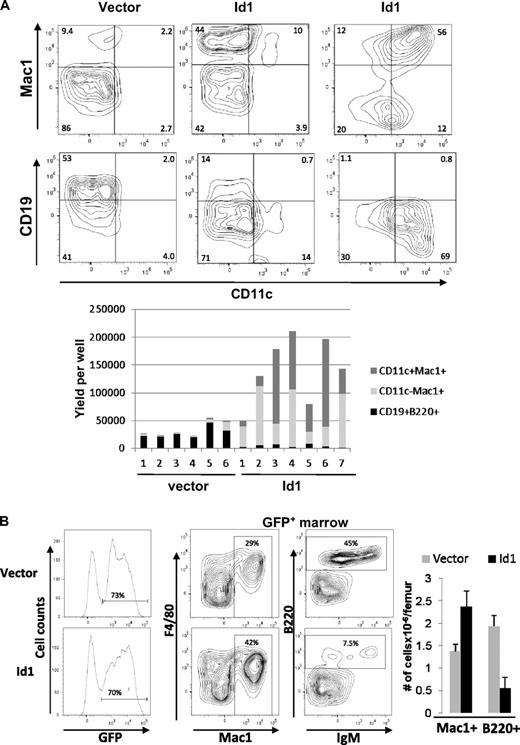
Because Id1 is expressed at early stages of myelopoiesis, we wanted to determine whether ectopic Id1 expression influenced myeloid differentiation. LSK progenitors were transduced with either vector or Id1-expressing retrovirus and assayed for their potential to differentiate along the myeloid and lymphoid lineages in vitro and in vivo. When cultured with SCF, Flt3-L, and IL-7, vector-transduced LSK progenitors gave rise to predominantly CD19+ B-lineage cells and some Mac-1+ myeloid lineage cells (Figure 3A left). In contrast, Id1-transduced LSK produced largely Mac-1+ cells and very few CD19+ lymphoid cells (Figure 3A center and right). In addition, a significant portion of wells containing Id1-transduced cells had robust production of cells expressing both Mac-1 and CD11c, which are characteristic of myeloid dendritic cells (Figure 3A right). Enumeration of lymphoid and myeloid cells produced in each of the individual wells are shown in the bar graph (Figure 3A bottom panel). On average, Id1 expression resulted in a 20-fold increase in myeloid production and a 7-fold decrease in B lymphoid differentiation.
Overexpression of Id1 in early HSCs drives myeloid differentiation at the expense of lymphoid development. (A) LSK progenitors were transduced with vector or Id1-expressing retrovirus. Transduced cells were sorted for GFP expression 36 hours later and plated at 2500 cells/well in X-VIVO15 media containing SCF, Flt3-L, and IL-7. After 14 days, cells were harvested and stained with the indicated antibodies and analyzed by flow cytometry. Representative plots are shown for wells containing vector (left) and Id1-transduced cells producing either predominantly Mac-1+CD11c− (center) or Mac-1+CD11c+ cells (right). The proportions of indicated cell populations produced from vector or Id1-transduced progenitors in individual wells are shown in the bar graph below the plots. Data are representative of 3 experiments. (B) CD45.1+ bone marrow cells from 5-FU–treated wild-type mice were transduced with vector or Id1-expressing retrovirus. Transduced cells were sorted for GFP expression, and equal numbers of transduced cells were injected into lethally irradiated CD45.2+ host mice. Three weeks later, bone marrow cells from transplanted hosts were isolated and analyzed using flow cytometry. GFP expression on donor-derived cells is shown in histograms on the left. Representatives of FACS analyses for indicated markers on gated GFP+ cells are shown, and the percentage of cells in each quadrant is indicated. Average numbers of Mac1+ and B220+ cells/femur derived from vector and Id1-transduced cells are shown in the bar graph with SD (n = 3). Data are representative of 3 experiments.
Overexpression of Id1 in early HSCs drives myeloid differentiation at the expense of lymphoid development. (A) LSK progenitors were transduced with vector or Id1-expressing retrovirus. Transduced cells were sorted for GFP expression 36 hours later and plated at 2500 cells/well in X-VIVO15 media containing SCF, Flt3-L, and IL-7. After 14 days, cells were harvested and stained with the indicated antibodies and analyzed by flow cytometry. Representative plots are shown for wells containing vector (left) and Id1-transduced cells producing either predominantly Mac-1+CD11c− (center) or Mac-1+CD11c+ cells (right). The proportions of indicated cell populations produced from vector or Id1-transduced progenitors in individual wells are shown in the bar graph below the plots. Data are representative of 3 experiments. (B) CD45.1+ bone marrow cells from 5-FU–treated wild-type mice were transduced with vector or Id1-expressing retrovirus. Transduced cells were sorted for GFP expression, and equal numbers of transduced cells were injected into lethally irradiated CD45.2+ host mice. Three weeks later, bone marrow cells from transplanted hosts were isolated and analyzed using flow cytometry. GFP expression on donor-derived cells is shown in histograms on the left. Representatives of FACS analyses for indicated markers on gated GFP+ cells are shown, and the percentage of cells in each quadrant is indicated. Average numbers of Mac1+ and B220+ cells/femur derived from vector and Id1-transduced cells are shown in the bar graph with SD (n = 3). Data are representative of 3 experiments.
To examine the effects of Id1 overexpression in a more physiologic setting, we transplanted 5-FU–treated bone marrow cells transduced with vector or Id1-expressing retrovirus into lethally irradiated recipients. Three weeks later, the hosts were humanely killed and bone marrow cells were analyzed by flow cytometry. Because the retroviral constructs also express GFP from an internal ribosome entry site, transduced cells were analyzed by gating on GFP-expressing cells. Differentiation was then scored by Mac-1 and F4/80 expression for myeloid and B220 and IgM expression for lymphoid cells. Similar to the results from in vitro assays, Id1-expressing progenitors produced greater percentages and numbers of myeloid cells compared with vector-transduced progenitors (Figure 3B). In contrast, lymphoid differentiation from Id1-transduced progenitors was significantly reduced relative to vector-transduced progenitors (Figure 3B). These results strongly suggest that ectopic expression of Id1 biases hematopoietic progenitors to the myeloid lineage and suppresses lymphoid development.
Expression of a dominant-negative mutant with unopposed E-protein activity
Although gain of Id1 function clearly promotes myeloid differentiation, ablation of the Id1 gene in Id1G/G mice did not appear to impair this process in any hematopoietic compartment (Figure S1).31 We reasoned that expression of functionally redundant Id as well as SCL/Tal1 proteins during myelopoiesis could compensate for the loss of Id1, resulting in apparently normal hematopoiesis. A common and well-established function of these molecules is to inhibit the activity of E proteins. Because it is impossible to disrupt the genes encoding all Id proteins because loss of 2 or more Id isoforms has been shown to result in embryonic lethality,36 a dominant-negative approach had to be taken to examine the collective roles of these inhibitory molecules in myeloid differentiation. We previously constructed a chimeric protein, termed ET2, which is capable of neutralizing the function of both Id and SCL/Tal proteins.37,38 ET2 consists of the transactivation domains of E47 and the bHLH domain of SCL/Tal1 (diagrammed in Figure 4C). Because the bHLH domain of SCL/Tal1 does not mediate homodimerization but has high affinity for E proteins, ET2 can compete with Id and SCL/Tal1 proteins to bind to endogenous E proteins. We have shown that ET2 does not exhibit transcriptional activity by itself but acts as a potent transcription activator when associated with wild-type E47.37 ET2 was able to compete with Tal1 to form complexes with endogenous E2A proteins in Jurkat T cells.38 This is important because overexpression of wild-type E47 is known to be detrimental to cells, whereas ET2 is tolerable because of the limitation of its activity by levels of endogenous E proteins. These properties of ET2 offered an excellent opportunity for examining the effect of gain of E-protein function or loss of Id and SCL/Tal1 function on the myeloid-versus-lymphoid lineage decision.
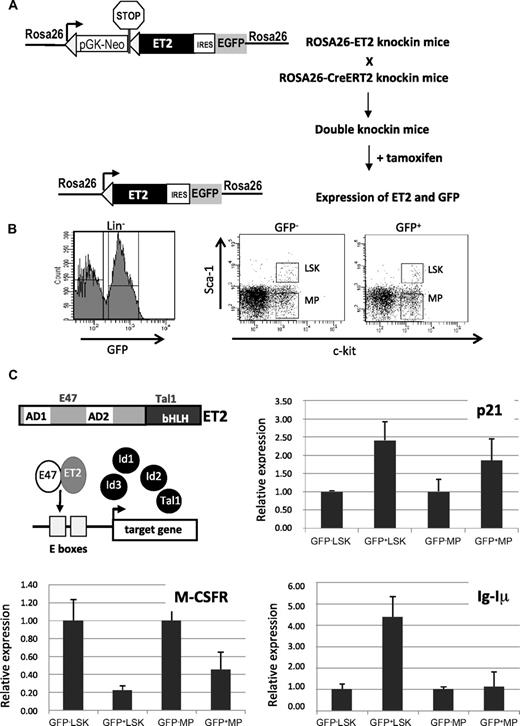
Tamoxifen-inducible ET2 expression and its effects on gene expression. (A) Diagram for induction of ET2 expression in vivo. In ROSA26-ET2 knockin mice, a construct was inserted downstream of the promoter of the ROSA26 gene. The construct contains a selection marker (pGK-NeoPGK-pA) and a transcriptional stop sequence (shown as a stop sign) flanked by 2 loxP sites (triangles), which are followed by the ET2 sequence plus the GFP sequence whose translation is mediated by an internal ribosome binding site (IRES). ROSA26-ET2 mice were crossed with ROSA26-CreERT2 mice, which produce a tamoxifen-inducible Cre. The double knockin mice were then treated with 5 doses of tamoxifen at 3 mg/mouse over 4 days to excise the selection marker and stop sequence, allowing expression of both ET2 and GFP. (B) Bone marrow cells from treated double knockin mice were harvested and lineage-depleted cells were obtained. Lineage-negative cells were sorted based on expression of GFP along with c-kit and Sca-1 markers. GFP-negative and -positive LSK and MP were collected based on the gates illustrated on the right. (C) Effect of ET2 on gene expression. A diagram shows the structure of ET2 protein and its proposed mechanism of action, which involves competition with Id proteins to dimerize with endogenous E proteins and subsequent binding to E boxes facilitating target gene transcription. To assess the function of ET2, real-time PCR assays were performed using total RNA isolated from cell populations sorted as described in panel B. Average levels of expression of indicated genes relative to that of β-actin in each indicated cell population are shown in bar graphs with SD. GFP-positive and -negative cells are considered to represent cells with or without ET2 expression.
Tamoxifen-inducible ET2 expression and its effects on gene expression. (A) Diagram for induction of ET2 expression in vivo. In ROSA26-ET2 knockin mice, a construct was inserted downstream of the promoter of the ROSA26 gene. The construct contains a selection marker (pGK-NeoPGK-pA) and a transcriptional stop sequence (shown as a stop sign) flanked by 2 loxP sites (triangles), which are followed by the ET2 sequence plus the GFP sequence whose translation is mediated by an internal ribosome binding site (IRES). ROSA26-ET2 mice were crossed with ROSA26-CreERT2 mice, which produce a tamoxifen-inducible Cre. The double knockin mice were then treated with 5 doses of tamoxifen at 3 mg/mouse over 4 days to excise the selection marker and stop sequence, allowing expression of both ET2 and GFP. (B) Bone marrow cells from treated double knockin mice were harvested and lineage-depleted cells were obtained. Lineage-negative cells were sorted based on expression of GFP along with c-kit and Sca-1 markers. GFP-negative and -positive LSK and MP were collected based on the gates illustrated on the right. (C) Effect of ET2 on gene expression. A diagram shows the structure of ET2 protein and its proposed mechanism of action, which involves competition with Id proteins to dimerize with endogenous E proteins and subsequent binding to E boxes facilitating target gene transcription. To assess the function of ET2, real-time PCR assays were performed using total RNA isolated from cell populations sorted as described in panel B. Average levels of expression of indicated genes relative to that of β-actin in each indicated cell population are shown in bar graphs with SD. GFP-positive and -negative cells are considered to represent cells with or without ET2 expression.
To express ET2 in vivo, we created knockin mice where the coding sequence of ET2 was introduced into the ROSA26 locus (Figure 4A). In this construct, the neomycin resistance gene and transcriptional termination sequence flanked by lox-P sites were placed upstream of ET2, so that ET2 was only expressed when these sequences were excised by Cre recombinases. In addition, GFP was also produced from the same transcript via an internal ribosome entry site and served as an indicator for ET2 expression. We crossed these mice with ROSA26-CreERT2 mice,32 in which constitutive expression of a tamoxifen-inducible Cre recombinase is driven by the ROSA26 promoter. Double knockin mice were then treated with 5 doses of tamoxifen, and bone marrow cells were harvested and sorted 3 days after induction. Lineage-negative bone marrow cells were fractionated based on c-kit and Sca-1 expression, as well as GFP expression (Figure 4B). The lin−c-kit+Sca-1+ (LSK) population consists of hematopoietic stem cells and multipotent progenitors, whereas the Lin−Sca-1−c-kit+ subset includes various myeloid progenitors (MPs) such as MEP, CMP, and GMP. GFP-positive and -negative LSK and MP fractions represent cells with or without ET2 expression, respectively (Figure 4B).
The function of ET2 proteins in these progenitors was verified by examining the expression of a known target gene of E2A proteins, the cyclin-dependent kinase inhibitor p21 (Figure 4C). Expression of the endogenous p21 gene has been shown to be highly responsive to E-protein activity mediated by E47 homodimers or E47/ET2 heterodimers in transient transfection assays.37,39 Therefore, p21 serves as an excellent indicator of E-protein function. Indeed, using real-time PCR assays, we detected a significant increase in p21 expression in GFP+ ET2-expressing LSK progenitors and, to a lesser extent, in MP cells (Figure 4C). Furthermore, we used the same samples to test the expression of known myeloid- and lymphoid-specific genes, M-CSF receptor (M-CSFR) and the sterile transcript, Iμ, of the immunoglobulin heavy chain gene (IgH) locus, respectively (Figure 4C). ET2 expression led to a down-regulation of M-CSFR in both LSK and MP subsets, which may be a result of a direct or indirect repression of M-CSFR transcription by ET2 or a result of the fact that different percentages of M-CSFR-producing myeloid progenitors were present in the ET2/GFP+ LSK and MP fractions compared with their ET2/GFP− counterparts. Expression of Iμ is driven by the intronic enhancer of the IgH gene and known to be activated by E proteins.40 Therefore, it is not surprising that ET2 could enhance its expression in LSK progenitors (Figure 4C bottom right). Interestingly, Iμ expression was not activated by ET2 in the MP population, suggesting that the IgH locus is shut off after myeloid specification. This is intriguing because E2A can stimulate Iμ expression in nonhematopoietic fibroblasts.41 Collectively, data from these gene expression studies suggest that E-protein function can be enhanced by ET2 expression, which could then influence myeloid and lymphoid differentiation by modulating lineage-specific gene programs.
Gain of E-protein function in LSK but not MP cells results in suppression of myelopoiesis
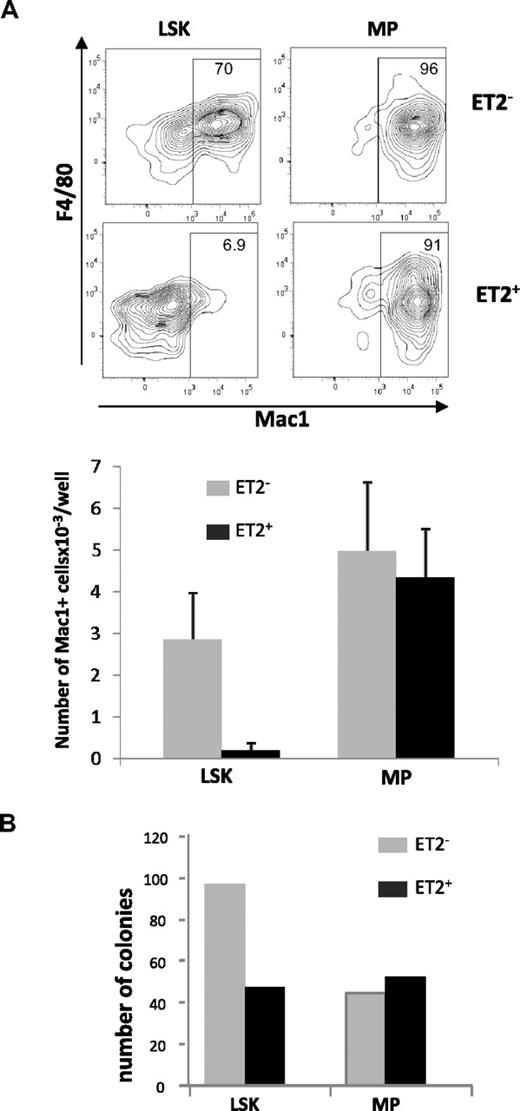
Because Id1 is preferentially expressed in myeloid lineage cells and overexpression of Id1 promoted myeloid differentiation, it is reasonable to hypothesize that gain of E-protein function diminishes myelopoiesis. To test this hypothesis, we isolated GFP-positive and -negative LSK or MP populations from ROSA26-ET2 mice, where ET2 expression was activated by tamoxifen-inducible Cre. These cells were seeded in multiple wells and cultured in media containing SCF, G-CSF, IL-3, and IL-6. Cells were harvested on day 4, stained for myeloid-specific cell surface markers, and analyzed by flow cytometry. ET2-expressing LSK progenitors produced 10-fold lower percentages and numbers of myeloid lineage cells compared with cells derived from GFP-negative LSKs that do not express ET2 (Figure 5A). In contrast, production of Mac-1+ cells by MP cells was not affected by ET2 expression (Figure 5A).
ET2 expression at the LSK but not the MP stage suppresses myeloid potential. (A) GFP-positive (ET2+) and -negative (ET2−) LSK and myeloid progenitor (MP) populations were isolated from mice as described for Figure 4 and seeded at 200 cells/well in media containing SCF, IL-3, IL-6, and G-CSF. Four days later, cells were stained with antibodies against F4/80 and Mac-1 and analyzed by flow cytometry. Percentages of Mac-1+ cells are indicated within the gate. Average numbers of Mac1+ cells/well (n = 6) produced are shown in the bar graphs below the plots. Data are representative of 2 experiments. (B) Colony formation assays. Additional aliquots of the cells as used for panel A were placed into myeloid-promoting methylcellulose media at a density of 1000 cells/plate and grown for 6 days. Colonies were then enumerated, and the average number of colonies per plate is shown in the bar graph (n = 2).
ET2 expression at the LSK but not the MP stage suppresses myeloid potential. (A) GFP-positive (ET2+) and -negative (ET2−) LSK and myeloid progenitor (MP) populations were isolated from mice as described for Figure 4 and seeded at 200 cells/well in media containing SCF, IL-3, IL-6, and G-CSF. Four days later, cells were stained with antibodies against F4/80 and Mac-1 and analyzed by flow cytometry. Percentages of Mac-1+ cells are indicated within the gate. Average numbers of Mac1+ cells/well (n = 6) produced are shown in the bar graphs below the plots. Data are representative of 2 experiments. (B) Colony formation assays. Additional aliquots of the cells as used for panel A were placed into myeloid-promoting methylcellulose media at a density of 1000 cells/plate and grown for 6 days. Colonies were then enumerated, and the average number of colonies per plate is shown in the bar graph (n = 2).
To verify findings obtained in liquid cultures, GFP-negative and -positive LSK and MP progenitors were also placed into myeloid-supportive methylcellulose cultures. After culturing for 6 days, colonies formed were enumerated. Compared with GFP-negative cells, GFP-positive ET2-expresssing LSK progenitors produced fewer myeloid cell colonies (Figure 5B). However, ET2-expressing MP cells generated similar numbers of colonies (Figure 5B). Taken together, these results suggest that ET2 can suppress myelopoiesis at the multipotent LSK progenitor stage but not at lineage-restricted myeloid progeni-tor stages.
E-protein activity at the Flt3HiLSK (LMPP) stage instructs myeloid-versus-lymphoid lineage fate
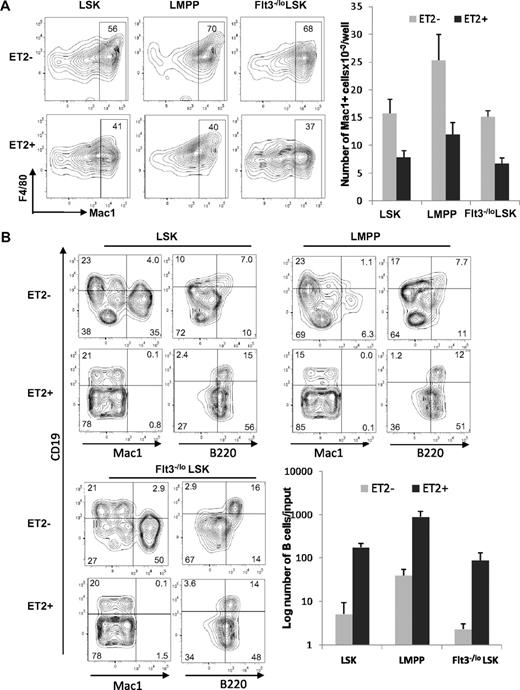
To further pinpoint the stage at which E proteins function to influence myeloid-versus-lymphoid lineage decisions, we determined the differentiation potential of ET2-expressing LMPP. LMPP is a subset of LSK progenitors and thought to have both myeloid and lymphoid potential. We isolated GFP-negative and -positive LSK progenitors, as demonstrated in Figure 4, and separated them into Flt3−/lo (containing HSC and MPP) and Flt3hi (representing LMPP) subsets. The GFP-positive populations were considered to express ET2, whereas the GFP-negative counterparts served as negative controls. When these progenitors were placed into cultures containing myeloid-promoting cytokines, ET2 expression in all 3 subsets significantly inhibited myeloid differentiation, as indicated by both the percentages and total numbers of Mac-1+ cells produced after 4 days (Figure 6A). Because of a higher density of cells seeded in this experiment, myeloid differentiation from ET2− LSK might have reached a plateau, thus making the difference in the number of Mac-1+ cell produced by ET2− and ET2+ cells slightly less dramatic than data shown in Figure 5A.
ET2 expression at the LMPP stage inhibits myeloid development and enhances B-cell production. (A) GFP-positive (ET2+) and -negative (ET2−) total LSK, Flt3HiLSK (LMPP), and Flt3−/loLSK were isolated from mice as described for Figure 4 and seeded at 300 cells/well in media containing SCF, IL-3, IL-6, and G-CSF. Four days later, cells were analyzed with the indicated antibodies. The number of Mac-1+ cells produced from each progenitor subset is shown in the bar graph on the right (n = 3). (B) The same subsets of progenitors were also placed at 1250 cells/well in media containing SCF, Flt3-L, and IL-7 and cultured for 16 days. Cells were stained with indicated antibodies and analyzed by flow cytometry. Average numbers of B220+CD19+ cells are shown in bar graphs with SD (n = 3).
ET2 expression at the LMPP stage inhibits myeloid development and enhances B-cell production. (A) GFP-positive (ET2+) and -negative (ET2−) total LSK, Flt3HiLSK (LMPP), and Flt3−/loLSK were isolated from mice as described for Figure 4 and seeded at 300 cells/well in media containing SCF, IL-3, IL-6, and G-CSF. Four days later, cells were analyzed with the indicated antibodies. The number of Mac-1+ cells produced from each progenitor subset is shown in the bar graph on the right (n = 3). (B) The same subsets of progenitors were also placed at 1250 cells/well in media containing SCF, Flt3-L, and IL-7 and cultured for 16 days. Cells were stained with indicated antibodies and analyzed by flow cytometry. Average numbers of B220+CD19+ cells are shown in bar graphs with SD (n = 3).
When LSK subsets were placed into culture conditions that support lymphoid differentiation, ET2 expression led to a robust production of B cells at the expense of myelopoiesis (Figure 6B). Although the percentage of CD19+B220+ cells did not change in cultures seeded with ET2-expressing progenitors, the numbers of B cells produced increased 20- to 40-fold compared with ET2/GFP− controls (Figure 6B). Furthermore, ET2-expressing progenitors also generated a large fraction of CD19−B220+ B-lineage cells (Figure 6B). These cells did not express CD11c (data not shown) and were thus doubtful to be plasmacytoid dendritic cells. However, they might represent early pro–B-cell populations before acquisition of the CD19 marker.42 Collectively, these data suggest that ET2 expression at the LMPP stage of development can modulate the myeloid-versus-lymphoid divergence, possibly by regulating downstream targets of E2A.
Discussion
A precise understanding of expression profiles of transcription factors is essential for discerning how they influence lineage decisions. Using Id1G/G knockin mice, we were able to examine Id1 expression on an individual cell basis. We have previously demonstrated that Id1 is selectively expressed by the rare LT-HSCs rather than the more abundant ST-HSCs or multipotent progenitors.31 We have now shown that the Id1 gene is turned back on in GMPs and subsequent myeloid lineage cells, but not in MEP, CMP, and CLP subsets. This result is in contrast to those obtained by Leeanansaksiri et al,28 who detected Id1 mRNA in CMP, GMP, and MEP populations. This discrepancy may be explained by the difference in sensitivity of detection; namely, higher levels of expression are required to detect GFP fluorescence, but excessive PCR amplification may indiscriminately score for Id1 expression even when only low levels of Id1 are present in some cell populations. Indeed, Jankovic et al performed similar RT-PCR assays and concluded that Id1 is expressed at higher levels in GMPs than MEPs and CMPs.43 Therefore, it is safe to conclude that higher levels of Id1 expression are found in GMP and differentiated myeloid lineage cells. This result is somewhat surprising in light of the popular model depicting the myeloid-versus-lymphoid branch point as the bifurcation of CLPs and CMPs.4 In addition, GMPs are thought to be derived from CMPs. If Id1 expression plays a role in myeloid specification, it should be expressed in CMPs according to these schemes. However, our finding agrees with a recent model where LMPPs are suggested to give rise to GMPs and CLPs without passing through the CMP stage.6 It is thus possible that up-regulation of Id1 in GMPs plays a role in myeloid specification by shutting off the lymphoid potential. Consistent with this proposed function of Id1, 2 cytokines known to promote myeloid differentiation, IL-3 and GM-CSF, are potent activators of the Id1 gene. Moreover, other studies have shown that both Id1 and Id2 expression can be induced by IL-3 in cells derived from hematopoietic progenitors.27,44
The Id family consists of 4 members, which all function to inhibit the DNA-binding activity of E proteins.17,18 In addition, the SCL/Tal1 protein also serves as an inhibitor of E proteins by forming DNA-binding heterodimers that are incapable of activating transcription.37 Because E proteins are well known to be crucial for lymphopoiesis,21,22 it is understandable that forced expression of Id or SCL inhibits the production of CD19+ B cells from multipotent progenitor cells of mouse bone marrow.28,45 Their expression also drives the differentiation of these cells into Mac1+ myeloid cells. In our studies, Id1 expression in LSK progenitors dramatically increased myeloid differentiation and completely inhibited lymphopoiesis under conditions supporting the growth of both lineages (Figure 3). Furthermore, a significant portion of the myeloid lineage cells produced under these conditions were Mac-1+CD11c+, which is characteristic of myeloid dendritic cells, although additional criteria are necessary to ascertain their identity. Despite these dramatic effects of Id1 on myeloid differentiation when overexpressed, loss of Id1 function has not been shown to cause any appreciable deficiency in myelopoiesis either in culture or in animals. This is probably the result of compensation of Id1 function by other members of the Id family or by the SCL/Tal1 protein.
To investigate the collective function of E protein inhibitors in myeloid-versus-lymphoid lineage decisions, we took a dominant-negative approach by using an E47 variant, ET2.37 ET2 is a chimeric protein composed of the N-terminal transcription activation domains of E47 and the bHLH domain of SCL/Tal1. This molecule does not form homodimers but has high affinities to E proteins and potent transcriptional activities when coupled with E proteins. The unique properties of ET2 enable it to compete with all Id and SCL/Tal1 proteins to bind endogenous E proteins without directly associating with either Id or SCL/Tal1 proteins.23,37,38 Therefore, examination of the effect of ET2 expression allowed us to address questions, such as what the collective roles of Id and SCL/Tal1 proteins are and whether their functions are mediated by inhibition of E proteins.
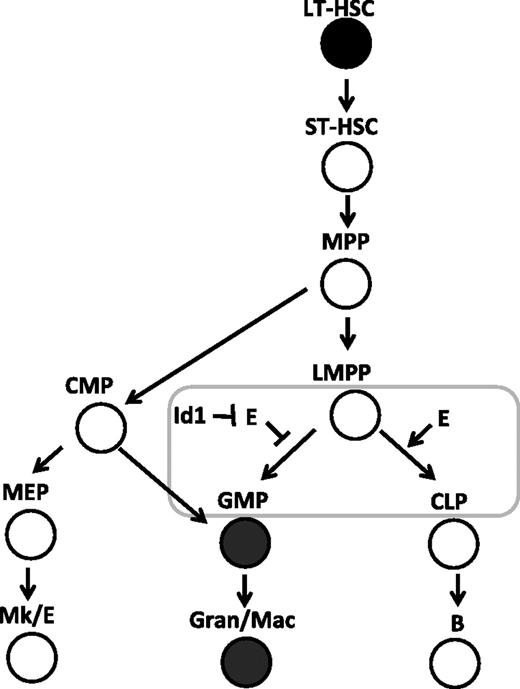
Inducible ET2 expression in mice was achieved by introducing the ET2 sequence into the ROSA26 locus along with a floxed stop sequence and by expression of tamoxifen-induble Cre recombinases.32,46 Using defined progenitors isolated from these mice, we were able to pinpoint the developmental stage at which the balance between Id and E proteins could influence the outcome of myeloid and lymphoid differentiation. We found that expression of ET2 at the Flt3HiLSK (LMPP) stage could suppress the myeloid fate and promote the lymphoid fate. However, once the cells have committed to the myeloid lineages, such as those included in the lin−c-kit+Sca-1− (MP) population, ET2 could no longer prevent them from differentiating into Mac-1+ cells. These data, combined with the fact that Id1 is expressed in GMPs and Id1 expression stimulates myeloid differentiation, lead us to propose that Id1 up-regulation and subsequent E-protein inhibition are involved in promoting myeloid differentiation while shutting off the lymphoid option as cells transit from the LMPP to MP compartments (Figure 7). Our findings also support the notion that LMPP represents a branch point for myeloid and lymphoid differentiation6 and E-protein activities play an important role in controlling this process.
E-protein activity regulates the balance between myeloid- and lymphoid-lineage specification at the LMPP stage. The developmental scheme of hematopoiesis is drawn according to Adolfsson et al.6 The stage at which the balance of E proteins (E) and Id proteins is thought to play a role in myeloid-versus-lymphoid decisions is shown with a rectangle. Cells expressing high levels of Id1 are shown in filled circles.
E-protein activity regulates the balance between myeloid- and lymphoid-lineage specification at the LMPP stage. The developmental scheme of hematopoiesis is drawn according to Adolfsson et al.6 The stage at which the balance of E proteins (E) and Id proteins is thought to play a role in myeloid-versus-lymphoid decisions is shown with a rectangle. Cells expressing high levels of Id1 are shown in filled circles.
How E proteins carry out this function remains to be elucidated. As transcription activators, E proteins are known to stimulate expression of B cell–specific genes.19 E proteins might also actively repress transcription of myeloid-specific genes. One potential candidate gene to be repressed is the gene encoding M-CSF receptor (M-CSFR), also called c-fms, which is known to be essential for macrophage differentiation.47,48 We have shown that expression of ET2 reduces the levels of M-CSFR in LSK and committed MPs. Consistently, loss of E2A function in Hodgkin lymphoma cell lines results in increases in M-CSFR expression.49 An active role of E proteins in suppressing myeloid differentiation is also suggested by the finding that the abilities of E2A in promoting B-cell differentiation and suppressing myeloid development depend on different functional domains in E2A proteins.50 If myelopoiesis were blocked by E proteins simply by inducing lymphoid-specific genes, identical structural requirements would have been found. Therefore, understanding how E proteins suppress myeloid fates would be an interesting area of investigation, which might lead to a realization that E proteins not only play an important role in lymphopoiesis but also directly regulate myelopoiesis. It will be interesting to determine whether E proteins do so by functionally interacting with other transcription factors, such as PU.1, C/EBP, and Ikaros. Understanding the molecular mechanisms governing myeloid and lymphoid differentiation may be of great significance in considering the mechanism of leukemogenesis and may allow for the development of therapeutic strategies that can target inappropriate gene expression in neoplastic cells, such as Reed-Sternberg cells in Hodgkin lymphoma.49
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Paul Kincade and Darryl Dudley for critical reading of the manuscript, Dr S. Scott Perry for advice, and the flow cytometry facility at the Oklahoma Medical Research Foundation for technical support.
This work was supported by National Institutes of Health grants AI56129 and AI33759 (to X.-H.S.). X.-H.S. holds the Eli Lilly Distinguished Chair in Biomedical Research.
National Institutes of Health
Authorship
Contribution: S.W.C. designed and performed research, analyzed data, and wrote the paper; Y.Z. and R.S.W. performed research and analyzed data; and X.-H.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiao-Hong Sun, Program in Immunobiology and Cancer Research, Oklahoma Medical Research Foundation, 825 NE 13th Street, Oklahoma City, OK 73104; e-mail: sunx@omrf.ouhsc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal