Abstract
Adoptive transfer of T cells expressing transgenic T-cell receptors (TCRs) with antitumor function is a hopeful new therapy for patients with advanced tumors; however, there is a critical bottleneck in identifying high-affinity TCR specificities needed to treat different malignancies. We have developed a strategy using autologous dendritic cells cotransfected with RNA encoding an allogeneic major histocompatibility complex molecule and a tumor-associated antigen to obtain allo-restricted peptide-specific T cells having superior capacity to recognize tumor cells and higher functional avidity. This approach provides maximum flexibility because any major histocompatibility complex molecule and any tumor-associated antigen can be combined in the dendritic cells used for priming of autologous T cells. TCRs of allo-restricted T cells, when expressed as transgenes in activated peripheral blood lymphocytes, transferred superior function compared with self-restricted TCR. This approach allows high-avidity T cells and TCR specific for tumor-associated self-peptides to be easily obtained for direct adoptive T-cell therapy or for isolation of therapeutic transgenic TCR sequences.
Introduction
T-cell responses against tumors are often directed against self-major histocompatibility complex (MHC) molecules presenting peptides derived from overexpressed self-proteins. In general, high-avidity T cells specific for self-peptide/self-MHC ligands are eliminated by negative selection to prevent autoimmunity. The T-cell receptor (TCR) affinity of remaining T cells specific for self-ligands is normally low; however, high-avidity T cells are needed to effectively eradicate tumors. Because negative selection is limited to self-MHC molecules, T cells that recognize allogeneic MHC molecules have not undergone negative selection. Thus, peptides presented by allogeneic MHC molecules can stimulate high-avidity T cells specific for tumor-associated ligands derived from overexpressed self-proteins.1 T cells that recognize allogeneic MHC molecules irrespective of specific peptide can be distinguished from peptide-specific T cells at the clonal level and excluded.2
Several approaches have been used to obtain allo-restricted peptide-specific cytotoxic T lymphocytes (CTLs). For example, peptide-loaded T2 cells were used to generate HLA-A2 allo-restricted T cells.3,4 This system is normally limited to HLA-A2 and CTLs induced with exogenous peptides often fail to effectively kill tumor cells.5 Activated B cells coated with allogeneic peptide-MHC (pMHC) monomers were also used to prime allo-restricted CD8+ T cells.6 For broad application, this method would require development of many different pMHC monomers. Furthermore, allo-restricted peptide-specific T cells were obtained using T cells and stimulating cells derived from HLA partial-mismatched donors.7 Here, necessary donor pairs that differ by single HLA allotypes are rare. Therefore, each of these methods has strong limitations.
We developed an approach that provides maximum flexibility to obtain allo-restricted peptide-specific T cells using RNA-transfected dendritic cells (DCs) as stimulating cells. The use of DCs as antigen-presenting cells is particularly valuable because of their superior capacity to prime naive lymphocytes. In this approach, T cells are stimulated with autologous DCs cotransfected with RNA that encode an allogeneic MHC molecule and a tumor-associated antigen (TAA), bypassing the need to search for appropriate HLA-mismatched donors for the DC source. These DCs coexpress both proteins and present allo-pMHC complexes that activate allo-restricted peptide-specific T cells. In this approach, high-avidity T cells that recognize self-peptides are more readily found because they were not subjected to negative selection in vivo.
Selected allogeneic high-avidity T cells can be used directly for adoptive T-cell therapy in HLA-haploidentical stem cell transplantation.8 Alternatively, their TCR sequences can be used to develop designer lymphocytes.4,9,10 Thereby, autologous T cells from tumor patients in general, or from MHC-matched donors in the case of stem cell transplantation, can be used for TCR transduction, bypassing the need for direct transfer of allogeneic T cells. Improved ability to isolate T cells with high-affinity TCR recognizing common tumor-associated ligands will allow quicker development of this therapy for more patients.
Methods
Cell lines and effector CTLs
The human melanoma cell lines, Mel-A375 (HLA-A2+, tyrosinase−; CRL-1619, ATCC), Mel-93.04A12 (HLA-A2+, tyrosinase+; gift of P. Schrier, Department of Immunohematology, Leiden University Hospital, Leiden, The Netherlands), Mel-624.3811 and SK-Mel-23 (HLA-A2+, tyrosinase+; gift of M. C. Panelli, National Institutes of Health, Bethesda, MD), SK-Mel-28 (HLA-A2−, tyrosinase+; MTB-72; ATCC), SK-Mel-29 (HLA-A2+, tyrosinase+, gift of P. Rieber, Institute of Immunology, Technical University Dresden, Dresden, Germany), WM-266-4 (HLA-A2+, tyrosinase+; CRL-1676; ATCC) and primary cultures of a human melanoma (passage 6-12) and MaCa1 (HLA-A2−, tyrosinase−, gift of Dr R. Wank, Immunotherapy Center, Munich, Germany), stable HLA-A*0201 transfectant of MaCa1 (MaCa1/A2; HLA-A2+, tyrosinase−, gift of E. Noessner, Institute of Molecular Immunology, Helmholtz Zentrum München, Munich, Germany), RCC-2612 (HLA-A2+, tyrosinase−), PancTu1 (HLA-A2+, tyrosinase−, gift of P. Nelson, Department for Biological Chemistry, University Hospital LMU Munich, Munich, Germany), UTS CC 1588 (HLA-A2+, tyrosinase−, gift of M. Schmitz, Institute of Immunology, Technical University Dresden, Dresden, Germany) as well as the lymphoid cell line T2 (CRL-1992; ATCC) were cultured in RPMI 1640 medium supplemented with 12% fetal bovine serum, 2 mM l-glutamine, and 1 mM sodium pyruvate and nonessential amino acids.
The HLA-A2 allo-reactive CTL JB4,13 the HLA-A*0201–restricted tyrosinase peptide-specific CTL Tyr-F8,14 which were primed using stimulating cells pulsed with exogenous peptide (gift of P. Schrier, Department of Immunohematology, Leiden University Hospital, Leiden, The Netherlands), the HLA-A*0201–restricted tyrosinase peptide-specific, melanoma patient–derived IVS-B15 CTLs (gift of T. Wölfel, Third Department of Medicine, Hematology, and Oncology, Johannes Gutenberg-University of Mainz, Mainz, Germany), and the HLA-A*0201–restricted melan-A peptide–specific A4211 CTLs (gift of M. C. Panelli) were cultured as described.11,13-15
Production of tyrosinase, melan-A, and HLA-A2 ivt-RNA
The plasmid pCDM8-HLA-A2 with HLA-A*0201 cDNA (gift of E. Weiß, Department of Biology II, Ludwig-Maximilians-University, Munich, Germany), pZeoSV2+/huTyr with tyrosinase cDNA (gift of I. Drexler, Institute of Molecular Virology, Helmholtz Zentrum München, Munich, Germany), and pcDNAI/Amp/Aa1 with melan-A cDNA (gift of T. Wölfel) were linearized and used as in vitro transcription templates to produce RNA with the aid of the mMESSAGE mMACHINE T7 kit (Ambion) according to the manufacturer's instructions.
Fluorescence-activated cell sorter analysis
HLA-A2 molecules were stained with BB7.2 monoclonal antibody (HB82; ATCC) followed by a secondary antibody conjugated with phycoerythrin (PE; goat anti–mouse IgG; Jackson ImmunoResearch Laboratories). The intracellular tyrosinase expression in RNA-transfected DCs was detected using tyrosinase-specific primary monoclonal antibody (clone T311; Novocastra Laboratories) and Cy5-conjugated secondary antibody (rat anti–mouse IgG; Jackson ImmunoResearch Laboratories) as described.16
De novo priming of T cells with RNA-pulsed DCs
Blood samples from healthy donors were collected with donors' informed consent in accordance with the Declaration of Helsinki and after approval by the Institutional Review Board of the University Hospital of the Ludwig-Maximilians-University. Donor HLA types are listed in Table 1. Mature DCs were prepared from adherent monocytes and transfected with in vitro transcribed (ivt)–RNA via electroporation as previously described.16 DCs of HLA-A2+ donors were loaded with 24 μg tyrosinase ivt-RNA, and DCs of HLA-A2− donors were cotransfected with 24 μg tyrosinase ivt-RNA and 48 μg HLA-A2 ivt-RNA. On the same day, autologous CD8+ T lymphocytes were enriched from peripheral blood mononuclear cells (PBMCs) via negative selection using a commercial kit according to the manufacturer's instructions (CD8+ T Cell Isolation Kit II [human]; Miltenyi Biotec). Cocultures were initiated 10 hours after DC electroporation in 24-well plates (Techno Plastic Products) by adding 105 RNA-pulsed DCs to 106 CD8+ T cells in RPMI 1640, supplemented with 10% heat-inactivated human serum, 4 mM l-glutamine, 12.5 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 50 μM β-mercaptoethanol, and 100 U/mL penicillin/streptomycin (T-cell medium). Interleukin-7 (IL-7, 5 ng/mL; Promokine) was added on day 0 and 50 U/mL IL-2 (Chiron Behring) after 2 days and then on every third subsequent day. Addition of IL-2 was delayed to decrease proliferation of nonspecific CD8+ T cells.17 The second stimulation of primed T cells was made after 7 days using freshly prepared RNA-pulsed DCs.
HLA types of donors used in these studies
| Donor . | Antigen . | HLA types . |
|---|---|---|
| A | Tyr | HLA-A*01/*02, -B*08/*44, -Cw*05/*07 |
| D | Tyr | HLA-A*02/*03, -B*35/*37, -Cw*04/*06 |
| T | Tyr | HLA-A*01/*26, -B*07/*57, -Cw*06/*07 |
| E | Tyr | HLA-A*24/*26, -B*07/*44, -Cw*07/*16 |
| Donor . | Antigen . | HLA types . |
|---|---|---|
| A | Tyr | HLA-A*01/*02, -B*08/*44, -Cw*05/*07 |
| D | Tyr | HLA-A*02/*03, -B*35/*37, -Cw*04/*06 |
| T | Tyr | HLA-A*01/*26, -B*07/*57, -Cw*06/*07 |
| E | Tyr | HLA-A*24/*26, -B*07/*44, -Cw*07/*16 |
HLA multimer sorting
Seven days after the second stimulation of CD8-enriched T cells with RNA-pulsed DCs, HLA-A2–restricted tyrosinase-specific T cells were stained with a PE-labeled HLA-A2 tyrosinase369-377 (A2-tyr) multimer,18 CD8-specific antibody (clone RPA-T8; BD Pharmingen) and propidium iodide (2 μg/mL) for sorting. Up to 5 × 106 cells were incubated with 12 μg multimer in 100 μL phosphate-buffered saline plus 0.5% human serum. Allophycocyanin-labeled, CD8-specific antibody was then added for an additional 25 minutes. After staining, cells were washed and sorted on a FACSAria cell sorter (BD Biosciences) as described.19
Culture of peptide-specific CTLs
Multimer-sorted T cells were cloned by limiting dilution in round-bottom 96-well plates (TPP). IL-2 (50 IU/mL) was supplemented every 3 days with 5 ng/mL IL-7 and 10 ng/mL IL-15 (PeproTech) every 7 days. CTLs were stimulated nonspecifically with CD3-specific antibody (0.1 μg/mL; OKT-3; gift of E. Kremmer, Institute of Molecular Immunology, Helmholtz Zentrum München, Munich, Germany) and provided with 105 feeder cells per 96-well, consisting of irradiated (50 Gy) PBMCs derived from a pool of 5 unrelated donors and 104 irradiated (150 Gy) Epstein-Barr virus–transformed allogeneic B-lymphoblastoid cell line every 2 weeks. Proliferating T cells were cultured in 24-well plates (TPP) in T-cell medium plus cytokines and feeder cells. Clonality was determined by TCR–β-chain receptor analysis, as described.20
Peptide loading of T2 cells, PBMCs, and tumor cells
For exogenous peptide pulsing, 106 T2 cells were incubated at 37°C and 5% CO2 for 2 hours with 10 μg/mL human β2-microglobulin (Calbiochem) and titrating amounts, ranging from 10−5 M to 10−11 M, of the tyrosinase peptide YMD (tyrosinase369-377 YMDGTMSQV; Metabion). T2 cells pulsed with 10−5 M influenza peptide GIL (flu: influenza matrix protein58-66 GILGFVFTL; Metabion) served as the negative control. PBMCs were loaded with tyrosinase peptide as for T2 cells with titrating amounts ranging from 10−6 to 10−11 M. Tumor cells were loaded with either 10−5 M flu peptide or 10−5 M tyrosinase peptide YMD as described for T2 cells. After washing, peptide-loaded T2 cells, PBMCs, or tumor cells were used as target cells in cytotoxicity or as stimulating cells in interferon-γ (IFN-γ) release assays.
IFN-γ release assay
To demonstrate the stimulatory capacity of RNA-pulsed DC 10 hours after electroporation, 100 μL of JB4, Tyr-F8, IVS-B, or A42 T-cell suspensions (2 × 104 cells in 100 μL) was added to RNA-loaded DCs (4 × 104 cells in 100 μL) in round-bottom 96-well plates. T cells with mock-transfected DCs and without stimulator cells served as controls and showed negligible IFN-γ secretion.
For investigation of specificity, CTLs (2 × 103 cells in 100 μL) were incubated with various tumor cell lines (104 cells in 100 μL), with or without peptide pulsing, as described in “Peptide loading of T2 cells, PBMCs, and tumor cells.” Culture supernatants were harvested after 24 hours of coculture and assessed by a standard enzyme-linked immunosorbent assay (ELISA) using the OptEIA Human IFN-γ Set (BD Biosciences). Data represent mean values with corresponding mean deviations calculated from duplicate determinations. For the calculation of percentage relative IFN-γ release, the maximum IFN-γ release was set to the reference value of 100% and corresponding values were calculated corresponding to this reference.
Cytotoxicity assay
Cytotoxic activity of CTLs was analyzed in a standard 4-hour chromium release assay. Melanoma cell lines and peptide-loaded T2 cells were used as target cells. T cells were cocultured with 103 melanoma target cells/well at effector cell to target cell (E:T) ratios of 5:1 or 10:1. For determination of functional avidity, 104 T cells were added to 103 peptide-pulsed T2 cells loaded with titrated amounts of peptide, giving a constant E:T of 10:1.
The percentage of specific lysis was calculated as: 100 × (experimental release − spontaneous release)/(maximum release − spontaneous release). Spontaneous release was assessed by incubating target cells in the absence of effector cells and was generally less than 15%. For the calculation of percentage relative lysis, the maximum percentage specific lysis was set to the reference value of 100%, and corresponding values were calculated corresponding to this reference. To determine half-maximum lysis, percentage relative lysis was plotted against peptide concentration. The peptide concentration at which the curve crossed 50% relative lysis was taken as the value of half-maximum lysis.21
Retroviral TCR gene transfer
For TCR identification of tumor-specific CTLs, regions of the TCR–α- and TCR–β-chains encoding CDR3 were amplified by polymerase chain reaction using a panel of TCRV-α and TCRV-β primers in combination with respective constant region primers as described.22 The full TCR–α- and TCR–β-chain genes of CTL clones T58 and D115 were amplified by polymerase chain reaction using cDNA as template. Primer sequences will be provided on request. The constant regions of both TCR chains were exchanged by the murine counterparts to increase the stability of the TCR.23 The TCR chains were linked by a 2A peptide linker (TCR-β-P2A-TCR-α),24 codon-optimized (Geneart),25 and cloned into the retroviral vector MP71PRE via NotI and EcoRI restriction sites.24 Retroviral vector plasmids were cotransfected into 293T cells with expression plasmids encoding Moloney MLV gag/pol and MLV-10A1 env gene, respectively, to produce amphotropic MLV-pseudotyped retroviruses as described.24 Ten days after the second transduction, peripheral blood lymphocytes (PBLs) were stained using PE-labeled A2-tyr multimer and fluorescein isothiocyanate-labeled CD8-specific antibody. Multimers presenting peptides derived from cytomegalovirus pp65 were used as controls: PE-labeled HLA-B7 pp65417-427 (B7-pp65) multimers served as the HLA control, and HLA-A2 pp65495-503 multimers as a peptide-specificity control. On day 15, an IFN-γ release assay was performed using T2 cells or autologous PBMCs loaded with graded amounts of tyrosinase peptide (10−12 M to 10−5 M) and the tumor cell lines MaCa1, SK-Mel-28, Mel-A375, RCC-26, PancTu 1, MaCa1/A2, UTS CC 1588, Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 as stimulating cells at an E:T of 2:1.
Statistical analysis
The nonparametric, 2-tailed Mann-Whitney U test was used to evaluate statistical differences between datasets derived from self- and allo-restricted CTLs in cytotoxicity and IFN-γ release assays. For statistical analysis of differences between paired datasets obtained from either self- or allo-restricted CTLs in cytotoxicity and IFN-γ release assays, the nonparametric, 2-tailed Wilcoxon signed rank test was used. Values of P less than .05 were considered statistically significant. All analyses were performed using the SPSS statistical software (Version 15.0; SPSS Inc).
Results
Activation of CD8+ T cells with RNA-pulsed DCs
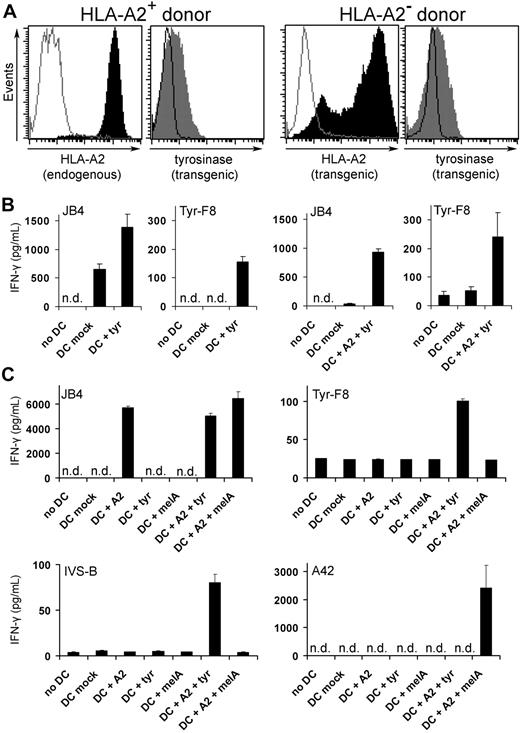
The first step in generating CTLs was the preparation of stimulating DCs that coexpressed allogeneic HLA molecules and a selected TAA. For proof of principle, mature DCs were loaded with ivt-RNA for HLA-A2 and tyrosinase via electroporation, as described.16 Tyrosinase ivt-RNA was transferred alone into HLA-A2+ DCs or in combination with HLA-A2 ivt-RNA into HLA-A2− DCs. Protein coexpression was detected by flow cytometry with specific monoclonal antibodies (Figure 1A). Uniform surface expression of endogenous HLA-A2 molecules was detected on DCs of HLA-A2+ donors, whereas transgenic HLA-A2 expression was variable on DCs of HLA-A2− donors, with some strongly positive DCs. Both DC populations coexpressed intracellular tyrosinase protein at comparable levels. Such DCs were tested for their capacity to stimulate effector CTL to secrete IFN-γ: JB4 cells recognize allogeneic HLA-A2 molecules irrespective of specific peptide,13 whereas Tyr-F8 cells recognize tyrosinase369-377 peptide presented by HLA-A2.14 JB4 and Tyr-F8 cells released IFN-γ in an HLA-A2–dependent manner because mock-transfected DCs of HLA-A2+ but not HLA-A2− donors activated JB4, whereas Tyr-F8 only recognized DCs expressing both HLA-A2 and tyrosinase (Figure 1B). Further analysis of CTL activation by RNA-pulsed DCs loaded with HLA-A2 and a selected TAA (tyrosinase or melan-A) is depicted in Figure 1C. JB4 recognized DCs that were loaded with HLA-A2 RNA alone and they also could respond to DC coexpressing either tyrosinase or melan-A RNA, demonstrating their allo-specificity for HLA-A2. In contrast, Tyr-F8 and IVS-B T cells, both of which recognize HLA-A2-tyrosinase369-377 peptide complexes, only responded specifically to DCs coexpressing HLA-A2 and tyrosinase RNA. Antigen specificity was demonstrated with A42 cells, which recognize a melan-A–derived peptide presented by HLA-A2; these CTLs only responded to DCs coexpressing the 2 corresponding RNA.
De novo priming of CD8+ T cells with RNA-pulsed DCs. (A) Coexpression of HLA-A2 and tyrosinase proteins was detected by flow cytometry in DCs of an HLA-A2+ donor transfected with 24 μg tyrosinase ivt-RNA (left histograms) and DCs of an HLA-A2− donor transfected with 48 μg HLA-A2 and 24 μg tyrosinase ivt-RNA (right histograms). Stained samples are represented by filled curves and corresponding controls by empty curves. Data are representative of 9 independent experiments, demonstrating that all DCs used for T-cell priming coexpressed both proteins. (B) Columns represent the amount of IFN-γ (picograms per milliliter) secreted by a tyrosinase-independent HLA-A2 allo-reactive T-cell clone (JB4) and an HLA-A2–restricted tyrosinase peptide–specific T-cell clone (Tyr-F8) after coincubation with RNA-pulsed DCs, 10 hours after electroporation. IFN-γ was quantified in culture supernatants by ELISA. Mean values and mean deviations represent duplicates. n.d. indicates not detectable. (C) DCs of an HLA-A2− donor were transfected with HLA-A2 (48 μg) and tyrosinase (24 μg) or melan-A (48 μg) ivt-RNA alone and in combination and used for coincubation with the HLA-A2 allo-reactive CTL clone (JB4), 2 HLA-A2–restricted tyrosinase peptide–specific CTL clones (Tyr-F8 and IVS-B), and an HLA-A2–restricted melan-A peptide–specific CTL clone (A42) 10 hours after electroporation. IFN-γ was quantified in culture supernatants by ELISA and presented as picograms per milliliter. Mean values and mean deviations represent duplicates. n.d. indicates not detectable.
De novo priming of CD8+ T cells with RNA-pulsed DCs. (A) Coexpression of HLA-A2 and tyrosinase proteins was detected by flow cytometry in DCs of an HLA-A2+ donor transfected with 24 μg tyrosinase ivt-RNA (left histograms) and DCs of an HLA-A2− donor transfected with 48 μg HLA-A2 and 24 μg tyrosinase ivt-RNA (right histograms). Stained samples are represented by filled curves and corresponding controls by empty curves. Data are representative of 9 independent experiments, demonstrating that all DCs used for T-cell priming coexpressed both proteins. (B) Columns represent the amount of IFN-γ (picograms per milliliter) secreted by a tyrosinase-independent HLA-A2 allo-reactive T-cell clone (JB4) and an HLA-A2–restricted tyrosinase peptide–specific T-cell clone (Tyr-F8) after coincubation with RNA-pulsed DCs, 10 hours after electroporation. IFN-γ was quantified in culture supernatants by ELISA. Mean values and mean deviations represent duplicates. n.d. indicates not detectable. (C) DCs of an HLA-A2− donor were transfected with HLA-A2 (48 μg) and tyrosinase (24 μg) or melan-A (48 μg) ivt-RNA alone and in combination and used for coincubation with the HLA-A2 allo-reactive CTL clone (JB4), 2 HLA-A2–restricted tyrosinase peptide–specific CTL clones (Tyr-F8 and IVS-B), and an HLA-A2–restricted melan-A peptide–specific CTL clone (A42) 10 hours after electroporation. IFN-γ was quantified in culture supernatants by ELISA and presented as picograms per milliliter. Mean values and mean deviations represent duplicates. n.d. indicates not detectable.
The DCs shown in Figure 1A-B were used to prime purified autologous CD8+ T cells using 2 rounds of stimulation with freshly prepared DCs. Peptide-specific T cells were sorted using specific HLA-A2 tyrosinase369-377 multimer (A2-tyr multimer),26 cloned in limiting dilution cultures and expanded using antigen-independent stimulation.17
Superior function of HLA-A2 allo-restricted T cells
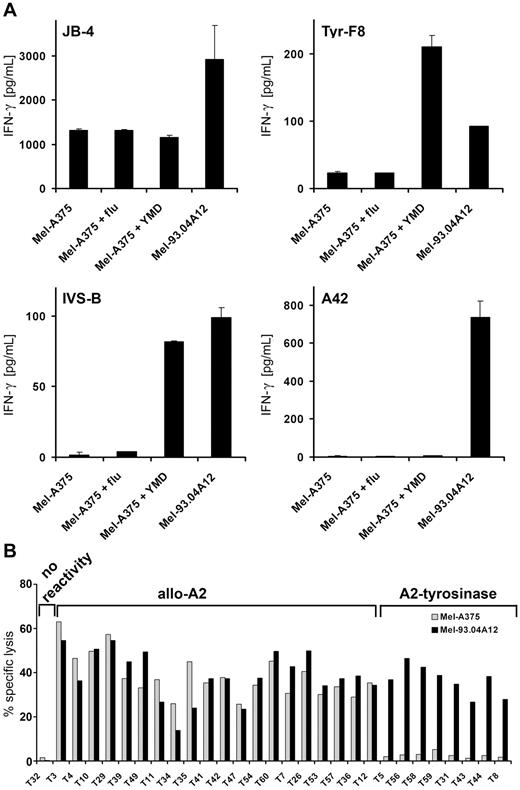
Isolated clones were analyzed in IFN-γ release and cytotoxicity assays for function and specificity. Initial screening was done using the Mel-A375 (HLA-A2+ tyrosinase−) and Mel-93.04A12 (HLA-A2+ tyrosinase+) tumor cell lines as negative and positive controls, respectively. The capacity of these tumor lines to specifically stimulate JB4, Tyr-F8, IVS-B, and A42 is shown in Figure 2A, demonstrating that both lines can be recognized by HLA-A2 allo-specific CTL (ie, JB4 cells). However, Mel-A375 cells could not activate Tyr-F8 and IVS-B CTL unless they were pulsed with tyrosinase peptide, whereas Mel-93.04A12 cells, which expressed both endogenous tyrosinase and melan-A proteins, could activate Tyr-F8, IVS-B, and A42 directly. Screening results for exemplary clones primed against DCs derived from an HLA-A2− donor are shown in Figure 2B, depicting either tyrosinase-independent allo-reactivity for HLA-A2 or presumed specificity for HLA-A2-tyrosinase–derived ligands. In total, 38 self-restricted clones from 2 HLA-A2+ donors were screened: 21 clones had no reactivity, no clones recognized HLA-A2 without tyrosinase peptide, and 17 clones recognized HLA-A2-tyrosinase-peptide ligands (Table 2). Of 51 clones isolated from 2 HLA-A2− donors, 8 were nonreactive; 27 were allo-reactive, recognizing HLA-A2 independent of tyrosinase, and 16 were specific for HLA-A2-tyrosinase ligands.
Recognition of peptide-loaded Mel-A375 and exemplary screening data from the DC priming. (A) HLA-A2+tyrosinase− Mel-A375 were exogenously loaded with irrelevant flu peptide, the tyrosinase-peptide YMD, and compared with HLA-A2+tyrosinase+ Mel-93.04A12 for capacity to stimulate CTLs. IFN-γ secretion of the tyrosinase-independent HLA-A2 allo-reactive T-cell clone (JB4), 2 HLA-A2–restricted tyrosinase peptide–specific T-cell clones (Tyr-F8 and IVS-B), and an HLA-A2–restricted melan-A peptide–specific T-cell clone (A42) was measured by ELISA and given as picograms per milliliter. (B) Exemplary screening data of clones derived from a priming using DCs derived from an HLA-A2− donor loaded with HLA-A2 and tyrosinase RNA. Cytotoxic activity was assessed in a standard 4 hours chromium release assay using HLA-A2+tyrosinase− Mel-A375 and HLA-A2+tyrosinase+ Mel-93.04A12 melanoma cells as target cells at different E:T ratios. Data are given as percentage-specific lysis.
Recognition of peptide-loaded Mel-A375 and exemplary screening data from the DC priming. (A) HLA-A2+tyrosinase− Mel-A375 were exogenously loaded with irrelevant flu peptide, the tyrosinase-peptide YMD, and compared with HLA-A2+tyrosinase+ Mel-93.04A12 for capacity to stimulate CTLs. IFN-γ secretion of the tyrosinase-independent HLA-A2 allo-reactive T-cell clone (JB4), 2 HLA-A2–restricted tyrosinase peptide–specific T-cell clones (Tyr-F8 and IVS-B), and an HLA-A2–restricted melan-A peptide–specific T-cell clone (A42) was measured by ELISA and given as picograms per milliliter. (B) Exemplary screening data of clones derived from a priming using DCs derived from an HLA-A2− donor loaded with HLA-A2 and tyrosinase RNA. Cytotoxic activity was assessed in a standard 4 hours chromium release assay using HLA-A2+tyrosinase− Mel-A375 and HLA-A2+tyrosinase+ Mel-93.04A12 melanoma cells as target cells at different E:T ratios. Data are given as percentage-specific lysis.
Overview of specificity of CTL from 2 HLA-A2+ and 2 HLA-A2− donors
| Reactivity . | Self-restricted (HLA-A2+ donors), no. (%) . | Allo-restricted (HLA-A2− donors), no. (%) . |
|---|---|---|
| No reactivity* | 21 (55) | 8 (16) |
| Allo-A2† | 0 (0) | 27 (53) |
| A2-tyrosinase‡ | 17 (45) | 16 (31) |
| Total no. | 38 (100) | 51 (100) |
| Reactivity . | Self-restricted (HLA-A2+ donors), no. (%) . | Allo-restricted (HLA-A2− donors), no. (%) . |
|---|---|---|
| No reactivity* | 21 (55) | 8 (16) |
| Allo-A2† | 0 (0) | 27 (53) |
| A2-tyrosinase‡ | 17 (45) | 16 (31) |
| Total no. | 38 (100) | 51 (100) |
Clones failed to recognize HLA-A2-tyrosinase ligands.
Clones recognized HLA-A2 without peptide.
Clones recognized HLA-A2-tyrosinase-peptide ligands.
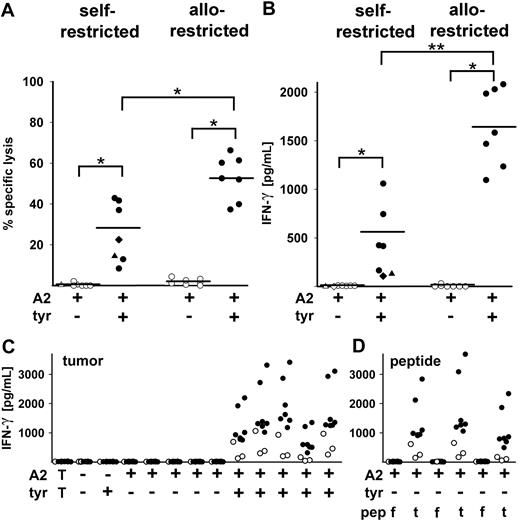
After discarding clones with tyrosinase-independent, HLA-A2 allo-reactivity, a set of strongly proliferating allo-restricted clones with presumed tyrosinase-derived peptide specificity was further analyzed compared with HLA-A2 self-restricted clones. All selected clones showed cytotoxic activity and IFN-γ release directed against Mel-93.04A12 (HLA-A2+tyrosinase+) but not Mel-A375 (HLA-A2+tyrosinase−) melanoma cell lines (Figure 3A-B). The allo-restricted CTLs had a significantly higher mean level of specific lysis than the self-restricted CTLs (52.8% vs 28.5%) and a significantly higher mean level of IFN-γ secretion (1639 pg/mL vs 561 pg/mL), demonstrating their general superior function. The self-restricted melanoma patient-derived CTLs IVS-B and the self-restricted peptide-primed Tyr-F8 CTLs were included for comparison and showed low-intermediate killing (22.5% and 14.4%, respectively) and very low IFN-γ release (106 pg/mL and 129 pg/mL, respectively). A majority of these clones were further analyzed for IFN-γ release after coculture with a panel of tumor lines derived from several different tumor types (Figure 3C). Although all the clones failed to recognize HLA-A2− and HLA-A2+tyrosinase− tumor cells, most clones secreted detectable amounts of IFN-γ after stimulation with the 5 HLA-A2+tyrosinase+ melanoma tumor lines. Furthermore, the allo-restricted peptide-specific CTLs showed higher levels of IFN-γ secretion compared with the self-restricted CTLs after stimulation with each of these 5 melanoma cell lines (range, 317-3417 pg/mL vs 43-1065 pg/mL). HLA-A2+tyrosinase− tumor cell lines could be recognized after exogenous pulsing with specific tyrosinase YMD peptide but not with irrelevant flu peptide (Figure 3D). Again, more IFN-γ was released by allo-restricted CTL compared with self-restricted CTLs (range, 494-3693 pg/mL vs 55-655 pg/mL, respectively, Figure 3D).
CTL clones from multimer-sorted cells of HLA-A2+ and HLA-A2− donors primed with RNA-pulsed DCs. (A) Cytotoxic activity of 5 CTL clones from HLA-A2+ and 7 CTL clones from HLA-A2− donors is shown against Mel-A375 (HLA-A2+tyrosinase−) and Mel-93.04A12 (HLA-A2+tyrosinase+) melanoma lines at E:T of 5:1. CTL clones showed significant differences in lysis of HLA-A2+tyrosinase− vs HLA-A2+tyrosinase+ melanoma lines (nonparametric Wilcoxon signed rank test, *P < .05). Allo-restricted clones displayed greater mean percentage-specific lysis than self-restricted clones (52.8% vs 28.5%; nonparametric Mann-Whitney U test; *P < .05). The patient-derived IVS-B clone showed 22.5% specific lysis (♦), and the peptide-primed CTL clone Tyr-F8 showed 14.4% specific lysis (▲). (B) IFN-γ secretion (picograms per milliliter) by the same clones after coculture with Mel-A375 (HLA-A2+tyrosinase−) and Mel-93.04A12 (HLA-A2+tyrosinase+) lines was measured by ELISA. Allo-restricted clones showed higher mean IFN-γ secretion than self-restricted clones (1639 pg/mL vs 561 pg/mL; nonparametric Mann-Whitney U test; **P < .005). The IVS-B clone (♦) released 106 pg/mL IFN-γ and Tyr-F8 (▲) released 129 pg/mL. (C) IFN-γ secretion (picograms per milliliter) by 7 allo-restricted (●) and 3 self-restricted (○) T-cell clones in coculture with a panel of tumor cell lines shown from left to right: a breast carcinoma line MaCa1 (HLA-A2−tyrosinase−); a melanoma line SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375 (HLA-A2+tyrosinase−), a renal cell carcinoma line RCC-26 (HLA-A2+tyrosinase−), a pancreas carcinoma line PancTu 1 (HLA-A2+tyrosinase−), a stable HLA-A*0201 transfectant of MaCa1/A2 (HLA-A2+tyrosinase−), and a tongue carcinoma line UTS CC 1588 (HLA-A2+tyrosinase−); and the melanoma cell lines Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA-A2+tyrosinase+). The leftmost values designated with T show the background levels of cytokine secreted by the CTL in the absence of stimulating cells. (D) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t) and IFN-γ secretion was measured by ELISA and given as picograms per milliliter.
CTL clones from multimer-sorted cells of HLA-A2+ and HLA-A2− donors primed with RNA-pulsed DCs. (A) Cytotoxic activity of 5 CTL clones from HLA-A2+ and 7 CTL clones from HLA-A2− donors is shown against Mel-A375 (HLA-A2+tyrosinase−) and Mel-93.04A12 (HLA-A2+tyrosinase+) melanoma lines at E:T of 5:1. CTL clones showed significant differences in lysis of HLA-A2+tyrosinase− vs HLA-A2+tyrosinase+ melanoma lines (nonparametric Wilcoxon signed rank test, *P < .05). Allo-restricted clones displayed greater mean percentage-specific lysis than self-restricted clones (52.8% vs 28.5%; nonparametric Mann-Whitney U test; *P < .05). The patient-derived IVS-B clone showed 22.5% specific lysis (♦), and the peptide-primed CTL clone Tyr-F8 showed 14.4% specific lysis (▲). (B) IFN-γ secretion (picograms per milliliter) by the same clones after coculture with Mel-A375 (HLA-A2+tyrosinase−) and Mel-93.04A12 (HLA-A2+tyrosinase+) lines was measured by ELISA. Allo-restricted clones showed higher mean IFN-γ secretion than self-restricted clones (1639 pg/mL vs 561 pg/mL; nonparametric Mann-Whitney U test; **P < .005). The IVS-B clone (♦) released 106 pg/mL IFN-γ and Tyr-F8 (▲) released 129 pg/mL. (C) IFN-γ secretion (picograms per milliliter) by 7 allo-restricted (●) and 3 self-restricted (○) T-cell clones in coculture with a panel of tumor cell lines shown from left to right: a breast carcinoma line MaCa1 (HLA-A2−tyrosinase−); a melanoma line SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375 (HLA-A2+tyrosinase−), a renal cell carcinoma line RCC-26 (HLA-A2+tyrosinase−), a pancreas carcinoma line PancTu 1 (HLA-A2+tyrosinase−), a stable HLA-A*0201 transfectant of MaCa1/A2 (HLA-A2+tyrosinase−), and a tongue carcinoma line UTS CC 1588 (HLA-A2+tyrosinase−); and the melanoma cell lines Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA-A2+tyrosinase+). The leftmost values designated with T show the background levels of cytokine secreted by the CTL in the absence of stimulating cells. (D) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t) and IFN-γ secretion was measured by ELISA and given as picograms per milliliter.
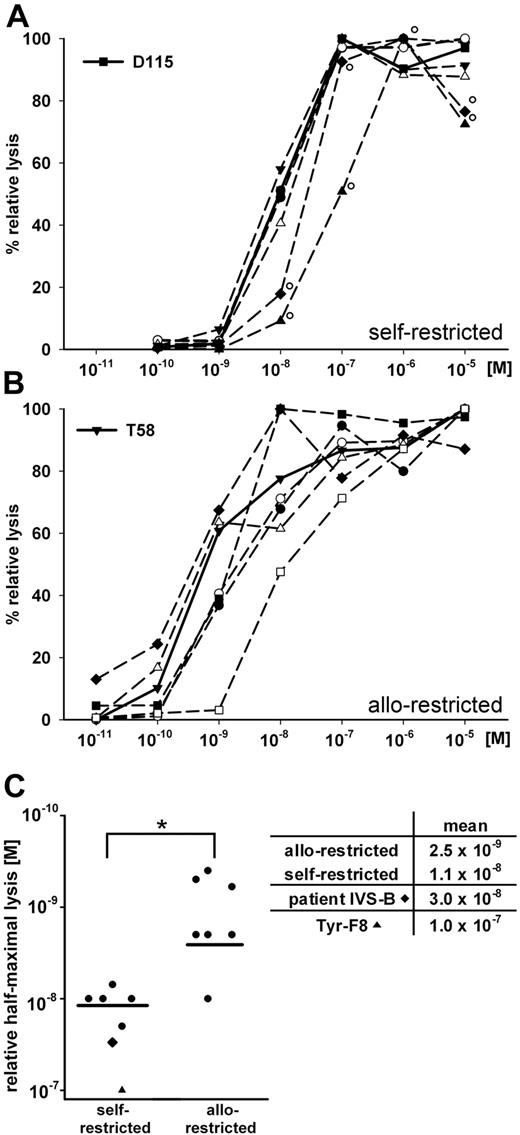
Functional T-cell avidity for tyrosinase-peptide recognition was measured in a 51Cr-release assay using HLA-A2+ T2 cells pulsed with graded amounts of exogenous tyrosinase peptide as target cells. The peptide concentration needed for 50% relative lysis defined the value of half-maximal lysis.21 Self-restricted clones had half-maximal values near 10−8 M (mean = 1.1 × 10−8 M; Figure 4A,C). Allo-restricted clones displayed a range of values and several clones responded to significantly less peptide (mean = 2.5 × 10−9 M; Figure 4B-C). This value differed significantly from self-restricted clones (Figure 4C). The patient-derived IVS-B clone and the Tyr-F8 clone required the most peptide (3.0 × 10−8 M and 1.0 × 10−7 M, respectively; Figure 4A,C).
Comparisons of self-restricted and allo-restricted CTLs. (A-B) The cytotoxic activity of individual CTL was measured against T2 cells loaded with graded amounts of tyrosinase peptide (10−11 M to 10−5 M) at an E:T of 10:1. Peptide concentration and percentage relative lysis are displayed on the x- and y-axes, respectively. Shown are (A) patient-derived CTL IVS-B (♦°), CTL Tyr-F8 derived via T2 peptide priming (▲°), and the 5 DC-primed self-restricted CTL and (B) DC-primed allo-restricted CTL. Bold lines represent CTL D115 and T58 in panels A and B, respectively. (C) Relative values of half-maximal lysis are presented for CTL IVS-B, Tyr-F8, self-restricted CTLs (n = 5), and allo-restricted CTLs (n = 7). Means are represented as black bars. Significant differences were found between the 2 groups (nonparametric Mann-Whitney U test; *P < .05).
Comparisons of self-restricted and allo-restricted CTLs. (A-B) The cytotoxic activity of individual CTL was measured against T2 cells loaded with graded amounts of tyrosinase peptide (10−11 M to 10−5 M) at an E:T of 10:1. Peptide concentration and percentage relative lysis are displayed on the x- and y-axes, respectively. Shown are (A) patient-derived CTL IVS-B (♦°), CTL Tyr-F8 derived via T2 peptide priming (▲°), and the 5 DC-primed self-restricted CTL and (B) DC-primed allo-restricted CTL. Bold lines represent CTL D115 and T58 in panels A and B, respectively. (C) Relative values of half-maximal lysis are presented for CTL IVS-B, Tyr-F8, self-restricted CTLs (n = 5), and allo-restricted CTLs (n = 7). Means are represented as black bars. Significant differences were found between the 2 groups (nonparametric Mann-Whitney U test; *P < .05).
One self-restricted T-cell clone (D115) and 1 allo-restricted T-cell clone (T58) were selected for further characterization based on their highest rank in killing of melanoma cell lines (data not shown). Responses to peptide-pulsed T2 cells showed that they localized to the middle of their respective groups with relative half-maximal lysis at lower peptide concentrations (T58: 6 × 10−10 M vs D115: 10−8 M; Figure 4A-B).
A side-by-side comparison of the patient-derived IVS-B CTL with self-restricted D115 and allo-restricted T58 CTL revealed that IVS-B showed the poorest response, whereas D115 showed intermediate and T58 the best recognition of HLA-A2+ tumor cells that endogenously expressed tyrosinase (Figure 5A). Although all 3 CTLs failed to recognize native HLA-A2+tyrosinase− tumor cell lines, these tumor lines were recognized after exogenous pulsing with specific tyrosinase YMD peptide but not with irrelevant flu peptide (Figure 5B).
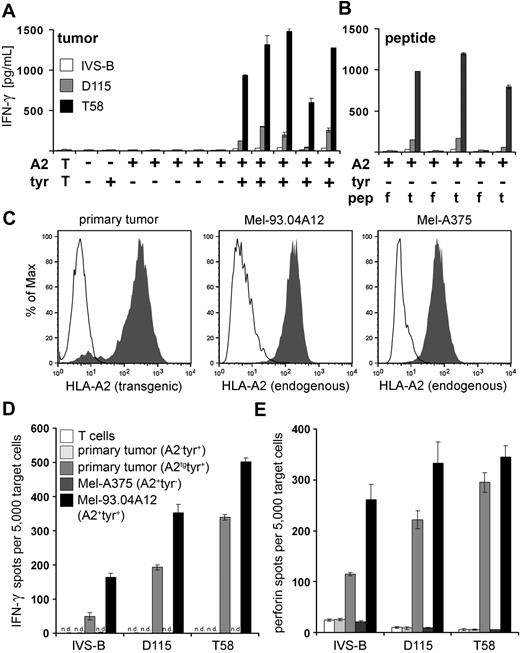
Tyrosinase peptide-specific CTL recognition of tumor cell lines and primary melanoma tumor cells. (A) Columns represent the amount of IFN-γ (picograms per milliliter) secreted by patient-derived IVS-B CTL, self-restricted D115 CTL, and allo-restricted T58 CTL in coculture with a panel of tumor cell lines from left to right: MaCa1 (HLA-A2−tyrosinase−); SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375, RCC-26, PancTu 1, MaCa1/A2, and UTS CC 1588 (all HLA-A2+tyrosinase−); Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA-A2+tyrosinase+). T indicates CTL without stimulating cells. (B) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t), and IFN-γ secretion was measured by ELISA and given as picograms per milliliter. (C) HLA-A2 expression on primary tumor cells (passage 12) of an HLA-A2− melanoma patient after transfection with 50 μg HLA-A2 ivt-RNA and on established melanoma cell lines Mel-93.04A12 (HLA-A2+tyrosinase+), and Mel-A375 (HLA-A2+tyrosinase−) was measured by flow cytometry after staining with HLA-A2–specific monoclonal antibody. Histograms represent stained samples (filled curves) and control samples (empty curves): control curves show untransfected primary tumor cells stained with HLA-A2–specific monoclonal (left histogram) or isotype control antibodies used with the melanoma cell lines (middle and right histograms). HLA-A2 expression on primary tumor cells was detected 10 hours after electroporation. (D) The capacity of the patient-derived CTLs (IVS-B), the representative self-restricted CTLs (D115), and the representative allo-restricted CTLs (T58) to secrete IFN-γ or (E) release perforin in coculture with the melanoma cells shown above was measured in ELISPOT assays. n.d. indicates not detectable.
Tyrosinase peptide-specific CTL recognition of tumor cell lines and primary melanoma tumor cells. (A) Columns represent the amount of IFN-γ (picograms per milliliter) secreted by patient-derived IVS-B CTL, self-restricted D115 CTL, and allo-restricted T58 CTL in coculture with a panel of tumor cell lines from left to right: MaCa1 (HLA-A2−tyrosinase−); SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375, RCC-26, PancTu 1, MaCa1/A2, and UTS CC 1588 (all HLA-A2+tyrosinase−); Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA-A2+tyrosinase+). T indicates CTL without stimulating cells. (B) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t), and IFN-γ secretion was measured by ELISA and given as picograms per milliliter. (C) HLA-A2 expression on primary tumor cells (passage 12) of an HLA-A2− melanoma patient after transfection with 50 μg HLA-A2 ivt-RNA and on established melanoma cell lines Mel-93.04A12 (HLA-A2+tyrosinase+), and Mel-A375 (HLA-A2+tyrosinase−) was measured by flow cytometry after staining with HLA-A2–specific monoclonal antibody. Histograms represent stained samples (filled curves) and control samples (empty curves): control curves show untransfected primary tumor cells stained with HLA-A2–specific monoclonal (left histogram) or isotype control antibodies used with the melanoma cell lines (middle and right histograms). HLA-A2 expression on primary tumor cells was detected 10 hours after electroporation. (D) The capacity of the patient-derived CTLs (IVS-B), the representative self-restricted CTLs (D115), and the representative allo-restricted CTLs (T58) to secrete IFN-γ or (E) release perforin in coculture with the melanoma cells shown above was measured in ELISPOT assays. n.d. indicates not detectable.
To answer the question whether allo-restricted T58 CTL would also show superior recognition of primary melanoma cells, they were compared with D115 CTL and patient-derived IVS-B CTLs. Early passage tumor cells expressing tyrosinase from an HLA-A2− melanoma patient were transfected with HLA-A2 ivt-RNA to create a pair of matched cell lines, with and without the specific pMHC ligand (Figure 5C). ELISPOT assays of IFN-γ and perforin showed a hierarchy similar to that observed with established melanoma cell lines, with poor recognition by IVS-B CTLs, intermediate recognition by D115 CTLs, and the strongest IFN-γ and perforin secretion by T58 CTLs (Figure 5D-E). Recognition was HLA-A2–restricted because the primary tumor cells that were not transfected with HLA-A2 ivt-RNA were not recognized. Mel-A375 (A2+tyr−) were not recognized, and Mel-93.04A12 (A2+tyr+) were recognized by the 3 CTL clones.
Allo-restricted transgenic TCRs confer superior function
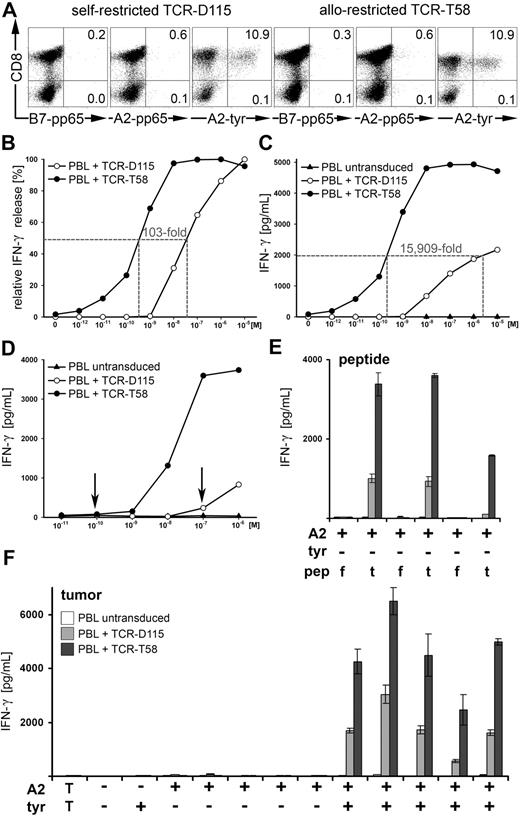
It was important to demonstrate that the superior functional avidity of allo-restricted T cells resided directly in the TCR; therefore, the TCR cDNA of D115 and T58 CTLs was isolated, modified for optimal expression, and introduced by retrovirus transfer into human PBLs of healthy donors. Multimer staining of untransduced PBLs was not seen (0.1%, data not shown). After transduction, no notable multimer staining could be detected with the irrelevant multimers B7-pp65 and A2-pp65 (0.2%-0.6%), and equal numbers of CD8+ PBLs bound the specific A2-tyr multimer (TCR-D115, 10.9%; and TCR-T58, 10.9%; Figure 6A). The TCR-transduced PBLs were tested without enrichment for IFN-γ release after stimulation with peptide-pulsed T2 cells, whereby PBLs expressing TCR-T58 required 100-fold less peptide for half-maximal release compared with PBL expressing TCR-D115 (Figure 6B). Untransduced PBLs showed no specific IFN-γ secretion (data not shown). The PBLs with TCR-T58 also released substantially higher amounts of cytokine compared with PBLs with TCR-D115 and the concentration of peptide required to induce release of 2000 pg/mL IFN-γ differed by more than 15 000-fold (TCR-T58: 2.2 × 10−10 M vs TCR-D115: 3.5 × 10−6 M; Figure 6C). Because T2 cells have a genetic defect in peptide loading, they fail to present most endogenous peptides in their endogenous HLA-A2 molecules; thus, they express highly uniform pMHCs after exogenous loading of tyrosinase peptide. To analyze the sensitivity of TCR recognition of specific pMHC ligands within a heterogeneous pMHC setting, which occurs in cells not having defects in endogenous peptide loading, we created new stimulating cells using autologous HLA-A2+ PBMCs loaded with graded amounts of tyrosinase peptide. In this case, tyrosinase peptide can only displace some endogenous peptides in HLA-A2 molecules, thereby strongly limiting these specific pMHC ligands on PBMCs. After coculture with these PBMCs, TCR-T58 PBLs secreted considerably higher amounts of IFN-γ. In addition, the lowest amount of peptide required to induce IFN-γ secretion above background levels differed substantially from TCR-D115 PBL (10−10 M vs 10−7 M; Figure 6D). Specificity was retained in the 2 TCR-transduced PBL populations, which only recognized HLA-A2+tyrosinase− tumor cell lines after pulsing with specific YMD but not irrelevant flu peptide. Furthermore, HLA-A2+tyrosinase+ tumor cell lines, but not HLA-A2− or HLA-A2+tyrosinase− tumor lines led to specific activation of both CTLs (Figure 6E-F), whereby PBL-T58 secreted significantly higher amounts of IFN-γ. Thus, by all these parameters, the allo-restricted TCR-T58 was found to be far superior to self-restricted TCR-D115 in its capacity to recognize MHC-tyrosinase-peptide complexes.
Transfer of antigen specificity by retroviral transfer of TCR-D115 and TCR-T58. (A) PBLs of a healthy donor were transduced with TCR-D115 or TCR-T58. Unsorted TCR-transduced PBLs were analyzed on day 10 for transgenic TCR expression using irrelevant B7-pp65 and A2-pp65 multimers and specific A2-tyr multimers. Untransduced PBLs showed no multimer binding (0.1%, data not shown). Percentages of multimer+CD8+ T cells are displayed in the top right quadrant. (B-C) The IFN-γ release of unsorted TCR-transduced PBLs after stimulation with T2 cells loaded with graded amounts of tyrosinase peptide (10−12 M to 10−5 M) at a ratio of 2:1. (B) The relative IFN-γ release is displayed in percentage. (C) The specific IFN-γ release is presented as picograms per milliliter. (D) Functionality of unsorted TCR-transduced PBLs was measured by IFN-γ release using autologous HLA-A2+ PBMCs loaded with tyrosinase peptide (10−11 M to 10−6 M) as stimulating cells at ratio of 2:1. Untransduced PBLs (▲) showed no peptide-specific IFN-γ release. (E) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t) and IFN-γ secretion was measured by ELISA and given as picograms per milliliter. (F) Specificity of recognition was assessed by IFN-γ release after coculture with the tumor cell lines from left to right: MaCa1 (HLA-A2−tyrosinase−); SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375, RCC-26, PancTu 1, MaCa1/A2, and UTS CC 1588 (all HLA-A2+tyrosinase−); Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA−A2+tyrosinase+). T indicates CTL without stimulating cells.
Transfer of antigen specificity by retroviral transfer of TCR-D115 and TCR-T58. (A) PBLs of a healthy donor were transduced with TCR-D115 or TCR-T58. Unsorted TCR-transduced PBLs were analyzed on day 10 for transgenic TCR expression using irrelevant B7-pp65 and A2-pp65 multimers and specific A2-tyr multimers. Untransduced PBLs showed no multimer binding (0.1%, data not shown). Percentages of multimer+CD8+ T cells are displayed in the top right quadrant. (B-C) The IFN-γ release of unsorted TCR-transduced PBLs after stimulation with T2 cells loaded with graded amounts of tyrosinase peptide (10−12 M to 10−5 M) at a ratio of 2:1. (B) The relative IFN-γ release is displayed in percentage. (C) The specific IFN-γ release is presented as picograms per milliliter. (D) Functionality of unsorted TCR-transduced PBLs was measured by IFN-γ release using autologous HLA-A2+ PBMCs loaded with tyrosinase peptide (10−11 M to 10−6 M) as stimulating cells at ratio of 2:1. Untransduced PBLs (▲) showed no peptide-specific IFN-γ release. (E) The HLA-A2+tyrosinase− tumor cell lines Mel-A375, RCC-26, and MaCa1/A2 were exogenously loaded with either 10−5 M irrelevant flu peptide (f) or 10−5 M tyrosinase peptide YMD (t) and IFN-γ secretion was measured by ELISA and given as picograms per milliliter. (F) Specificity of recognition was assessed by IFN-γ release after coculture with the tumor cell lines from left to right: MaCa1 (HLA-A2−tyrosinase−); SK-Mel-28 (HLA-A2−tyrosinase+); Mel-A375, RCC-26, PancTu 1, MaCa1/A2, and UTS CC 1588 (all HLA-A2+tyrosinase−); Mel-624.38, Mel-93.04A12, SK-Mel-23, SK-Mel-29, and WM-266-4 (all HLA−A2+tyrosinase+). T indicates CTL without stimulating cells.
Discussion
This systematic comparison of self-restricted and allo-restricted peptide-specific CTLs clearly demonstrated that DCs that coexpressed allogeneic MHC molecules and tumor-associated antigen induced allo-restricted peptide-specific T cells with superior antitumor activity. Three features of the procedure are of particular relevance. First, we took advantage of the well-documented knowledge that allo-restricted T cells can express high-affinity TCR specific for peptide ligands of self-proteins because they were not deleted by negative selection.1,2 Thereby, T cells that more effectively recognized tumor cells were more readily obtained. Second, we used T cells of healthy donors, taking advantage of the optimal priming capacity of DCs for naive cells. This allowed us to avoid use of patient lymphocytes, which often show poor proliferation and function. Third, we used ivt-RNA as the source of both allogeneic MHC molecules and TAAs. This provided maximum flexibility for creating allo-ligands specific for any MHC class I allele and any selected antigen. Furthermore, priming was not limited to known peptides because the whole TAA was available for processing and presentation within the DCs.27 Here we used HLA multimers to isolate peptide-specific T cells, but alternative methods are available to select a broader array of T cells, such as the IFN-γ capture assay.28 Indeed, CTLs that released high levels of IFN-γ after tumor cell stimulation often had the best killing capacity. It should also be noted that this same strategy can probably be extended to identify high-avidity allo-restricted CD4+ T cells by providing DCs with ivt-RNA encoding MHC class II alleles in combination with TAAs. In this case, the ivt-RNA for TAAs can be modified to shuttle the proteins into the endosomal pathway to allow better peptide loading of class II molecules.27
Significant tumor regression can occur after adoptive transfer of T cells with antitumor specificity.29 However, patient-derived T cells may have suboptimal activity, as seen with IVS-B CTLs of a melanoma patient. Furthermore, most self-restricted T cells with high-affinity TCR specific for self-peptides have undergone negative selection, and remaining T cells may be negatively controlled in the periphery. For these reasons, CTLs with appropriate specificity and function are often missing in patients with rapidly progressing tumors. Therefore, there is current interest in using precharacterized TCR genes to create designer lymphocytes for adoptive cell therapies.4,9 Expression of TCR transgenes in activated PBLs could imbue recipient lymphocytes with antitumor activities comparable with the original CTLs,4,9 as seen here with TCR-D115 and TCR-T58. Furthermore, some transgenic TCRs can displace endogenous TCRs, yielding lymphocytes with monoclonal TCRs.10,30 If more TCRs with this feature can be identified, the safety of adoptive cell therapy would be improved.
Our studies comparing self-restricted and allo-restricted CTL clones specific for the same MHC-tyrosinase ligand revealed that there were inherent differences in the TCR themselves. Far superior recognition of peptide-pulsed cells was seen with PBLs expressing the allo-restricted TCR-T58 compared with the self-restricted TCR-D115. The improved capacity of TCR-T58 PBLs to respond to substantially lower amounts of peptide indicates that these T cells should be better able in vivo to effectively recognize tumor cells that express only limited amounts of specific pMHC ligand. This is supported by the far superior recognition of melanoma tumor cell lines by PBL-T58.
The first clinical trials of adoptive transfer of TCR-transgenic T cells in melanoma patients achieved clinical responses in a number of patients with advanced disease.31,32 These results demonstrated the therapeutic potential of this approach. It would be an advantage to provide patients with mixtures of TCR-transgenic lymphocytes that target their tumors via several different pMHC ligands to avoid immune selection of tumor cell variants that lack expression of individual TAAs. Our priming strategy should allow better access to high-avidity CTLs specific for different TAAs that are prevalent in melanomas and other tumors, which could serve as sources of therapeutic TCR sequences. Future adoptive therapy of more patients will be feasible if useful TCR sequences are available as “off-the-shelf” reagents. Our approach helps to close the technologic gap in identifying suitable TCR with specificity for common TAA ligands that have sufficient affinity to effectively recognize tumor cells.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Th. Blankenstein and E. Simpson for helpful discussions; A. Zobywalski for advice on DC; B. Mosetter, S. Eichenlaub, B. Stadlbauer, S. Kresse, and A. Slusarski for auxiliary work; S. Krauss-Etschmann and C. Heumann for help with statistical analysis; F. Anderl and M. Odendahl for support with the multimer technology; J. Ellwart and K. Ebelt for help with flow cytometry; and E. Noessner and M. Javorovic for technical advice.
This work was supported by the German Research Council (grants SFB-455, SFB-456, and SFB-TR36) and the Helmholtz Society (Alliance for Immunotherapy of Cancer, HA-202).
Authorship
Contribution: S.W. designed and performed the experiments and helped in drafting the manuscript; M.S. performed cell sorting; B.F. helped in the characterization of the TCR; D.S. and W.U. contributed retroviral transduction technology; S.S. helped in cytotoxic assays; S.M. cloned ivt-plasmids; H.P. provided ELISPOT technology; D.H.B. contributed analytical tools and data analysis; D.J.S. designed the concept and prepared the final manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests. However, a patent for specific TCRs has been submitted by the Helmholtz Zentrum München, German Research Center for Environmental Health, Munich, Germany.
Correspondence: Dolores J. Schendel, Institute of Molecular Immunology, Helmholtz Zentrum München, Marchioninistrasse 25, 81377 Munich, Germany; e-mail: schendel@helmholtz-muenchen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal