Abstract
In the revised National Cancer Institute Working Group (NCI-WG)/International Workshop on Chronic Lymphocytic Leukemia (IWCLL) guidelines for CLL, CLL-like monoclonal B lymphocytosis (MBL) is defined as the presence of less than 5 × 109/L B lymphocytes in the peripheral blood. However, the concentration of MBL in the blood is extremely variable. MBL in subjects with lymphocytosis require treatment at a rate of 1.1% per year and present immunoglobulin (IG) gene features and similar to good prognosis CLL. Little is known about low-count MBL cases, accidentally found in the general population. We analyzed IGHV-D-J rearrangements in 51 CLL-like MBL cases from healthy individuals, characterized by few clonal B cells. Seventy percent of the IGHV genes were mutated. The most frequent IGHV gene was IGHV4-59/61, rarely used in CLL, whereas the IGHV1–69 gene was lacking and the IGHV4-34 gene was infrequent. Only 2 of 51 (3.9%) MBL cases expressed a CLL-specific stereotyped HCDR3. Therefore, the IG gene repertoire in low-count MBL differs from both mutated and unmutated CLL, suggesting that the detection of MBL in an otherwise healthy subject is not always equivalent to a preleukemic state. Detailed IG analysis of individual MBL may help to identify cases that necessitate continuous clinical monitoring to anticipate disease progression.
Introduction
The existence of monoclonal B lymphocytes in the blood of otherwise healthy individuals has long been known,1-4 but only recently has the frequency of this phenomenon been fully appreciated, thanks to more advanced flow cytometry techniques.5,6 Monoclonal B-cell expansions, now named monoclonal B lymphocytosis (MBL),7 are rather heterogeneous in terms of phenotype.5 They are classified on the basis of the presence or absence of the CD5 molecule on the cell surface. Cases that do not express CD5 are named CD5− MBL, those expressing CD5 are subdivided into atypical CLL-like (with bright CD20 expression) and CLL-like (with low levels of CD20) MBL.7 The latter also express CD23 and low levels of both surface immunoglobulins (Ig) and CD79 and attracted the attention of investigators because of their close phenotypic similarity with chronic lymphocytic leukemia (CLL) cells. CLL-like MBL is the most frequent MBL type, being detected in more than 3.5% of all individuals older than 40 years, and its frequency increases with age, becoming greater than 7% in individuals older than 75 years.5,6
The interest in CLL-like MBL increased after this entity was included in the revised National Cancer Institute Working Group (NCI-WG)/International Workshop on Chronic Lymphocytic Leukemia (IWCLL) guidelines for the diagnosis and management of CLL8 and defined as “the presence of fewer than 5 × 109/L B lymphocytes” in the peripheral blood. However, the concentration of MBL in the blood of any given individual is extremely variable, accounting in some cases for the vast majority of circulating B cells while being a rather limited portion of them in others.5,6 It is then conceivable that molecular differences could exist between low- versus high-count MBL, with the latter likely being more advanced on the way to becoming CLL.9 In this context, it was recently reported10 that subjects with less than 5 × 109/L CLL-like monoclonal B lymphocytes but with lymphocytosis require treatment at a rate of 1.1% per year. The absolute B-cell count turned out to be the only independent prognostic factor associated with progressive lymphocytosis, as all MBL cases studied had immunoglobulin (IG) gene features and cytogenetic abnormalities similar to good prognosis CLL.10 In contrast, very little is known about low-count MBL accidentally found in the general population. The possibility exists that these cases may not be necessarily a preleukemic condition but represent an aspect of immune-senescence and/or the outcome of a persistent immune stimulation. This hypothesis appears to be plausible, as a progressive restriction of the immune repertoire with the appearance of monoclonality is well-known in the normal aging immune system.11,12 MBL is at least 100 times more frequent than CLL and the transition into CLL, even among high-count cases with lymphocytosis, is not an inevitable fate, occurring only in 1.1% of the cases per year.10
The high incidence of MBL, if coupled with the possibility of evolution into a frank leukemic state, poses evident clinical and health system problems and creates the need for a better characterization of MBL, especially of low-count cases, aiming at identifying molecular and biologic features that may define which cases are more prone to progress toward a clinically overt CLL. This approach should also help to avoid the social and psychologic burden of prolonged follow-ups in a large number of persons who are extremely unlikely to develop CLL simply because they have MBL.
We have investigated the occurrence, frequency, and IG molecular features of MBL detected in 128 of 1725 healthy individuals older than 18 years of age who belong to a population living in a rural community in Northern Italy, the Val Borbera. These individuals are currently involved in a study not aimed at elucidating any particular health problem but rather to dissect potential genetic components of common diseases thanks to their degree of genetic homogeneity.13 We here report that, overall, the IG molecular features of low-count MBL, in terms of IGHV gene repertoire and HCDR3 stereotypy, differ significantly from those detected in both mutated and unmutated CLL cases, suggesting that MBL does not necessarily represent a preleukemic condition, especially when present at a low concentration. We also show that in some cases the acquisition of a CLL-like phenotype is independent from the appearance of monoclonality, as polyclonal CLL-like B cells may be detected albeit at a low frequency. Finally, we document that the development of MBL, though having an important familial/genetic component, is independent of the IGHV gene repertoire, thereby suggesting the importance of other (eg, environmental) factors.
Methods
Study population
We studied 1725 healthy individuals belonging to the Val Borbera population, a rural valley in Northern Italy that is isolated from the surrounding areas by geographic barriers and is likely to derive from a limited number of common ancestors (ie, “genetically isolated”).14 The whole population is currently involved in a study not aimed at elucidating any particular health problem but rather at dissecting potential genetic components of common diseases.13
The individuals studied were older than 18 years of age (967 women and 758 men) with a mean age of 55.2 years (range, 18-102 years). The study population has been carefully investigated from the clinical and genealogic point of view. Clinical records were collected on all 1725 individuals who underwent routine blood tests including a complete blood count with differential. No individual had a known history of CLL. One male was affected by a splenic marginal zone lymphoma whose leukemic phase was detected by our analysis as atypical CLL-MBL (see next paragraph), and was not included in the analysis. The study was performed over 3 years (2005 to 2007).
The research protocol was approved by the Institutional Ethics Committee at San Raffaele Scientific Institute, Milano, Italy, and all participants gave written informed consent in accordance with the Declaration of Helsinki.
Pedigrees were reconstructed from the genealogic tree (G. Milani, C. Larizza, I. Buetti, C. Masciullo, C.S., D.T., and B. Bellazzi, manuscript in preparation) using the PedHunter software package.15 Kinship coefficient was calculated by KinInbcoef program,16 and measures the family relationships between every 2 individuals in a family tree. It ranges from 0.25 for siblings and child-parent relations to 0 for unrelated people.
Blood samples, cell preparation, staining, and FACS analysis
EDTA (ethylenediaminetetraacetic acid) peripheral blood (PB) samples obtained from all individuals enrolled were processed within 24 hours after blood withdrawal, incubated with the proper antibodies, followed by NH4Cl (8.6 g/L in distilled water), and washed repeatedly in phosphate-buffered saline (PBS) plus 0.3% bovine serum albumin (BSA). The following antibody mixes were used: fluorescein isothiocyanate (FITC)–conjugated F(ab)2–anti-κ, phycoerythrin (PE)–anti-λ light chain (Dako Cytomation), PE-cyanin7 (Cy7)–labeled anti-CD20 (Becton Dickinson), PE-cyanin5 (Cy5)–conjugated anti-CD5 (Caltag Laboratories), and PE-Texas Red (ECD)–conjugated anti-CD19. For each sample, up to 500 000 events were acquired on a FC500 equipped with 488 argon ion laser and 635 red HeNe laser and analyzed with the CXP software system (Beckman Coulter) according to the following gating strategy: low forward and side scatter (FSC/SSC) CD19+ cells were gated and further divided into CD5− and CD5+ subsets and the κ/λ ratio was evaluated in both populations (indicating the presence of atypical CLL or non-CLL MBL).5 The κ/λ ratio was considered abnormal when it was more than 3:1 or less than 1:3. Concomitantly, we used a dot plot showing CD5 versus CD20 expression on gated CD19+ cells to identify CLL-like MBL (ie, CD19+cells, CD20dim, CD5bright).17
For quality control purposes, we daily used 0.4 mL Flow-Check Fluorospheres (Beckman Coulter) mixed with 0.2 mL Flow-Check 770 (Beckman Coulter PC7 [770/488] Setup Kit) to assess flow cytometer optical alignment and fluidics system. In addition, we daily controlled light scatter intensity, fluorescence intensity, and hydrodynamics using 0.4 mL Flow-Set Fluorospheres (Beckman Coulter) mixed with 0.2ml Flow-Set 770 (Beckman Coulter PC7 (770/488) SETUP KIT), to assess optimal conditions for quantitative analysis of human leukocytes.
PCR amplification of IGHV-D-J rearrangements and sequence analysis
Genomic DNA was extracted from whole-blood samples using the QIAmp blood kit (QIAGEN). Polymerase chain reaction (PCR) amplification of germ line IGHV1-69 and IGHV4-34 sequences was performed in 51 CLL-like MBL cases and 30 nonaffected individuals with the appropriate primer sets. In particular, the upstream primers were complementary to leader or framework 1 sequences whereas the downstream primers were reverse complementary to the 3′ untranslated region (3′UTR) of the corresponding genes. IGHV-D-J rearrangements were amplified with a nested PCR approach, using consensus FR1 or FR2 primers with 2 consensus IGHJ primers, and run on a polyacrylamide gel (PAGE 6%) to confirm monoclonality. Low-melting-point agarose gel-purified monoclonal PCR products were subjected to sequencing on an automated ABI sequencer either directly (34 of 51 cases) or after cloning (17 of 51 cases) using the TA Cloning kit (Invitrogen). In the latter case, at least 10 colonies have been analyzed for each PCR amplification product. All cases in which more than 20% (up to 100%) of the clones were identical (14 cases) were considered monoclonal. In the remaining 3 cases (using the IGHV4-59/61, IGHV3-30, and IGHV4-34 genes, respectively), the rearrangement was assigned when appearing in at least 2 different clones, with all the others being different. The sequencing reaction was performed using (1) an antisense IGHJ gene consensus oligonucleotide; (2) a sense consensus FR1- or FR2-specific oligonucleotide.18
Sequence data were analyzed using the international ImMunoGeneTics information system (IMGT) database and tools.19 All nonproductive rearrangements have been excluded from the repertoire analysis. The sequences with a germ line identity of 98% or higher were considered unmutated, and those with an identity less than 98% were considered mutated.20,21 Rearrangements using the IGHV4-59 and IGHV4-61 genes were considered one group because the usage of an FR2-primer for PCR amplification precluded a clear assignment to either one or the other IGHV gene that differ mainly in the HFR1-HCDR1.
Our sequences were aligned to a panel of 1939 sequences from patients with CLL previously reported from our group18 and 5303 non-CLL IGHV-D-J sequences available from the literature and/or retrieved from the IMGT/LIGM-DB sequence database (including 278 sequences from B cells of healthy elderly individuals).22
In order to assess stereotypy within the HCDR3 sequences and to define subsets with stereotyped receptors, we followed previously described criteria.23
Results
Phenotypic characterization of MBL cases in a healthy population
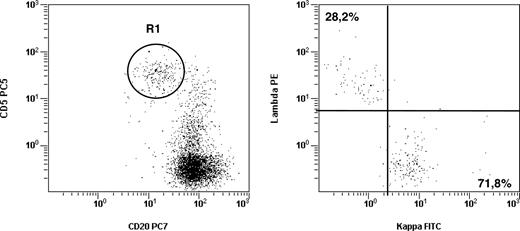
To identify the MBL population we used 5-color flow cytometric analysis and adhered to standard criteria.5 Accordingly, the definition of CD5− MBL and atypical CLL-MBL was based on the occurrence of an unbalanced κ/λ ratio (> 3:1 or < 1:3) within CD5− or CD5+ B lymphocytes, respectively. In contrast, the third type of MBL, CLL-like MBL, was defined based on the distinct CD5brightCD20dim expression pattern on CD19+ B cells. Following this strategy, we identified all 3 MBL subtypes.5 Monoclonal B lymphocytes were detected in 128 of 1725 individuals (7.4%) and included 20 CD5− MBL cases, 19 atypical CLL MBL cases, and 89 CLL-like MBL cases, the latter being detected in 5.2% of the whole population studied. When we analyzed the light chain expression on CLL-like MBL, the vast majority (83/89) had an unbalanced ratio (> 3:1 or < 1:3), suggestive of monoclonality, but unexpectedly, 6 of 89 cases showed a normal κ/λ ratio (< 3:1; range 1.3:1 to 2.9:1; Figure 1).
CLL-like MBL may show a normal κ/λ ratio. Flow cytometric analyses from a representative sample from 6 individuals carrying a CLL-like MBL phenotype with a normal (< 3:1 or > 1:3) κ/λ ratio. (A) Dot plot was obtained by gating on CD19+ lymphocytes (identified on forward-scatter vs CD19 profiles, not shown). The intensity of CD20 and CD5 is shown on the x-axis and y-axis, respectively. R1 gate indicates the CLL-like MBL cells. (B) Dot plot shows κ and λ light chain expression of the B-cell clone as identified on panel A. All other B-cell populations present in the dot plot (A) are polyclonal (data not shown).
CLL-like MBL may show a normal κ/λ ratio. Flow cytometric analyses from a representative sample from 6 individuals carrying a CLL-like MBL phenotype with a normal (< 3:1 or > 1:3) κ/λ ratio. (A) Dot plot was obtained by gating on CD19+ lymphocytes (identified on forward-scatter vs CD19 profiles, not shown). The intensity of CD20 and CD5 is shown on the x-axis and y-axis, respectively. R1 gate indicates the CLL-like MBL cells. (B) Dot plot shows κ and λ light chain expression of the B-cell clone as identified on panel A. All other B-cell populations present in the dot plot (A) are polyclonal (data not shown).
The median age of all individuals with MBL was 66.9 years (range 18-98 years) and the frequency was increased in males (9.9% as compared with 5.5% in females; CLL-like MBL occurred in 6.7% males and 3.9% females).
Overall, the frequency of MBL appeared to increase with age (Table 1). However, this phenomenon was mainly accounted for by CLL-like MBL, whose frequency was negligible below 40 years, became more evident above 40 years and peaked at > 9,0% above 80 years of age (table 1). The frequency of the other types of MBL was less affected by aging.
Frequency of all types of MBL and of CLL-like MBL in different age groups
| Age group, y . | All individuals . | All MBL (%) . | CLL-like MBL (%) . | Non–CLL-like MBL (%)* . |
|---|---|---|---|---|
| <40 | 403 | 7 (1.7) | 1 (0.2) | 6 (1.5) |
| 40-49 | 235 | 6 (2.5) | 4 (1.7) | 2 (0.9) |
| 50-59 | 317 | 19 (6.0) | 15 (4.7) | 4 (1.3) |
| 60-69 | 355 | 37 (10.4) | 31 (8.7) | 6 (1.7) |
| 70-79 | 264 | 34 (12.9) | 24 (9.1) | 10 (3.8) |
| ≥80 | 151 | 25 (16.5) | 14 (9.3) | 11 (7.3) |
| Total | 1725 | 128 (7.4) | 89 (5.2) | 39 (2.3) |
| Age group, y . | All individuals . | All MBL (%) . | CLL-like MBL (%) . | Non–CLL-like MBL (%)* . |
|---|---|---|---|---|
| <40 | 403 | 7 (1.7) | 1 (0.2) | 6 (1.5) |
| 40-49 | 235 | 6 (2.5) | 4 (1.7) | 2 (0.9) |
| 50-59 | 317 | 19 (6.0) | 15 (4.7) | 4 (1.3) |
| 60-69 | 355 | 37 (10.4) | 31 (8.7) | 6 (1.7) |
| 70-79 | 264 | 34 (12.9) | 24 (9.1) | 10 (3.8) |
| ≥80 | 151 | 25 (16.5) | 14 (9.3) | 11 (7.3) |
| Total | 1725 | 128 (7.4) | 89 (5.2) | 39 (2.3) |
Includes CD5– and atypical CLL-MBL.
All 128 individuals with any type of MBL had a normal absolute lymphocyte count (ALC) (mean value 2.3 × 109/L [2256/μL]; range, 1.0 × 109/L-3.4 × 109/L [1040/μL-3420/μL]), except for 5 cases whose lymphocyte counts were slightly above the 4.0 × 109/L value (range 4.1 × 109/L-5.8 × 109/L). The mean absolute B lymphocyte count (BALC) was 0.17 × 109/L (165/μL) and monoclonal B cells represented a variable proportion of total CD19+ B cells, with a mean of 18.7%, and reached virtually 100% of all B lymphocytes in only one case.
When we considered CLL-like MBL only, all 89 individuals had a normal ALC (lymphocyte mean value, 2.3 × 109/L [2250/μL]; range, 1.0 × 109/L-4.3 × 109/L [1040/μL-4250/μL]), except for 3 cases above the 4.0 × 109/L value. In no case was the mean BALC above 5.0 × 109/L, (average 0.17 × 109/L [170/μL], range 0.01 × 109/L-1.92 × 109/L [10/μL-1920/μL]). Interestingly, the 3 cases with slight lymphocytosis had only 0.1, 1.9, and 0.6 × 109 B cells/L, respectively, while all others presented with less than 0.45 × 109 B cells/L. Monoclonal B cells represented a variable, though small, proportion of total CD19+ B cells, with an average of 6.9% (mean MB lymphocyte cell count was 34/μL); only 13 cases had more than 10% MB lymphocytes. For this reason, we consider this MBL population as representative of what we defined as low-count MBL.
IGHV gene repertoire and mutational status in low-count CLL-like MBL
Analysis of IGHV-D-J rearrangements was performed only in the monoclonal CLL-like MBL cases (excluding the 6 polyclonal cases) and demonstrated a distinct monoclonal rearrangement on acrylamide gels in all 83 cases. However, this was usually admixed with a polyclonal background as the MBL monoclonal cells represented a minor proportion of total B lymphocytes (mean 6.9%, with only 13 cases > 10% and 34 cases < 10% but > 1%). From this analysis, we were able to either directly sequence or clone 51 IGHV-D-J rearrangements among which IGHV3 subgroup genes predominated (32/51, 62.7%), followed by IGHV4 subgroup genes, according to the expected frequency of the same subgroups in the normal repertoire (Table 2). Following the 98% identity cut-off value,20,21 36 of 51 sequences (70.5%) were defined as “mutated,” whereas the remainder (15/51 sequences; 29.5%) had “unmutated” IGHV genes; 10 of 15 unmutated sequences had 100% identity to germ line (Table 2). No significant intraclonal variation could be observed in the 17 IGHV genes that were sequenced after cloning.
IGHV-D-J rearrangements detected in CLL-like MBL
| Case no. . | IGHV gene . | IGHD gene . | IGHJ gene . | %* . |
|---|---|---|---|---|
| 4VB | IGHV3-15 | IGHD1-26 | IGHJ3 | 98.2 |
| 60VB | IGHV3-15 | ND | ND | 93.5 |
| 2VB | IGHV3-21 | IGHD2-15 | IGHJ6 | 88.2 |
| 113VB | IGHV3-21 | IGHD3-22 | IGHJ3 | 95.1 |
| 19VB | IGHV3-21 | IGHD1-1 | IGHJ4 | 96.4 |
| 20VB | IGHV3-23 | IGHD2-15 | IGHJ2 | 92 |
| 24VB | IGHV3-23 | ND | ND | 95.2 |
| 22VB | IGHV3-23 | IGHD5-12 | IGHJ3 | 100 |
| 27VB | IGHV3-23 | IGHD1-26 | IGHJ6 | 100 |
| 14VB | IGHV3-30 | IGHD7-27 | IGHJ2 | 91.1 |
| 43VB | IGHV3-30 | IGHD3-3 | IGHJ4 | 97.5 |
| 6VB | IGHV3-30 | IGHD6-13 | IGHJ3 | 100 |
| 96VB | IGHV3-30 | ND | ND | 97.8 |
| 7VB | IGHV3-30 | IGHD2-21 | IGHJ4 | 97.5 |
| 92VB | IGHV3-30 | ND | ND | 97.7 |
| 90VB | IGHV3-48 | IGHD3-10 | IGHJ4 | 100 |
| 35VB | IGHV3-48 | IGHD1-20 | IGHJ4 | 88.3 |
| 97VB | IGHV3-48 | IGHD5-12 | IGHJ3 | 93.9 |
| 13VB | IGHV3-49 | IGHD2-21 | IGHJ4 | 100 |
| 99VB | IGHV3-53 | ND | ND | 95 |
| 68VB | IGHV3-53 | IGHD3-9 | IGHJ2 | 96.8 |
| 72VB | IGHV3-53 | IGHD6-19 | IGHJ4 | 100 |
| 3VB | IGHV3-64 | IGHD3-10 | IGHJ3 | 100 |
| 8VB | IGHV3-64 | IGHD4-17 | IGHJ2 | 93.9 |
| 48VB | IGHV3-64 | IGHD3-9 | IGHJ5 | 86.8 |
| 105VB | IGHV3-64 | IGHD3-3 | IGHJ3 | 98.1 |
| 1VB | IGHV3-66 | IGHD2-2 | IGHJ6 | 93.7 |
| 87VB | IGHV3-7 | IGHD3-22 | IGHJ3 | 92.6 |
| 63VB | IGHV3-7 | IGHD2-2 | IGHJ5 | 95.7 |
| 66VB | IGHV3-7 | IGHD2-2 | IGHJ5 | 96.9 |
| 29VB | IGHV3-7 | ND | ND | 97 |
| 9VB | IGHV3-73 | IGHD4-4 | IGHJ4 | 92.6 |
| 34VB | IGHV4-30-4 | IGHD3-16 | IGHJ5 | 94.4 |
| 10VB | IGHV4-30-4 | IGHD5-24 | IGHJ4 | 91.1 |
| 16VB | IGHV4-31 | IGHD3-10 | IGHJ5 | 93.3 |
| 102VB | IGHV4-34 | IGHD3-3 | IGHJ4 | 97.7 |
| 108VB | IGHV4-34 | IGHD3-10 | IGHJ4 | 88.1 |
| 57VB | IGHV4-39 | IGHD3-22 | IGHJ4 | 100 |
| 58VB | IGHV4-39 | IGHD3-22 | IGHJ4 | 100 |
| 5VB† | IGHV4-59 | IGHD1-26 | IGHJ2 | 93.5 |
| 101VB | IGHV4-59/-61 | IGHD5-5 | IGHJ1 | 100 |
| 115VB | IGHV4-59/-61 | IGHD3-22 | IGHJ4 | 100 |
| 12VB | IGHV4-59/-61 | IGHD1-26 | IGHJ3 | 100 |
| 15VB | IGHV4-59/-61 | IGHD4-23 | IGHJ4 | 89.4 |
| 17VB | IGHV4-59/-61 | IGHD4-17 | IGHJ6 | 97 |
| 18VB | IGHV4-59/-61 | IGHD4-4 | IGHJ6 | 95.8 |
| 21VB | IGHV4-59/-61 | IGHD5-24 | IGHJ4 | 90.6 |
| 47VB | IGHV4-59/-61 | IGHD2-21 | IGHJ4 | 94.4 |
| 80VB | IGHV4-59/-61 | IGHD5-12 | IGHJ4 | 94.1 |
| 89VB | IGHV4-59/-61 | IGHD2-15 | IGHJ2 | 95 |
| 95VB | IGHV4-59/-61 | IGHD3-3 | IGHJ5 | 100 |
| Case no. . | IGHV gene . | IGHD gene . | IGHJ gene . | %* . |
|---|---|---|---|---|
| 4VB | IGHV3-15 | IGHD1-26 | IGHJ3 | 98.2 |
| 60VB | IGHV3-15 | ND | ND | 93.5 |
| 2VB | IGHV3-21 | IGHD2-15 | IGHJ6 | 88.2 |
| 113VB | IGHV3-21 | IGHD3-22 | IGHJ3 | 95.1 |
| 19VB | IGHV3-21 | IGHD1-1 | IGHJ4 | 96.4 |
| 20VB | IGHV3-23 | IGHD2-15 | IGHJ2 | 92 |
| 24VB | IGHV3-23 | ND | ND | 95.2 |
| 22VB | IGHV3-23 | IGHD5-12 | IGHJ3 | 100 |
| 27VB | IGHV3-23 | IGHD1-26 | IGHJ6 | 100 |
| 14VB | IGHV3-30 | IGHD7-27 | IGHJ2 | 91.1 |
| 43VB | IGHV3-30 | IGHD3-3 | IGHJ4 | 97.5 |
| 6VB | IGHV3-30 | IGHD6-13 | IGHJ3 | 100 |
| 96VB | IGHV3-30 | ND | ND | 97.8 |
| 7VB | IGHV3-30 | IGHD2-21 | IGHJ4 | 97.5 |
| 92VB | IGHV3-30 | ND | ND | 97.7 |
| 90VB | IGHV3-48 | IGHD3-10 | IGHJ4 | 100 |
| 35VB | IGHV3-48 | IGHD1-20 | IGHJ4 | 88.3 |
| 97VB | IGHV3-48 | IGHD5-12 | IGHJ3 | 93.9 |
| 13VB | IGHV3-49 | IGHD2-21 | IGHJ4 | 100 |
| 99VB | IGHV3-53 | ND | ND | 95 |
| 68VB | IGHV3-53 | IGHD3-9 | IGHJ2 | 96.8 |
| 72VB | IGHV3-53 | IGHD6-19 | IGHJ4 | 100 |
| 3VB | IGHV3-64 | IGHD3-10 | IGHJ3 | 100 |
| 8VB | IGHV3-64 | IGHD4-17 | IGHJ2 | 93.9 |
| 48VB | IGHV3-64 | IGHD3-9 | IGHJ5 | 86.8 |
| 105VB | IGHV3-64 | IGHD3-3 | IGHJ3 | 98.1 |
| 1VB | IGHV3-66 | IGHD2-2 | IGHJ6 | 93.7 |
| 87VB | IGHV3-7 | IGHD3-22 | IGHJ3 | 92.6 |
| 63VB | IGHV3-7 | IGHD2-2 | IGHJ5 | 95.7 |
| 66VB | IGHV3-7 | IGHD2-2 | IGHJ5 | 96.9 |
| 29VB | IGHV3-7 | ND | ND | 97 |
| 9VB | IGHV3-73 | IGHD4-4 | IGHJ4 | 92.6 |
| 34VB | IGHV4-30-4 | IGHD3-16 | IGHJ5 | 94.4 |
| 10VB | IGHV4-30-4 | IGHD5-24 | IGHJ4 | 91.1 |
| 16VB | IGHV4-31 | IGHD3-10 | IGHJ5 | 93.3 |
| 102VB | IGHV4-34 | IGHD3-3 | IGHJ4 | 97.7 |
| 108VB | IGHV4-34 | IGHD3-10 | IGHJ4 | 88.1 |
| 57VB | IGHV4-39 | IGHD3-22 | IGHJ4 | 100 |
| 58VB | IGHV4-39 | IGHD3-22 | IGHJ4 | 100 |
| 5VB† | IGHV4-59 | IGHD1-26 | IGHJ2 | 93.5 |
| 101VB | IGHV4-59/-61 | IGHD5-5 | IGHJ1 | 100 |
| 115VB | IGHV4-59/-61 | IGHD3-22 | IGHJ4 | 100 |
| 12VB | IGHV4-59/-61 | IGHD1-26 | IGHJ3 | 100 |
| 15VB | IGHV4-59/-61 | IGHD4-23 | IGHJ4 | 89.4 |
| 17VB | IGHV4-59/-61 | IGHD4-17 | IGHJ6 | 97 |
| 18VB | IGHV4-59/-61 | IGHD4-4 | IGHJ6 | 95.8 |
| 21VB | IGHV4-59/-61 | IGHD5-24 | IGHJ4 | 90.6 |
| 47VB | IGHV4-59/-61 | IGHD2-21 | IGHJ4 | 94.4 |
| 80VB | IGHV4-59/-61 | IGHD5-12 | IGHJ4 | 94.1 |
| 89VB | IGHV4-59/-61 | IGHD2-15 | IGHJ2 | 95 |
| 95VB | IGHV4-59/-61 | IGHD3-3 | IGHJ5 | 100 |
ND indicates not determined.
Percentage of nucleotide identity to germline.
Sequenced with FR1 primer.
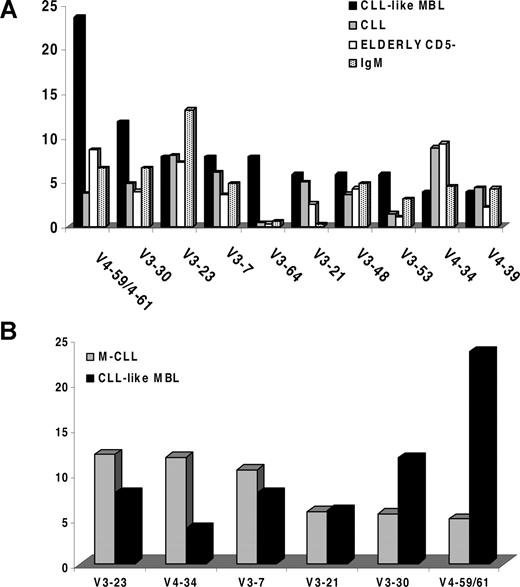
At the individual gene level, IGHV4-59 and IGHV4-61 were by far the most frequent genes (12/51 sequences; 23.5%; Figure 2A). The comparison with 1939 previously published CLL IG sequences18 showed that these 2 genes barely rank among the first 10 genes in the CLL repertoire (X2-test: P < .001) and are also rare in the normal elderly population (Figure 2A-B). In contrast, in comparison to CLL, the CLL-like MBL repertoire was conspicuous for the lack of the IGHV1-69 gene (the most frequently used gene in unmutated CLL; frequency ∼ 25%) and was also characterized by a frequency of the IGHV4-34 gene (2/51 sequences, 3.9%), which is lower than in CLL, where IGHV4-34 is the most frequent gene among mutated cases (frequency ∼ 12%18 ; P < .001), though similar to that of the normal B-cell repertoire (3%-6%).25 PCR amplification of germ line IGHV1-69 and IGHV4-34 sequences in all 51 cases showed that both genes were present in the genome (data not shown).
Frequency of IGHV genes in CLL-like MBL cases. (A) The diagram shows the relative frequency of each IGHV gene in CLL-like MBL cases, CLL patients, CD19+ CD5− lymphocytes in the elderly and IgM+ B lymphocytes, ranked based on the usage in CLL-like MBL. (B) Frequency of IGHV genes in mutated CLL as compared with mutated CLL-like MBL.
Frequency of IGHV genes in CLL-like MBL cases. (A) The diagram shows the relative frequency of each IGHV gene in CLL-like MBL cases, CLL patients, CD19+ CD5− lymphocytes in the elderly and IgM+ B lymphocytes, ranked based on the usage in CLL-like MBL. (B) Frequency of IGHV genes in mutated CLL as compared with mutated CLL-like MBL.
HCDR3 features and stereotypy in CLL-like MBL
Analysis of the HCDR3 region was possible in 44 of 51 rearrangements (Table 2). To explore HCDR3 sequence homology (“stereotypy”), we performed cluster analysis of MBL HCDR3 sequences. Though the size of the MBL cohort was admittedly small, MBL cases did not usually share similar or identical HCDR3s, in contrast to CLL which is characterized by more than 20% HCDR3-stereotyped cases.18,23 However, we identified 2 MBL cases with an HCDR3 similar to that previously described in CLL cases.18,23 Both cases had a normal lymphocyte count (2.46 × 109/L and 2.82 × 109/L, respectively). One of the 2 cases belongs to the large family reported in Table 3. Intriguingly, one of these MBL cases, expressing the IGHV4–59 gene, had an HCDR3 very similar to the stereotyped HCDR3s of 4 CLL cases expressing mutated IGHV4-59 IG, 2 originating from France, 1 from Greece, and 1 from Italy (subsets 1323 ). These sequences are remarkably similar to that of a mutated rheumatoid factor from a healthy donor immunized with mismatched red blood cells (U85234)26 as well as to the sequence carried by a hepatitis C virus–infected male patient with IGHV4-59–expressing CLL/SLL developing in a setting of type II cryoglobulinemia (AF303917).24 Our MBL case was also positive for the presence of serum antibodies against HCV.
IGHV gene usage by CLL-like MBL cases in the same family
| Family 1 . | Family 2 . | Family 3 . | Family 4 . | Family 5 . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . |
| XX016077 | ND | XX140361 | IGHV4-39 | XX771989 | IGHV3-23 | XX881669 | IGHV3-49 | XX622852 | Polyclonal |
| XX932341 | IGHV4-59 | XX180728 | ND | XX320954 | IGHV3-64 | XX531948 | IGHV3-23 | XX448277 | ND |
| XX275343 | IGHV3-53 | XX199142 | IGHV3-64 | XX832798 | ND | XX416415 | IGHV4-59 | XX939822 | IGHV3-30 |
| XX707818 | ND | XX331890 | ND | XX081769 | IGHV3-64 | ||||
| XX169450 | ND | XX846461 | IGHV3-30 | ||||||
| XX648253 | IGHV3-73 | ||||||||
| XX509891 | Polyclonal | ||||||||
| XX976825 | IGHV3-30 | ||||||||
| XX291337 | IGHV3-53 | ||||||||
| XX175926 | IGHV3-21 | ||||||||
| Family 1 . | Family 2 . | Family 3 . | Family 4 . | Family 5 . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . | Case no. . | Gene . |
| XX016077 | ND | XX140361 | IGHV4-39 | XX771989 | IGHV3-23 | XX881669 | IGHV3-49 | XX622852 | Polyclonal |
| XX932341 | IGHV4-59 | XX180728 | ND | XX320954 | IGHV3-64 | XX531948 | IGHV3-23 | XX448277 | ND |
| XX275343 | IGHV3-53 | XX199142 | IGHV3-64 | XX832798 | ND | XX416415 | IGHV4-59 | XX939822 | IGHV3-30 |
| XX707818 | ND | XX331890 | ND | XX081769 | IGHV3-64 | ||||
| XX169450 | ND | XX846461 | IGHV3-30 | ||||||
| XX648253 | IGHV3-73 | ||||||||
| XX509891 | Polyclonal | ||||||||
| XX976825 | IGHV3-30 | ||||||||
| XX291337 | IGHV3-53 | ||||||||
| XX175926 | IGHV3-21 | ||||||||
ND indicates not done.
Family analysis of CLL-like MBL
Pedigree reconstruction from all CLL-like MBL cases showed that 25 cases (28%) were clustered in 5 main families (Table 3). The largest family entails 7 generations and includes 10 members with CLL-like MBL, with a mean kinship coefficient of 0.0322. These results suggest the existence of a genetic component of the predisposition to develop MBL.
Analysis of the IGHV rearrangements expressed by MB lymphocytes present in members of the same family demonstrated that they do not share the same rearrangement (Table 3), thereby suggesting that the relevant familial/genetic component of the predisposition to develop MBL is not genetically determined by the type of IGHV rearrangement.
Discussion
We describe the occurrence, frequency, and molecular characterization of the MB lymphocytes detected in a large general population including individuals from 18 to over 100 years old living in a rural community in Northern Italy. In the past, MBL has been studied in the general populations as part of environmental health investigations3,27 or blood donor surveys.4 Mainly due to technical limits, these studies showed a prevalence largely less than 1%. More recently, thanks to a multicolor flow cytometric approach which allowed us to detect MBL as low as a few cells per microliter, a strikingly higher frequency of MBL (3.5%) has been detected in the blood of otherwise healthy individuals in both the United Kingdom6,10 and Italy.5 However, a potential drawback of these studies is that they were conducted on either in-patient or out-patient cohorts (ie, on subjects under medical attention for several, though nonneoplastic, health problems). For all these reasons, our present study can be considered as the first report of MBL in a general population who voluntarily participated in a study not aimed at elucidating any particular health problem. For this reason, we were able to collect a large number of cases allowing molecular studies in a meaningful number of MBL cases, especially those presenting with low-count monoclonal B cells.
The population we studied exhibits a high degree of genetic homogeneity (ie, it is genetically isolated).14 As shown in other populations of the same kind,28,29 the findings obtained in such populations are usually replicated in the general population. Accordingly, analyses of several clinical, pathologic, and laboratory parameters in the Val Borbera population demonstrated frequencies or sex prevalence similar to the general population in Northern Italy.13 Therefore, this type of population provides a unique opportunity to identify potential familial linkages that might be helpful to define the genetic components of the traits of interest (eg, the genetic component of the predisposition to develop MBL). Along this line, it was interesting to note that a sizeable number of CLL-like MBL cases (> 25%) clustered in families, thereby indicating a potentially strong familial predisposition to develop MBL, confirmed by the high mean kinship coefficient of the largest family. Our findings are in keeping with a previous study reporting a higher frequency of MBL among the relatives of patients with CLL.30 Interestingly, the IGHV-D-J rearrangements expressed by the monoclonal B lymphocytes present in members of the same family were different (Table 3), implying that the development of MBL, though having a strong familial/genetic component, is not genetically determined by the type of IG rearrangement (ie, it is independent of the IGHV gene repertoire). As genealogic data collection is still ongoing, the possibility exists that more cases will be further clustered in fewer families that at present do not appear to be linked because of lack of genealogic data. Linkage analysis may allow us to identify the potential genetic loci involved. Still, the fact that most MBL cases are actually sporadic cases indicates the importance of other (eg, environmental) events that might be involved in the onset of MBL.
Our results further support the notion that MBL is frequent in the general population and also phenotypically heterogeneous, with CLL-like MBL accounting for most cases. Compared with previous studies, the higher frequency reported here (7.2%) is likely due a higher sensitivity of the applied flow cytometry technique, as we analyzed a larger amount of events for each sample (500 000 vs 200 000 in the previous studies5,6 ) and used a 5-color protocol (vs 4-color).
CLL-like MBL is virtually nonexistent below the age of 40 years, while its frequency is almost 9% in the elderly. In clear contrast, the other types of MBL (CD5− and atypical CLL MBL) are already present in sizeable proportions among young individuals and their frequency is only relatively affected by aging (Table 1). These observations are also in keeping with the hypothesis that CLL-like MBL might be related to a process of physiologic senescence of the immune system, while this possibility does not appear to be applicable to the other types of MBL, which might be accounted for by likely self-limiting immune stimulations occurring randomly during one's lifetime. Long-term follow-up will help to define the persistence of these entities.
An intriguing observation of this study is the finding of few seemingly polyclonal CLL-like MBL cases (ie, cases that are not, strictly speaking, MBL; Figure 1). When a consensus on the term MBL was reached,7 it was believed that the distinct CD5brightCD20dim phenotype could be present exclusively on the surface of monoclonal CD19+ B cells. Our finding creates a semantic problem (polyclonal CD5+ B-cell expansion?) that needs to be properly addressed when confirmed in larger and different series. Even more importantly, it leads us to hypothesize that the acquisition of a CLL-like surface phenotype does not necessarily go along with the appearance of monoclonality, but it might reflect a functional state possibly due to prolonged/chronic B-cell stimulation and activation.
Along this line of reasoning, it is also interesting to note that the vast majority of CLL-like MBL observed in this population is limited in terms of number of monoclonal cells, with only 1 case having more than 1 × 103 but less than 2 × 103/L B cells circulating in the blood and all the remaining ones carrying less than or equal to 0.60/L CD19+ cells. Monoclonal B cells were actually a minority of the CD19+ lymphocytes, being more than 10% of all B lymphocytes in only 13 of 89 cases. This is somehow expected as we analyzed a healthy population of individuals, not followed for any particular disease. This provided us with the opportunity to pay particular attention to low-count CLL-like MBL.
It has been reported recently that CLL requiring treatment developed at a rate of 1.1% per year mainly in subjects with CLL-like MBL but with lymphocytosis; interestingly, the absolute B-cell count was the only independent prognostic factor associated with progressive lymphocytosis.10 These MBL cases had molecular features similar to good prognosis CLL in terms of immunoglobulin (IG) gene features and chromosomal abnormalities.10 These findings underscore the fact that the actual B-cell number has clinical implications. Though the numerical value may simply reflect a different duration time of each MBL at the time of detection, it is also possible that it may depend, at least in a portion of cases, from different biologic features of MBL with low amount of circulating B cells, that could make them less prone to expand and to evolve into a frank leukemia. The study of low-count MBL at molecular level has been so far hampered by the lack of real general population studies and the technical difficulties in analyzing such limited amount of cells.
In this study, the analysis of the IGHV genes expressed by low-count CLL-like MBL revealed a repertoire that is strikingly different from the typical CLL repertoire, in terms of gene usage (eg, absence of the IGHV1-69 gene and a lower frequency of the IGHV4-34 gene; overrepresentation of the IGHV4-59/61 genes),23 though the missing IGHV genes are present in the genome of our population (data not shown). Interestingly, though the majority of CLL-like MBL cases (70%) carry somatic mutations on the expressed IGHV genes, unmutated cases do exist in a sizeable fraction, accounting for almost one-third of cases, though at a lower frequency as compared with CLL (40%-50%).18 One might suggest that the differences in terms of IGHV repertoire shown in the present work are merely due to the increased number of mutated cases among MBL that may create a bias in the expressed repertoire. This would imply that the MBL repertoire is similar to that of mutated CLL rather than different from all CLL. Though plausible, this hypothesis is not supported by the data here presented where the genes most frequently used by CLL-like MBL (ie, IGHV4-59/61) are only rarely expressed by mutated CLL. Conversely, the most frequently used IGHV gene by mutated CLL (ie, IGHV4-34) is expressed at a lower frequency in our MBL series (Figure 2B), though similar to that of the normal B-cell repertoire (3%-6%25 ).
In addition, though the IGHV4-59/61 genes have been reported to be significantly increased in older individuals versus young adults,22 overall, the IGHV repertoire in MBL appears rather different also from the repertoire of normal IgM+ B cells in the elderly. All the above indicate a potential bias for the IGHV4-59/4-61 genes in MBL populations, which may suggest the existence of a latent or chronic antigenic element stimulating B lymphocytes over time. Nevertheless, the results of the comparison between the CLL-like MBL cohort described herein and healthy elderly individuals should be interpreted with caution, given the lack of a truly representative “normal” database that could be used for reference and the potential biases due to the donor sources, the cloning techniques, or the genetic material used.
Finally, it should be noted that MBL cases very rarely share similar HCDR3s sequences, in sharp contrast with the observed greater than 20% frequency of stereotyped HCDR3s in CLL.18,23 This result could be taken as confirmatory of the idea that MBL cells do not necessarily represent a preleukemic condition. However, the finding that the HCDR3s of 2 CLL-like MBL cases were highly similar to those expressed in patients with CLL from our previously published cohort,23 suggests that the potential to become an overt CLL may indeed be present, though at low frequency, also within low count MBL cells. This observation drives the attention to the specific IG receptor expressed as a sign of a potential predisposition to develop into a frank leukemia. This suggests that a distinct capacity to respond to particular (antigenic) signals might confer the cell the property of being more prone to evolve toward a transformed leukemic phenotype. A thorough molecular analysis and further characterization of IG stereotypy in selected well-defined MBL may help in finding a clue to identify which cases necessitate continuous monitoring, because they are at higher risk of evolving into a clinically relevant disease, and to avoid lengthy and expensive follow-ups in the vast number of persons who are extremely unlikely to develop CLL simply because they have MBL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, Milano, Italy), CLL Global Research Foundation, Progetti di Ricerca di Interesse Nazionale (PRIN), Fondo per gli Investimenti della Ricerca di Base (FIRB), Progetto Integrato Oncologia (PIO), Compagnia di San Paolo, Torino, Italy, Fondazione Ferrero, Alba, Italy, and Fondazione Anna Villa e Felice Rusconi, Varese, Italy.
Authorship
Contribution: A.D. performed IGHV-D-J molecular analysis; C.F. and V.C. performed flow-cytometric analyses; C.S. performed the cloning experiments; R.M. contributed vital reagents; C.S. and D.T. performed the pedigree reconstruction; F.C.C. supervised analysis and wrote the paper; K.S. performed the bio-informatic analyses; and P.G. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Federico Caligaris-Cappio, Department of Oncology, Istituto Scientifico San Raffaele, Via Olgettina 60, 20132, Milano, Italy; e-mail: caligaris.federico@hsr.it.
References
Author notes
*A.D. and C.F. share first authorship of this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal