Abstract
Acute promyelocytic leukemia (APL) is a hematopoietic malignant disease characterized by the chromosomal translocation t(15;17), resulting in the formation of the PML-RARA gene. Here, 47 t(15;17) APL samples were analyzed with high-density single-nucleotide polymorphism microarray (50-K and 250-K SNP-chips) using the new algorithm AsCNAR (allele-specific copy-number analysis using anonymous references). Copy-number-neutral loss of heterozygosity (CNN-LOH) was identified at chromosomes 10q (3 cases), 11p (3 cases), and 19q (1 case). Twenty-eight samples (60%) did not have an obvious alteration (normal-copy-number [NC] group). Nineteen samples (40%) showed either one or more genomic abnormalities: 8 samples (17%) had trisomy 8 either with or without an additional duplication, deletion, or CNN-LOH (+8 group); and 11 samples (23%) had genomic abnormalities without trisomy 8 (other abnormalities group). These chromosomal abnormalities were acquired somatic mutations. Interestingly, FLT3-ITD mutations (11/47 cases) occurred only in the group with no genomic alteration (NC group). Taken together, these results suggest that the pathway of development of APL differs in each group: FLT3-ITD, trisomy 8, and other genomic changes. Here, we showed for the first time hidden abnormalities and novel disease-related genomic changes in t(15;17) APL.
Introduction
Acute promyelocytic leukemia (APL) is a hematopoietic malignant disease characterized by the chromosomal translocation t(15;17), resulting in the fusion of the promyelocytic leukemia (PML) gene and retinoic acid receptor α (RARA) gene (PML-RARA).1,2 The fusion product PML-RARα homodimerizes, binds to DNA, and works as a transcriptional repressor together with corepressors including histone deacetylase.3 Therefore, reactivation of RARα-dependent transcription is one of the major strategies to treat APL patients. In fact, all-trans retinoic acid (ATRA), which binds to RARα and leads to the activation of the transcription factor, is a highly effective compound for the induction of remission of APL patients.4,5
Transgenic mice revealed that PML-RARα is necessary but not sufficient for the development of APL.6,7 APL occurred in these mice only after a long latency (8.5 to 12 months) and penetrance was 15% to 30%.6,7 These findings suggest that additional genetic mutations are also required for the development of APL. Candidate genes include the tyrosine kinase receptor gene, FLT3, and the oncogene, RAS. Activating FLT3 mutations occur in approximately 30% to 35% APL samples8,9 ; and NRAS and KRAS mutations are found in 4% to 5% and 5% to 10% of APL samples, respectively.9,10 Interestingly, transgenic mice coexpressing PML-RARα and either FLT3W51 (constitutively activated form of murine FLT3), FLT3-ITD, or K-Ras (K12D) develop APL with a short latency and a high penetrance.11-14
Comparative genomic hybridization (CGH) is one of the genome-wide screening methods to identify chromosomal abnormalities. However, CGH analysis cannot detect copy-number-neutral loss of heterozygosity (CNN-LOH). Single-nucleotide polymorphism microarray (SNP-chip) is a powerful method to examine genomic alterations including small copy-number changes and/or CNN-LOH in several cancers.15-17 SNP-chip analysis has been used for chronic lymphocytic leukemia (CLL),18,19 childhood acute lymphoblastic leukemia (ALL),20,21 acute myeloid leukemia (AML),22-26 and AML with normal karyotype (Gorletta et al27 ; Akagi et al28 ).
In the present study, we focused on t(15;17) APL and examined whether additional genomic alterations could be found to subcategorize this disease on the basis of genomic status. The use of CNAG (copy-number analysis for Affymetrix GeneChips; Santa Clara, CA) program15 and a new algorithm AsCNAR (allele-specific copy-number analysis using anonymous references)17 provides a highly sensitive technique to detect CNN-LOH as well as copy-number changes in APL.
Methods
Patient samples
DNA from the bone marrow of 47 anonymized cases of t(15;17) APL at diagnosis as well as 7 complete remission bone marrow samples were examined. Sample information including the form of PML-RARα (long, short, or variant), sex, age, white blood cell counts (WBCs), blast percentage in the bone marrow, mutational status of the FLT3 gene, FLT3-ITD level, and karyotype are shown in Table 1. This study received IRB approval from the Cedars-Sinai Medical Center and informed consent was obtained in accordance with the Declaration of Helsinki.
Baseline clinical characteristic of 47 t(15;17) APL patients
| Group/case no. . | PML-RARα . | FLT3 . | FLT3-ITD . | Chromosome . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Isoform . | Sex . | Age, y . | WBC, ×109/L . | Blast, % . | D835 . | ITD . | Level, % . | ||
| NC | |||||||||
| 5 | S | F | 43 | 1.9 | 88 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 48 | S | M | 45 | 1.3 | 76 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 23 | L | F | 38 | 0.4 | 84 | - | - | ND, RT-PCR(+) | |
| 28 | L | F | 36 | 9.9 | 87 | - | - | ND, RT-PCR(+) | |
| 35 | L | F | 60 | 1.0 | 94 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 40 | L | M | 32 | 0.9 | 75 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 12 | L | M | 54 | 1.3 | 97 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 55 | L | M | 36 | 2.0 | 76 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 56 | L | M | 17 | 1.9 | 85 | - | - | ND, RT-PCR(+) | |
| 24 | V | M | 32 | 0.9 | 77 | - | - | 46,XY,t(15;17)(q22;q12) | |
| 46 | V | M | 33 | 2.8 | 62 | - | - | ND, RT-PCR(+) | |
| 33 | S | M | 42 | 31.4 | 88 | +H | - | 46,XY,t(15;17)(q22;q21) | |
| 1 | L | F | 68 | 1.3 | 96 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 9 | L | F | 30 | 1.4 | 87 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 11 | L | M | 30 | 4.3 | 78 | +D | - | ND, RT-PCR(+) | |
| 53 | L | M | 36 | 36.2 | 96 | +E | - | 46,XY,t(15;17)(q22;q21) | |
| 61 | L | M | 32 | 0.3 | 93 | +E | - | ND, RT-PCR(+) | |
| 7 | S | F | 31 | 120.0 | 95 | - | + | 42 | 46,XX,t(15;17)(q22;q12) |
| 8 | S | M | 49 | 4.5 | 96 | - | + | 37 | 46,XY,t(15;17)(q22;q21) |
| 14 | S | M | 9 | 28.0 | 80 | - | + | 40 | 46,XY,t(15;17)(q22;q21) |
| 17 | S | F | 47 | 51.1 | 98 | - | + | 24 | 46,XX,t(15;17)(q22;q21) |
| 29 | S | M | 75 | 1.5 | 88 | - | + | 35 | ND, RT-PCR(+) |
| 63 | S | F | 57 | 9.7 | 96 | - | + | 42 | 46,XX,t(15;17)(q22;q21) |
| 64 | S | M | 28 | 24.8 | 90 | - | + | 57 | 46,XY,t(15;17)(q22;q21) [25/26] |
| 46,XY[1/26] | |||||||||
| 6 | L | F | 37 | 29.5 | 93 | - | + | 32 | ND, RT-PCR(+) |
| 42 | L | F | 24 | 1.2 | 87 | - | + | 4 | 46,XX,t(15;17)(q22;q21) |
| 51 | L | M | 51 | 45.4 | 78 | - | + | 47 | 46,XY,t(15;17)(q21;q12) |
| 62 | L | F | 46 | 45.0 | 89 | - | + | 42 | 46,XX,t(15;17) (q22;q21) |
| +8 | |||||||||
| 65 | S | F | 58 | 1.5 | 87 | - | - | 46,XX,RT-PCR(+) | |
| 38 | L | F | 22 | 7.0 | 78 | - | - | 47,XX,+8,t(15;17)(q22;q12) | |
| 2 | S | M | 42 | 2.4 | 79 | - | - | 47,XY,+8,t(15;17)(q22;q21) | |
| 50 | S | F | 58 | 19.8 | 84 | - | - | 47,XX,+8,t(15;17)(q22;q21) | |
| 60 | L | M | 22 | 9.9 | 90 | +H | - | 46,XY,t(15;17)(q22;q21) [16/21] | |
| 47XY,+8,t(15;17) (q22;q21) [4/21] | |||||||||
| 46,XY [1/21] | |||||||||
| 3 | V | M | 33 | 3.2 | 67 | - | - | 47,XY,+8,t(15;17)(p22;q12) | |
| 39 | L | M | 38 | 1.0 | 51 | - | - | 47,XY,+8,t(15;17)(q22;q21) | |
| 18 | L | M | 23 | 18.1 | 94 | - | - | ND, RT-PCR(+) | |
| Other | |||||||||
| 66 | S | F | 41 | 15.6 | 89 | +Y | - | 46,XX,t(15;17)(q22;q11.2) | |
| 20 | L | F | 51 | 77.9 | 88 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 13 | L | F | 7 | 1.6 | 99 | +Y | - | ND, RT-PCR(+) | |
| 4 | S | M | 68 | 0.7 | 91 | +E | - | 46,XY,t(15;17)(q22;q21) | |
| 37 | S | F | 26 | 2.0 | 75 | +Y | - | 46,XX,t(15;17)(q22;q12) | |
| 57 | L | F | 30 | 1.4 | 87 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 19 | L | M | 51 | 1.1 | 73 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 21 | L | M | 45 | 4.7 | 87 | - | - | ND, RT-PCR(+) | |
| 43 | L | M | 7 | 8.6 | 75 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 52 | L | M | 54 | 8.6 | 82 | - | - | 46,XY,t(15;17)(q21;q12) [19/20] | |
| 47,XY, +21, t(15;17)(q21;q12) [1/20] | |||||||||
| 58 | L | F | 27 | 6.9 | 90 | - | - | 46,XX,t(15;17)(q22;q21)ider(17) (q10)t(15;17) [23/26] | |
| 46,XX [3/26] | |||||||||
| Group/case no. . | PML-RARα . | FLT3 . | FLT3-ITD . | Chromosome . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Isoform . | Sex . | Age, y . | WBC, ×109/L . | Blast, % . | D835 . | ITD . | Level, % . | ||
| NC | |||||||||
| 5 | S | F | 43 | 1.9 | 88 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 48 | S | M | 45 | 1.3 | 76 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 23 | L | F | 38 | 0.4 | 84 | - | - | ND, RT-PCR(+) | |
| 28 | L | F | 36 | 9.9 | 87 | - | - | ND, RT-PCR(+) | |
| 35 | L | F | 60 | 1.0 | 94 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 40 | L | M | 32 | 0.9 | 75 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 12 | L | M | 54 | 1.3 | 97 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 55 | L | M | 36 | 2.0 | 76 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 56 | L | M | 17 | 1.9 | 85 | - | - | ND, RT-PCR(+) | |
| 24 | V | M | 32 | 0.9 | 77 | - | - | 46,XY,t(15;17)(q22;q12) | |
| 46 | V | M | 33 | 2.8 | 62 | - | - | ND, RT-PCR(+) | |
| 33 | S | M | 42 | 31.4 | 88 | +H | - | 46,XY,t(15;17)(q22;q21) | |
| 1 | L | F | 68 | 1.3 | 96 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 9 | L | F | 30 | 1.4 | 87 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 11 | L | M | 30 | 4.3 | 78 | +D | - | ND, RT-PCR(+) | |
| 53 | L | M | 36 | 36.2 | 96 | +E | - | 46,XY,t(15;17)(q22;q21) | |
| 61 | L | M | 32 | 0.3 | 93 | +E | - | ND, RT-PCR(+) | |
| 7 | S | F | 31 | 120.0 | 95 | - | + | 42 | 46,XX,t(15;17)(q22;q12) |
| 8 | S | M | 49 | 4.5 | 96 | - | + | 37 | 46,XY,t(15;17)(q22;q21) |
| 14 | S | M | 9 | 28.0 | 80 | - | + | 40 | 46,XY,t(15;17)(q22;q21) |
| 17 | S | F | 47 | 51.1 | 98 | - | + | 24 | 46,XX,t(15;17)(q22;q21) |
| 29 | S | M | 75 | 1.5 | 88 | - | + | 35 | ND, RT-PCR(+) |
| 63 | S | F | 57 | 9.7 | 96 | - | + | 42 | 46,XX,t(15;17)(q22;q21) |
| 64 | S | M | 28 | 24.8 | 90 | - | + | 57 | 46,XY,t(15;17)(q22;q21) [25/26] |
| 46,XY[1/26] | |||||||||
| 6 | L | F | 37 | 29.5 | 93 | - | + | 32 | ND, RT-PCR(+) |
| 42 | L | F | 24 | 1.2 | 87 | - | + | 4 | 46,XX,t(15;17)(q22;q21) |
| 51 | L | M | 51 | 45.4 | 78 | - | + | 47 | 46,XY,t(15;17)(q21;q12) |
| 62 | L | F | 46 | 45.0 | 89 | - | + | 42 | 46,XX,t(15;17) (q22;q21) |
| +8 | |||||||||
| 65 | S | F | 58 | 1.5 | 87 | - | - | 46,XX,RT-PCR(+) | |
| 38 | L | F | 22 | 7.0 | 78 | - | - | 47,XX,+8,t(15;17)(q22;q12) | |
| 2 | S | M | 42 | 2.4 | 79 | - | - | 47,XY,+8,t(15;17)(q22;q21) | |
| 50 | S | F | 58 | 19.8 | 84 | - | - | 47,XX,+8,t(15;17)(q22;q21) | |
| 60 | L | M | 22 | 9.9 | 90 | +H | - | 46,XY,t(15;17)(q22;q21) [16/21] | |
| 47XY,+8,t(15;17) (q22;q21) [4/21] | |||||||||
| 46,XY [1/21] | |||||||||
| 3 | V | M | 33 | 3.2 | 67 | - | - | 47,XY,+8,t(15;17)(p22;q12) | |
| 39 | L | M | 38 | 1.0 | 51 | - | - | 47,XY,+8,t(15;17)(q22;q21) | |
| 18 | L | M | 23 | 18.1 | 94 | - | - | ND, RT-PCR(+) | |
| Other | |||||||||
| 66 | S | F | 41 | 15.6 | 89 | +Y | - | 46,XX,t(15;17)(q22;q11.2) | |
| 20 | L | F | 51 | 77.9 | 88 | - | - | 46,XX,t(15;17)(q22;q21) | |
| 13 | L | F | 7 | 1.6 | 99 | +Y | - | ND, RT-PCR(+) | |
| 4 | S | M | 68 | 0.7 | 91 | +E | - | 46,XY,t(15;17)(q22;q21) | |
| 37 | S | F | 26 | 2.0 | 75 | +Y | - | 46,XX,t(15;17)(q22;q12) | |
| 57 | L | F | 30 | 1.4 | 87 | +Y | - | 46,XX,t(15;17)(q22;q21) | |
| 19 | L | M | 51 | 1.1 | 73 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 21 | L | M | 45 | 4.7 | 87 | - | - | ND, RT-PCR(+) | |
| 43 | L | M | 7 | 8.6 | 75 | - | - | 46,XY,t(15;17)(q22;q21) | |
| 52 | L | M | 54 | 8.6 | 82 | - | - | 46,XY,t(15;17)(q21;q12) [19/20] | |
| 47,XY, +21, t(15;17)(q21;q12) [1/20] | |||||||||
| 58 | L | F | 27 | 6.9 | 90 | - | - | 46,XX,t(15;17)(q22;q21)ider(17) (q10)t(15;17) [23/26] | |
| 46,XX [3/26] | |||||||||
Chromosomal translocation of t(15;17) was determined by karyotype studies and/or RT-PCR analysis specific for PML-RARα fusion products. Three types of PML-RARα (long, short, and variant) are shown as L, S, and V, respectively. The recorded number of white blood cells (WBCs) and bone marrow blast percentages were obtained at diagnosis. Mutations of FLT3 were either tyrosine kinase domain (TKD) at codon 835 or internal tandem repeat (ITD). The APL samples are divided into 3 groups based on SNP-chip analysis: normal-copy-number (NC), trisomy 8 including duplication of the MYC gene region (+8), and other abnormalities (other).
High-density SNP-chip analysis
Genomic DNA was isolated from bone marrow samples from t(15;17) APL patients at diagnosis and complete remission, as well as APL cell lines NB4 and PL-21. The DNA was subjected to GeneChip Human mapping 50-K or 250-K microarray (SNP-chip; Affymetrix) as described previously.15,17 Hybridization, washing, and signal detection were performed on GeneChip Fluidics Station 400 and GeneChip scanner 3000 according to the manufacturer's protocols (Affymetrix). Microarray data were analyzed for determination of both total and allelic-specific copy number (AsCN) using the CNAG program as previously described15,17 with minor modifications, where the status of copy numbers as well as CNN-LOH at each SNP was inferred using the algorithms based on hidden Markov models.15,17 For clustering of AML samples with regard to the status of copy-number changes, as well as CNN-LOH, GNAGraph software (Tokyo University, Tokyo, Japan) was used.21 Size, position, and location of genes were identified with UCSC Genome Browser (http://genome.ucsc.edu/). Germline copy-number changes previously described as copy-number variant at Database of Genomic Variants (http://projects.tcag.ca/variation/) and UCSC Genome Browser were excluded. This microarray data are available for public viewing in the Gene Expression Omnibus (GEO) database29 under accession number GSE14016.
Determination of SNP sequences in cases of CNN-LOH and FLT3 mutations
To validate CNN-LOH, 2 SNP sequences (rs10500648 and rs7937815) in chromosome 11p of case no. 39 at diagnosis and complete remission, and 6 SNP sequences (rs10491032, rs363221, rs2099803, rs2104543, rs7075893, and rs7918018) in chromosome 10q of case no. 18 at diagnosis were determined. The genomic region of each SNP site was amplified by genomic polymerase chain reaction (PCR) using specific primers (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), and PCR products were purified and sequenced. For determinations of FLT3-TKD and FLT3-ITD mutations, genomic PCR was performed as described previously.30
Cell culture, mRNA isolation, and quantitative real-time PCR
APL cell lines, NB4 and PL-21, were cultured in RPMI1640 medium (Invitrogen, Carlsbad, CA) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA). Total RNA was isolated from these cells and case no. 48 bone marrow sample at diagnosis using RNeasy kit (QIAGEN, Valencia, CA), and 1 μg total RNA was converted into cDNA by reverse transcription with Superscript III (Invitrogen). Gene expression of c-Myc mRNA was quantified with real-time quantitative PCR (iCycler; Bio-Rad, Hercules, CA) using Sybr Green. β-Actin was used as control.
Copy number of chromosome 11p15.4 in case no. 39, 10q24.31 in case no. 18, the MYC gene in cases no. 2, no. 18, and no. 65, and the ERG gene in case no. 43 were determined by quantitative real-time PCR (Bio-Rad) using Sybr Green. The region on chromosome 2p21 was used as control.21 Copy number of the 2p21 region was normal as determined by SNP-chip analysis in these samples. The delta threshold cycle value (ΔCt) was calculated from the given Ct value by the formula ΔCt = (Ct sample − Ct control). The fold change was calculated as 2−ΔCt. Primer sequences are shown in Table S2.
Results
SNP-chip analysis of t(15;17) APL samples
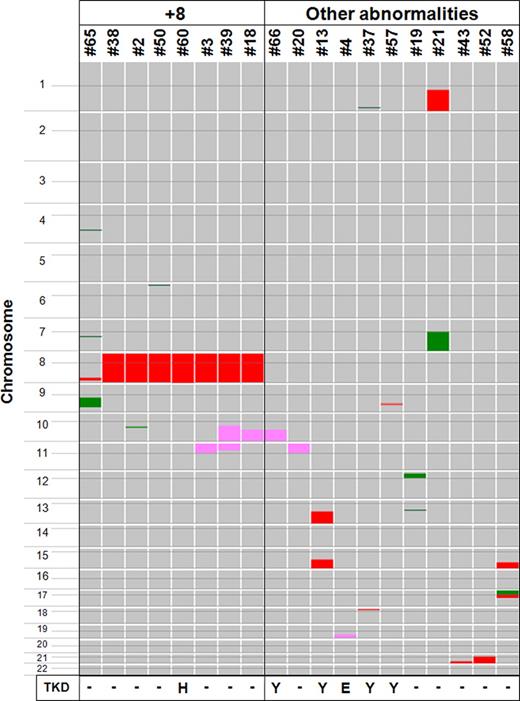
We examined the genomic changes in 47 samples of t(15;17) APL using 50-K and 250-K SNP-chip analyses. A total of 28 patients (60%) showed no detectable genomic abnormalities (normal-copy-number [NC] group). In contrast, 19 patients (40%) had one or more genomic abnormalities: 8 patients (17%) had trisomy 8 or duplication on chromosome 8 in the region of the MYC gene either with or without other genomic abnormalities (+8 group), and 11 patients (23%) had genomic abnormalities without trisomy 8 (other abnormalities group; Figure 1; Table 2).
Summary of genomic abnormalities in t(15;17) APL samples. Genomic DNA of 47 t(15;17) APL samples were subjected to SNP-chip analysis, and genomic abnormalities are summarized. Color boxes are used to denote the type and size of abnormalities: pink (copy-number-neutral loss of heterozygosity; CNN-LOH); green (deletion); and red (duplication including trisomy). A total of 28 patients (60%) showed no detectable genomic abnormalities (data not shown). In contrast, 19 patients (40%) had one or more genomic abnormalities: trisomy 8 or duplication of the MYC gene region either with or without genomic abnormalities was found in 8 patients (17%, referred as “+8”) and 11 patients (23%, referred as “other abnormalities”) had genomic abnormalities without trisomy 8. Six patients (13%) had CNN-LOH; and 1 sample in +8 group and 5 samples in other abnormalities group had FLT3 point mutations that are shown by their amino acid change at codon 835 from D (aspartic acid) to either Y (tyrosine), E (glutamic acid), or H (histidine).
Summary of genomic abnormalities in t(15;17) APL samples. Genomic DNA of 47 t(15;17) APL samples were subjected to SNP-chip analysis, and genomic abnormalities are summarized. Color boxes are used to denote the type and size of abnormalities: pink (copy-number-neutral loss of heterozygosity; CNN-LOH); green (deletion); and red (duplication including trisomy). A total of 28 patients (60%) showed no detectable genomic abnormalities (data not shown). In contrast, 19 patients (40%) had one or more genomic abnormalities: trisomy 8 or duplication of the MYC gene region either with or without genomic abnormalities was found in 8 patients (17%, referred as “+8”) and 11 patients (23%, referred as “other abnormalities”) had genomic abnormalities without trisomy 8. Six patients (13%) had CNN-LOH; and 1 sample in +8 group and 5 samples in other abnormalities group had FLT3 point mutations that are shown by their amino acid change at codon 835 from D (aspartic acid) to either Y (tyrosine), E (glutamic acid), or H (histidine).
Chromosomal alterations in t(15;17) APL samples
| Group/case no. . | Status . | Location . | Physical localization . | Size, Mb . | Gene(s) in the region . | |
|---|---|---|---|---|---|---|
| Proximal . | Distal . | |||||
| +8 | ||||||
| 65 | Del | 4q28.1 | 125 190 507 | 126 521 903 | 1.33 | KIAA1223 |
| Del | 7q21.11-q21.12 | 85 414 972 | 86 445 002 | 1.03 | GRM3, KIAA1324L, and DMTF1 | |
| Dup | 8q24.13-q24.22 | 122 607 785 | 132 092 760 | 9.48 | > 10 genes including MYC | |
| Del | 9q12-q31.3 | 64 207 745 | 111 479 523 | 47.27 | > 10 genes | |
| 38 | Tri | Trisomy 8 | ||||
| 2 | Tri | Trisomy 8 | ||||
| Del | 10q21.2-q21.3 | 62 496 958 | 68 046 104 | 5.55 | >10 genes | |
| 50 | Del | 6p25.1-p24.3 | 5 545 437 | 8 054 930 | 2.51 | > 10 genes |
| Tri | Trisomy 8 | |||||
| 60 | Tri | Trisomy 8 | ||||
| 3 | Tri | Trisomy 8 | ||||
| CNN-LOH | 11p.ter–p11.12 | 1 938 894 | 49 879 899 | 47.9 | > 10 genes including WT1, CDKN1C and HRAS | |
| 39 | Tri | Trisomy 8 | ||||
| CNN-LOH | 10q21.1–q-ter | 59 576 047 | 135 228 726 | 75.7 | > 10 genes including PTEN and FGFR2 | |
| CNN-LOH | 11p.ter–p14.1 | 1 938 894 | 30 627 880 | 28.7 | > 10 genes including CDKN1C and HRAS | |
| 18 | Tri | Trisomy 8 | ||||
| CNN-LOH | 10q22.2–q-ter | 76 995 152 | 135 228 726 | 58.2 | > 10 genes including PTEN and FGFR2 | |
| Other | ||||||
| 66 | CNN-LOH | 10q22.2–q-ter | 76 289 513 | 135 295 604 | 59.0 | > 10 genes including PTEN and FGFR2 |
| 20 | CNN-LOH | 11p.ter–p11.12 | 1 938 894 | 49 330 228 | 47.4 | > 10 genes including WT1, CDKN1C and HRAS |
| 13 | Dup | 13q21.1–q-ter | 56 784 440 | 114 051 465 | 57.27 | > 10 genes |
| Dup | 15q22.2–q-ter | 57 244 668 | 100 182 183 | 42.94 | > 10 genes including PML | |
| 4 | CNN-LOH | 19q13.2–q-ter | 46 160 099 | 63 437 743 | 17.3 | > 10 genes |
| 37 | Del | 1q42.2 | 227 843 862 | 227 867 765 | 0.02 | EGLN1 |
| Dup | 18p11.31-p11.23 | 7 192 739 | 7 657 575 | 0.46 | PTPRM | |
| 57 | Dup | 9q22.32 | 94 435 025 | 94 710 006 | 0.27 | FBP2, FBP1, and C9orf3 |
| 19 | Del | 12p13.31-p11.22 | 6 755 671 | 29 248 257 | 22.49 | > 10 genes including ETV6 and CDKN1B |
| Del | 13q14.2-q14.3 | 49 630 676 | 50 510 777 | 0.88 | FAM10A4, DLEU7, FLJ11712, and GUCY1B2 | |
| 21 | Dup | 1q21–q-ter | 142 487 224 | 245 120 412 | 102.63 | > 10 genes |
| Del | 7q11.21–q-ter | 61 522 282 | 158 554 645 | 97.03 | > 10 genes | |
| 43 | Dup | 21q22.12–q-ter | 36 234 195 | 46 924 583 | 10.69 | >10 genes including ERG |
| 52 | Tri | Trisomy 21 | ||||
| 58 | Dup | 15q24.1–q-ter | 72 224 840 | 100 192 115 | 27.97 | > 10 genes |
| Del | 17p-ter–p11.2 | 18 901 | 21 459 693 | 21.44 | > 10 genes including TP53 | |
| Dup | 17p11.2-q21.1 | 21 491 135 | 35 542 587 | 14.05 | > 10 genes including NF1 and ERBB2 | |
| Group/case no. . | Status . | Location . | Physical localization . | Size, Mb . | Gene(s) in the region . | |
|---|---|---|---|---|---|---|
| Proximal . | Distal . | |||||
| +8 | ||||||
| 65 | Del | 4q28.1 | 125 190 507 | 126 521 903 | 1.33 | KIAA1223 |
| Del | 7q21.11-q21.12 | 85 414 972 | 86 445 002 | 1.03 | GRM3, KIAA1324L, and DMTF1 | |
| Dup | 8q24.13-q24.22 | 122 607 785 | 132 092 760 | 9.48 | > 10 genes including MYC | |
| Del | 9q12-q31.3 | 64 207 745 | 111 479 523 | 47.27 | > 10 genes | |
| 38 | Tri | Trisomy 8 | ||||
| 2 | Tri | Trisomy 8 | ||||
| Del | 10q21.2-q21.3 | 62 496 958 | 68 046 104 | 5.55 | >10 genes | |
| 50 | Del | 6p25.1-p24.3 | 5 545 437 | 8 054 930 | 2.51 | > 10 genes |
| Tri | Trisomy 8 | |||||
| 60 | Tri | Trisomy 8 | ||||
| 3 | Tri | Trisomy 8 | ||||
| CNN-LOH | 11p.ter–p11.12 | 1 938 894 | 49 879 899 | 47.9 | > 10 genes including WT1, CDKN1C and HRAS | |
| 39 | Tri | Trisomy 8 | ||||
| CNN-LOH | 10q21.1–q-ter | 59 576 047 | 135 228 726 | 75.7 | > 10 genes including PTEN and FGFR2 | |
| CNN-LOH | 11p.ter–p14.1 | 1 938 894 | 30 627 880 | 28.7 | > 10 genes including CDKN1C and HRAS | |
| 18 | Tri | Trisomy 8 | ||||
| CNN-LOH | 10q22.2–q-ter | 76 995 152 | 135 228 726 | 58.2 | > 10 genes including PTEN and FGFR2 | |
| Other | ||||||
| 66 | CNN-LOH | 10q22.2–q-ter | 76 289 513 | 135 295 604 | 59.0 | > 10 genes including PTEN and FGFR2 |
| 20 | CNN-LOH | 11p.ter–p11.12 | 1 938 894 | 49 330 228 | 47.4 | > 10 genes including WT1, CDKN1C and HRAS |
| 13 | Dup | 13q21.1–q-ter | 56 784 440 | 114 051 465 | 57.27 | > 10 genes |
| Dup | 15q22.2–q-ter | 57 244 668 | 100 182 183 | 42.94 | > 10 genes including PML | |
| 4 | CNN-LOH | 19q13.2–q-ter | 46 160 099 | 63 437 743 | 17.3 | > 10 genes |
| 37 | Del | 1q42.2 | 227 843 862 | 227 867 765 | 0.02 | EGLN1 |
| Dup | 18p11.31-p11.23 | 7 192 739 | 7 657 575 | 0.46 | PTPRM | |
| 57 | Dup | 9q22.32 | 94 435 025 | 94 710 006 | 0.27 | FBP2, FBP1, and C9orf3 |
| 19 | Del | 12p13.31-p11.22 | 6 755 671 | 29 248 257 | 22.49 | > 10 genes including ETV6 and CDKN1B |
| Del | 13q14.2-q14.3 | 49 630 676 | 50 510 777 | 0.88 | FAM10A4, DLEU7, FLJ11712, and GUCY1B2 | |
| 21 | Dup | 1q21–q-ter | 142 487 224 | 245 120 412 | 102.63 | > 10 genes |
| Del | 7q11.21–q-ter | 61 522 282 | 158 554 645 | 97.03 | > 10 genes | |
| 43 | Dup | 21q22.12–q-ter | 36 234 195 | 46 924 583 | 10.69 | >10 genes including ERG |
| 52 | Tri | Trisomy 21 | ||||
| 58 | Dup | 15q24.1–q-ter | 72 224 840 | 100 192 115 | 27.97 | > 10 genes |
| Del | 17p-ter–p11.2 | 18 901 | 21 459 693 | 21.44 | > 10 genes including TP53 | |
| Dup | 17p11.2-q21.1 | 21 491 135 | 35 542 587 | 14.05 | > 10 genes including NF1 and ERBB2 | |
Physical localization, size (Mb), and gene(s) at the chromosomal regions were obtained from UCSC Genome Browser. If known gene(s) in the chromosomal regions are less than 10, all gene names are displayed.
Copy number changes as previously described as copy number variant at Database of Genomic Variants (http://projects.tcag.ca/variation/)29 and UCSC Genome Browser (http://genome.ucsc.edu/)32 were excluded.
Del indicates deletion; Dup, duplication; Tri, trisomy; ter, terminal; and CNN-LOH, copy-number-neutral loss of heterozygosity.
One case (case no. 65, 2%) had 4 chromosomally altered regions; 2 cases (4%; no. 39 and no. 58) had 3 chromosomally altered regions; 8 cases (17%; no. 2, no. 50, no. 3, no. 18, no. 13, no. 37, no. 19, and no. 21) had 2 chromosomally altered regions; and 8 cases (17%; no. 38, no. 60, no. 66, no. 20, no. 4, no. 57, no. 43, and no. 52) had 1 chromosomally altered region. Importantly, 6 patients (13%) had CNN-LOH.
Validation of SNP-chip analysis
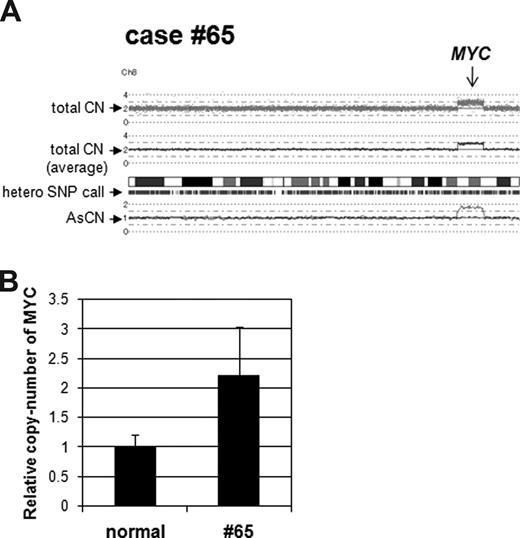
As proof of principal, we validated SNP-chip results using quantitative genomic real-time PCR (QG RT-PCR) and nucleotide sequencing of SNP sites. Case no. 65 had a duplicated region at chromosome 8, and this region contained the MYC gene (Figure 2A). QG RT-PCR showed that levels of the MYC copy number were approximately 2-fold higher than normal genomic DNA (Figure 2B). Other copy-number changes including duplication of the MYC gene in cases no. 2 and no. 18, and duplication of the ERG gene in case no. 43 were also confirmed by QG RT-PCR (data not shown).
Validation of copy-number change in case no. 65. (A) SNP-chip data of chromosome 8 in case no. 65. Top dots are SNP sites as probes and indicate total copy number (CN). Middle line is an average of the copy number and shows gene dosage. Bars are heterozygous (hetero) SNP calls. Bottom 2 lines show allele-specific copy number (AsCN). (B) Duplication of the MYC gene region in case no. 65. Copy number of the MYC gene in case no. 65 was compared with normal genomic DNA with quantitative genomic real-time PCR. Level of the copy number was determined as a ratio between the MYC gene and the reference genomic region 2p21. Results represent mean of 3 experiments plus or minus SD.
Validation of copy-number change in case no. 65. (A) SNP-chip data of chromosome 8 in case no. 65. Top dots are SNP sites as probes and indicate total copy number (CN). Middle line is an average of the copy number and shows gene dosage. Bars are heterozygous (hetero) SNP calls. Bottom 2 lines show allele-specific copy number (AsCN). (B) Duplication of the MYC gene region in case no. 65. Copy number of the MYC gene in case no. 65 was compared with normal genomic DNA with quantitative genomic real-time PCR. Level of the copy number was determined as a ratio between the MYC gene and the reference genomic region 2p21. Results represent mean of 3 experiments plus or minus SD.
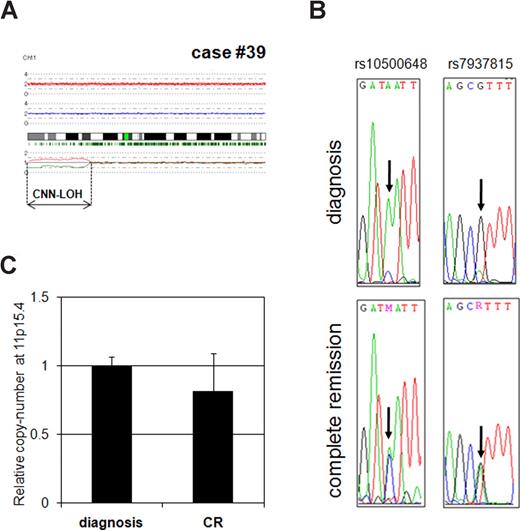
Next, we validated CNN-LOH detected by SNP-chip analysis (Figure 3). If a chromosome has LOH, SNP sequences in this region should have homozygosity at diagnosis but heterozygosity at complete remission. Therefore, we examined 2 independent SNP sequences in case no. 39 on chromosome 11p in the CNN-LOH region using diagnosis and complete remission samples. Two SNP sites (rs10500648 and rs7937815) clearly showed a single signal at diagnosis (homozygosity), whereas, the sites showed a double signal at complete remission (heterozygosity; Figure 3B). These results demonstrated that this region had LOH. Next, we determined copy number of the region to exclude the possibility of a hemizygous deletion. As shown in Figure 3C, level of DNA at the 11p15.4 region of case no. 39 at diagnosis was almost the same as level of the complete remission sample, indicating that this region had a normal copy number and the region represented CNN-LOH. CNN-LOH region of case no. 18 was also validated by SNP sequencing and QG RT-PCR (Figure S1). Taken together, these results indicated that SNP-chip analysis clearly reflected real chromosomal abnormalities.
Validation of CNN-LOH in case no. 39. (A) SNP-chip data of chromosome 11 in case no. 39. The samples had CNN-LOH on chromosome 11 (11p-terminal–p14.1, 28.7 Mb). (B) Determination of SNP sequence in 11p CNN-LOH region in case no. 39. 2 SNP sites (rs10500648 and rs7937815) were sequenced. Both SNP sites showed heterozygosity in the complete remission sample, whereas they showed homozygosity in the diagnosis sample. (C) Determination of copy number in the 11p15.4 region. Copy number of 11p15.4 (CNN-LOH region) in case no. 39 at diagnosis was compared with complete remission (CR) sample with quantitative genomic real-time PCR. Levels of the copy number were determined as a ratio between 11p15.4 and the reference genomic DNA, 2p21. Results represent the mean of 3 experiments plus or minus SD.
Validation of CNN-LOH in case no. 39. (A) SNP-chip data of chromosome 11 in case no. 39. The samples had CNN-LOH on chromosome 11 (11p-terminal–p14.1, 28.7 Mb). (B) Determination of SNP sequence in 11p CNN-LOH region in case no. 39. 2 SNP sites (rs10500648 and rs7937815) were sequenced. Both SNP sites showed heterozygosity in the complete remission sample, whereas they showed homozygosity in the diagnosis sample. (C) Determination of copy number in the 11p15.4 region. Copy number of 11p15.4 (CNN-LOH region) in case no. 39 at diagnosis was compared with complete remission (CR) sample with quantitative genomic real-time PCR. Levels of the copy number were determined as a ratio between 11p15.4 and the reference genomic DNA, 2p21. Results represent the mean of 3 experiments plus or minus SD.
Copy-number changes in t(15;17) APL samples
As shown in Table 2, several copy-number changes were detected by SNP-chip analysis. Deletions were found in 7 cases (15%) including case no. 65 (4q28.1, 1.33 Mb; 7q21.11-q21.12, 1.03 Mb; and 9q12-q31.3, 47.27 Mb), case no. 2 (10q21.2-q21.3, 5.55 Mb), case no. 50 (6p25.1-p24.3, 2.51 Mb), case no. 37 (1q42.2, 0.02 Mb), case no. 19 (12p13.31-p11.22, 22.49 Mb; and 13q14.2-q14.3, 0.88 Mb), case no. 21 (7q11.21–q-terminal, 97.03 Mb), and case no. 58 (17p-terminal–p11.2, 21.44 Mb). Of note, deleted region at 12p of case no. 19 and 17p of case no. 58 contained the ETV6/TEL and TP53 genes, respectively.
Duplications were found in 7 cases (15%) including case no. 65 (8q24.13-q24.22, 9.48 Mb), case no. 13 (13q21.1–q-terminal, 57.27 Mb; and 15q22.2–q-terminal, 42.94 Mb), case no. 57 (9q22.32, 0.27 Mb), case no. 37 (18p11.31–p11.23, 0.46 Mb), case no. 21 (1q21–q-terminal, 102.63 Mb), case no. 43 (21q22.12–q-terminal, 10.69 Mb), and case no. 58 (15q24.1–q-terminal, 27.97 Mb; and 17p11.2–q21.1, 14.05 Mb). The duplicated region at 21q of case no. 43 and at 17p of case no. 58 included the ERG and ERBB2 genes, respectively. Importantly, the duplicated region at 8q of case no. 65 contained the MYC gene (Figure 2A). Seven cases had trisomy 8, and one of the candidate genes on chromosome 8 is the MYC gene; therefore, we classified case no. 65 within the +8 group.
Of note, the NB4 APL cell line had amplification of the MYC gene region (8q24.21), whereas the PL-21 APL cell line showed duplication of the region (Figure S2A). We compared levels of c-Myc mRNA in these cell lines, and found that NB4 cells had approximately 6-fold higher expression of c-Myc than did PL-21 cells (Figure S2B). This result indicated that copy-number change was associated with mRNA levels of the target gene.
CNN-LOH in t(15;17) samples
Several cases had the same chromosomal region involved in CNN-LOH. Chromosome 10q CNN-LOH was found in 3 cases (6%) including case no. 18 (10q22.2–q-terminal), case no. 39 (10q21.1–q-terminal), and case no. 66 (10q22.2–q-terminal; Figure 1; Figure S3A). This region included the tyrosine kinase receptor gene FGFR2 and the tumor suppressor gene PTEN (Table 2). Cases no. 3, no. 20, and no. 39 had CNN-LOH on 11p-terminal–p11.12, 11p-terminal–p11.12, and 11p-terminal–p14.1, respectively; and the common region was 28.7 Mb (Figure 1; Figure S3B) containing the tumor suppressor genes WT1 and CDKN1C, and the oncogene HRAS (Table 2). CNN-LOH of 19q13.2–q-terminal (17.3 Mb) occurred in one case (no. 4, Table 2).
Comparison of chromosomal changes between diagnosis and complete remission samples
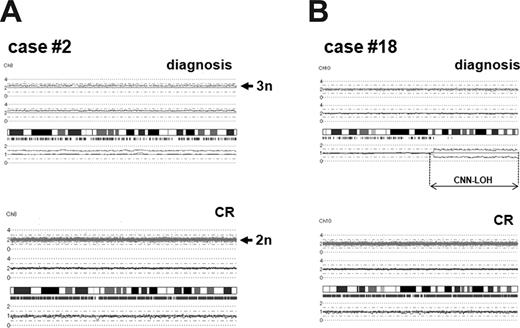
To assess whether chromosomal alterations detected by SNP-chip analysis were acquired abnormalities, germ-line mutations, or copy-number variants, we compared chromosomal changes between diagnosis and complete remission (CR) samples in the same patients. CR samples of cases no. 2, no. 3, no. 18, no. 19, no. 38, no. 39, and no. 50 were available, and these CR samples were subjected to SNP-chip analysis. As shown in Figure 4, trisomy 8 of case no. 2 and CNN-LOH of case no. 18 at diagnosis were not present in the samples obtained at CR. Other alterations including trisomy 8 (cases no. 3, no. 18, no. 38, no. 39, and no. 50), CNN-LOH at chromosomes 10 (case no. 39) and 11 (cases no. 3 and no. 39), and deletions (cases no. 2, no. 19, and no. 50) also were not present at CR (Figure S4). Taken together, these results showed that chromosomal alterations detected by SNP-chip analysis were acquired somatic changes.
Comparison of chromosomal changes between diagnosis and complete remission samples. (A) Trisomy 8 in case no. 2. Case no. 2 had trisomy 8 at diagnosis, whereas chromosome 8 was 2n at complete remission (CR). (B) CNN-LOH in case no. 18. Case no. 18 had CNN-LOH in chromosome 10 (10q22.2–q-terminal, 58.2 Mb), and the alteration was not present in the matched CR sample.
Comparison of chromosomal changes between diagnosis and complete remission samples. (A) Trisomy 8 in case no. 2. Case no. 2 had trisomy 8 at diagnosis, whereas chromosome 8 was 2n at complete remission (CR). (B) CNN-LOH in case no. 18. Case no. 18 had CNN-LOH in chromosome 10 (10q22.2–q-terminal, 58.2 Mb), and the alteration was not present in the matched CR sample.
Relationship between genomic abnormalities and FLT3 mutations
Finally, we compared genomic abnormalities and FLT3 status. Twenty-four samples (51%) had wild-type FLT3; whereas 12 samples (26%) had FLT3-TKD mutation (aspartic acid at codon 835, D835) and another 11 samples (23%) had FLT3-ITD form. Interestingly, all 11 samples with FLT3-ITD were found only in the normal-copy-number (NC) group (Tables 1 and 3). One sample in the trisomy 8 group had a FLT3-TKD mutation. Samples in the “other abnormalities” group did not have FLT3-ITD; and 6 samples in NC group and 5 samples in the “other abnormalities” group had a FLT3-TKD mutation. These results suggested that the pathway of development of APL differs in each group; in a mutually exclusive fashion, FLT3-ITD, trisomy 8, and unknown factor(s) were involved in each group.
Relationship between chromosomal abnormality and FLT3 mutations
| . | Group . | Total (%), 47 (100%) . | ||
|---|---|---|---|---|
| NC, 28 (60%) . | +8, 8 (17%) . | Other, 11 (23%) . | ||
| FLT3 WT | 11 | 7 | 6 | 24 (51) |
| FLT3 TKD | 6 | 1 | 5 | 12 (26) |
| FLT3 ITD | 11 | 0 | 0 | 11 (23) |
| . | Group . | Total (%), 47 (100%) . | ||
|---|---|---|---|---|
| NC, 28 (60%) . | +8, 8 (17%) . | Other, 11 (23%) . | ||
| FLT3 WT | 11 | 7 | 6 | 24 (51) |
| FLT3 TKD | 6 | 1 | 5 | 12 (26) |
| FLT3 ITD | 11 | 0 | 0 | 11 (23) |
Mutational status of the FLT3 gene is shown.
NC indicates normal-copy-number; +8, trisomy 8 or duplication of the MYC gene region; other, other abnormalities; and WT, wild-type.
Discussion
Our genome-wide SNP-chip analysis showed that 40% of t(15;17) APL samples had one or more genomic abnormalities including deletions, duplications, and/or CNN-LOH. Since the PML-RARα fusion protein is probably not sufficient to cause APL in murine model systems, the additional genetic changes that we found may be necessary to cause the leukemia.
Our analysis revealed that 6 samples (13%) of t(15;17) APL samples had CNN-LOH. Previously, we analyzed AML with normal karyotype samples and found that 32% of cases had CNN-LOH (Akagi et al28 ); and other investigators also demonstrated CNN-LOH in AML samples at a frequency of 15% to 20%.22-25,27 Of interest, approximately 40% of relapse AML had CNN-LOH.26 CNN-LOH in t(15;17) APL is about half as frequent as the other AMLs. Two CNN-LOH regions occurred in multiple samples: chromosomes 10q (58.2 Mb, 3 cases) and 11p (28.7 Mb, 3 cases). Of note, case no. 39 had both 10q and 11p CNN-LOHs, suggesting that 10q and 11p might contain novel APL-related gene(s). CNN-LOH is a genomic abnormality that normally cannot be detected by conventional cytogenetic analysis. These regions usually contain a mutation of a key gene. For example, a constitutively active form of either JAK2 V617F mutant, FLT3-ITD, AML1/RUNX1 frameshift, and/or mutations of WT1 and NPM1 were found in CNN-LOH regions in AML.22-25 CNN-LOH regions identified in this study contain genes coding for several tyrosine kinase and/or tumor suppressors. Further studies are required to identify the key dysregulated gene(s) in these regions. In addition to CNN-LOH, we also found several copy-number changes that may be sites containing novel disease-related genomic regions in t(15;17) APL. Although we cannot rule-out copy-number variants (CNVs) at several sites, we think it is unlikely. We had 7 genomic DNA at complete remission samples and confirmed for each that the chromosomal changes were only in the leukemia cells. Furthermore, for each of these sites, we interrogated a collated library of CNVs (Database of Genomic Variants and UCSC Genome Browser) to assure that these regions were not known CNVs.
FLT3 is a tyrosine kinase receptor involved in normal hematopoiesis, and mutations of the gene often occur in AML. Incidence of FLT3-ITD and FLT3-TKD was 23% and 26% in our samples, respectively. Experiments have shown that FLT3-ITD and FLT3-TKD have differences in their downstream signaling.33-35 Interestingly, bone marrow transplantation in mice showed that FLT3-ITD induced an oligoclonal myeloproliferative disease,33 whereas FLT3-TKD produced an oligoclonal lymphoid disorder with a long latency.34 Furthermore, only FLT3-ITD caused activation of STAT5 and repression of C/EBPα and PU.1.34,35
Here, t(15;17) APL samples were divided into 3 groups based on genomic status detected by SNP-chip analysis: normal-copy-number group (NC group, 28 samples); trisomy 8 group (+8 group, 8 samples); and other abnormalities group (11 samples). Notably, our subclassifications did reveal an interesting relationship between genomic status and FLT3 mutation. Eleven samples of NC group (39% of the NC group samples) had FLT3-ITD, whereas no FLT3-ITD occurred in samples from the other 2 groups. In contrast, one good candidate gene in the +8 cohort is the oncogene MYC. In fact, case no. 65 had duplication localized to 8q24.13-q24.22 that included the MYC gene. Previous karyotype analysis showed that the PL-21 cell line, which was established from an APL patient, had a polyploid male karyotype with 13q+ chromosome, but a translocation between chromosome 15 and 17 was not identified.36 NB4 cells are cytogenetically very complex, with a hypotetraploid karyotype and multiple chromosomal alterations.37,38 Our SNP-chip analysis of NB4 cells also showed ploidy = 3.67, indicating that the karyotype is hypotetraploid. Of interest, NB4 cells have amplification of the MYC gene. Importantly, expression of c-Myc mRNA is stimulated by FLT3-ITD39,40 ; and AML samples with FLT3-ITD have increased expression of c-Myc mRNA compared with normal bone marrow.41 These data indicate that the MYC gene may be dysregulated by either copy-number change or FLT3-ITD as a secondary abnormality to enhance the development of APL. Of course, our sample population is small, therefore, additional studies are needed to confirm these findings.
Our APL cohort “other abnormalities” group has neither FLT3-ITD nor trisomy 8, but has several other genomic changes including deletion of ETV6/TEL (case no. 19) and duplication of ERG (case no. 43). ETV6/TEL is a transcriptional repressor, and approximately 30% of AML patients have loss of expression of ETV6/TEL protein.42,43 In addition, mutation of the ETV6/TEL gene occurs in approximately 2% of AML samples, and these mutants behaved in a dominant-negative fashion.43 ERG is a member of the ETS family of transcription factors and is a proto-oncogene. Overexpression of ERG predicts a worse outcome in AML with normal karyotype.44 Taken together, our observed copy-number changes in these regions may be involved in development of APL.
Our findings extend those of Le Beau et al13 who recently reported elegant models of APL using transgenic mice coexpressing PML-RARα and either BCL2, IL3, activated IL3R, or activated murine FLT3 (FLT3W51). PML-RARα/BCL2 mice developed leukemia, and these cells had a complex karyotype including trisomy 15 (100% of these mice), where the oncogene MYC is located. In contrast, PML-RARα/FLT3W51 mice develop leukemia, and these cells had normal karyotype except for trisomy of either chromosomes 8 (29%), 10 (43%), or 15 (43%), and monosomy X (86%). These models suggest that different cooperating events are involved in the development of murine APL. Taken together, these findings strongly suggest that the pathway of development of APL differs in each of our cohorts; FLT3-ITD, MYC, and unknown factor(s) are involved in the development of APL; and these finding should facilitate the screening for novel therapeutic targets in each case.
Further studies in a larger cohort of patients will begin to stratify prognostically the APL patients in relation to the genomic changes of their leukemic cells; and new therapeutic targets, which are involved in the development of APL, should be discovered.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank members of our laboratory for helpful discussions. APL cell line PL-21 was kindly provided by Dr Ikezoe Takayuki (Kochi University, Kochi, Japan).
This work was supported by National Institutes of Health (NIH, Bethesda, MD) grant 5R01CA026038-30 (H.P.K.), the Inger Foundation, (Greenwich, CT), the Tom Collier Memorial Regatta Foundation (Los Angeles, CA), the Parker Hughes Fund (Los Angeles, CA), as well as grant NHRI-EX96-9434SI (National Health Research Institutes, Miaoli, Taiwan; L.-Y.S.) and grant MMH-E-96009 (Mackay Memorial Hospital, Taipei, Taiwan; D.-C.L.). H.P.K. is the holder of the Mark Goodson endowed Chair in Oncology Research and is a member of the Jonsson Cancer Center and the Molecular Biology Institute, UCLA. The study is dedicated to David Golde, a mentor and friend.
National Institutes of Health
Authorship
Contribution: T.A. performed research, analyzed the data, and wrote the paper; M.K., S.O., G.Y., and M.S. performed SNP-chip analysis and developed CNAG; N.K., R.O., and C.W.M. assisted in data analysis; L.-Y.S. and D.-C.L. provided APL samples and clinicohematologic data for all APL patients, and performed FLT3 mutation analysis; and H.P.K. directed the overall study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tadayuki Akagi, Department of Stem Cell Biology, Graduate School of Medical Science, Kanazawa University, 13-1 Takara-machi, Kanazawa, Ishikawa 920-8640, Japan; e-mail: tadayuki@staff.kanazawa-u.ac.jp.
References
Author notes
*T.A. and L.-Y.S. contributed equally to the study and should be considered joint first authors.