Abstract
Macrophages infiltrating solid tumors exhibit a tumor-supporting phenotype and are critical for tumor development. Little is known which tumor-derived signal provokes this phenotype shift and how these signals are interpreted in macrophages to support tumor growth. We used the supernatant of apoptotic cells and noticed transcriptional, nuclear factor of activated T cells-dependent up-regulation of hypoxia-inducible factor (HIF)–1α mRNA, subsequent protein expression, and HIF-1 activity. Blocking calcineurin with cyclosporine A attenuated nuclear factor of activated T cells binding during electrophoretic mobility shift assay analysis and circumvented the HIF-1α mRNA increase. Knockdown experiments, receptor analysis, and antibody neutralization pointed to sphingosine-1-phosphate and transforming growth factor-β as the initiators of the HIF-1 response. The use of macrophages from conditional HIF-1α knockout mice revealed that macrophages, under the impact of apoptotic cell supernatants, use HIF-1 to produce factors that induce CD31 expression in murine embryonic stem cells. Our study supports the notion that soluble factors produced from apoptotic tumor cells activate the HIF-1 system under normoxia in macrophages to enhance their tumor-promoting capacity by, for example, releasing vascular endothelial growth factor. This shows the importance of HIF-1–elicited responses in regulatory macrophages under normoxia.
Introduction
Tumor-associated macrophages (TAMs) infiltrate tumors, which often predicts a poor survival prognosis. TAMs apparently support tumor development by improving vascularization and cell survival rather than behaving tumoricidally (for review, see Lewis and Pollard1 ).
Features characterizing TAMs are often elicited by alternative activation, which polarizes cells toward regulatory macrophages, a subgroup of the so-called M2 phenotype. They share anti-inflammatory characteristic, for example, reduced interleukin (IL)-12 production2,3 compared with classical stimulation and M1 polarization.2 Unfortunately, it is only poorly understood which factors influence the TAM phenotype profile. The phenotype alteration occurs in the tumor microenvironment, with the likely contribution of tumor-derived molecules such as IL-4, IL-10, transforming growth factor (TGF)–β, prostaglandin E2 (PGE2), chemokines, or tumor hypoxia.1,2 Interestingly, apoptotic cells (AC) also release anti-inflammatory cytokines such as TGF-β and IL-104,5 and reduce tumor-directed macrophage cytotoxicity.6 It became apparent that phagocytosis of AC by macrophages is an immunoregulatory process that shapes the regulatory macrophage phenotype.7-10
Macrophages cocultured in vitro with AC or exposed to the supernatant of AC express a pattern of markers that overlaps with those established for TAMs.11-13 Furthermore, sphingosine-1-phosphate (S1P) provoked M2 macrophage polarization,13,14 and S1P is released from AC.13,15 S1P activates distinct G protein–coupled receptors,16 elicits a variety of immune cell responses,17 and promotes tumor angiogenesis.18,19
Hypoxia-inducible factor (HIF)–1 is known for its importance in tumorigenesis, and controls angiogenesis, proliferation, metabolism, metastasis, and differentiation.20 HIF-1, composed of a HIF-1α and HIF-1β subunit, senses and coordinates adaptation to low oxygen availability. Under normoxia (21% O2), HIF-1β is constitutively expressed, whereas HIF-1α is hydroxylated by prolyl hydroxylases, polyubiquitinated by an E3-ubiquitin ligase complex containing the von Hippel Lindau protein, and concomitantly degraded (for review, see Semenza21 ). Because hypoxia mainly affects HIF-1α protein stability, HIF-1α mRNA expression regulation only recently attracted some interest. In macrophages, lipopolysaccharide (LPS) induced nuclear factor (NF)–κB binding to the HIF-1α promoter and increased its mRNA expression.22 Nuclear factor of activated T cells (NFAT)c1 increased the HIF-α mRNA content in mast cells after stimulation with ionomycin,23 and signal transducer and activator of transcription (STAT)3 up-regulated HIF-1α mRNA in human tumor cells,24 substantiating the importance of transcriptional control mechanisms governing HIF-1 appearance/activity.
We provide evidence that HIF-1α under normoxia is transcriptionally activated via NFAT in response to apoptotic cell–derived S1P and TGF-β. As verified in cells from HIF-1α conditional knockout mice, activation of HIF-1 in polarized macrophages moderates differentiation of embryonic stem cells toward CD31-positive cells.
Methods
Materials
Medium, supplements, and fetal calf serum (FCS) came from either PAA or Biochrom. Leukemia-inhibitory factor (LIF) and anti-CD31 came from Millipore. Gelatin was purchased from StemCell Technologies. Staurosporine, 5-[4-phenyl-5(trifluoromethyl)-2-thienyl]-3-[3-(trifluoromethyl)pheyl]-1,2,4-oxadiazole (SEW2871), cyclosporine A (CsA), LPS (from Escherichia coli, serotype 0127:B8), and 4,6 diamidino-2-phenylindole (DAPI) were from Sigma-Aldrich. S1P and (R)-phosphoric acid mono-[2-amino-2(3-octyl-phenylcarbamoyl)-ethyl] ester (VPC23019) were from Avanti Polar Lipids. JTE-013 was purchased from BIOZOL. Murine macrophage colony–stimulating factor was from PeproTech. Reagents for Western analysis, including the HIF-1α antibody (AB), were previously described.25 Smad2 and phospho-Smad2 AB were from New England Biolabs; anti-NFATc1x was from Santa Cruz Biotechnology. Anti–TGF-β1 AB and mouse IgG1 isotype control were from R&D Systems. STA-21 and JetPEI were delivered by BIOMOL. Cy3-labeled anti–rat secondary AB was from Dianova.
Cell culture
The mouse monocyte/macrophage cell line RAW264.7, human MCF-7 breast carcinoma, mouse 4T1 mammary carcinoma, and human Jurkat lymphoma T cell lines were cultured in RPMI 1640 supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% heat-inactivated FCS. MCF-7 cells additionally received 2 μg/mL insulin. Stable transfected MCF-7-neo and MCF-7-sphingosine kinase 2 (SK2) knockdown (kd) were treated with 500 μg/mL neomycin. Primary mouse macrophages were cultured, as previously described.26 The mouse embryonic stem cell line CCE27,28 provided by Dr Heinrich Sauer (University of Giessen) was grown on gelatin-coated culture dishes in Dulbecco modified Eagle medium high glucose, supplemented with 50 U/mL penicillin, 50 μg/mL streptomycin, 10% heat-inactivated FCS, 8% active FCS, 1 mM sodium pyruvate, 2 mM glutamine, 100 μM β-mercaptoethanol, and 1% nonessential amino acids. In addition, 103 U/mL LIF was added. Cells were kept at 37°C in a humidified atmosphere with 5% CO2. For hypoxic exposure, cells were incubated at 1% O2 in a hypoxia workstation (Ruskin Technology).
Isolation of primary mouse macrophages
Mice with a conditional knockout of HIF-1α in the myeloid lineage were bred as described and provided by Dr Randall Johnson (Division of Biological Sciences, University of California, San Diego).29 Isolation of primary mouse macrophages from spleen was performed, as described.26 Animal experiments followed the guidelines of the Hessian animal care and use committee.
Generation of SK2 knockdown cells
For SK2 knockdown, sequence-specific small interfering RNA oligonucleotides (forward, 5′-GATCCACTAAACAAGCTTGGTACCTTCA-AGAGAGGTACCAAGCTTGTTAGTTTA-3′; reverse, 5′-AGCTTAAACTAAACAAGCTTGGTACCCTCTTGAAGGTACCAAGCTTGTTTAGTG-3′) were ligated into the pSilencer 4.1-CMV neo vector (Ambion), according to manufacturer's instructions. pSilencer-siSK2 was then transfected into MCF-7 cells using Nucleofector technology (Amaxa), as described previously.13 MCF-7-Neo cells were generated by transfecting MCF-7 cells with the pSilencer 4.1-CMV neo control vector.
Generation of conditioned media
To generate apoptotic Jurkat, MCF-7, or 4T1 cells, 2.5 × 106 cells/mL were used. Conditioned medium from AC (CMAC), necrotic cells, or viable cells was obtained, as described.30 CMAC from Jurkat cells is abbreviated CMAC-J, and CMAC from 4T1 cells is abbreviated CMAC-4T1. The control CMAC from MCF-7-neo cells is referred to as CMAC-MCF7, and CMAC from MCF-7 with SK2 knockdown as CMAC-SK2kd. CMAC was used at a ratio of 5:1 (AC:macrophages). For the generation of conditioned media from macrophages (CMPM), RAW264.7 or primary macrophages were exposed to CMAC for 16 hours. After 16 hours, CMAC was removed, cells were washed twice with phosphate-buffered saline (PBS), and further incubated with fresh media for 5 hours. Afterward, cells were centrifuged for 10 minutes at 1000g, and the supernatant was passed through a 0.2-μm cellulose syringe filter.
Western blotting
Western analysis (100 μg of protein/sample) was performed as described.25
RNA isolation and qRT-PCR
Isolation of RNA and quantitative real-time polymerase chain reaction (qRT-PCR) was performed, as described.26 RNA data were normalized to ribosomal 16S protein or actin RNA. Primers (biomers.net) are given in Table 1.
Primers for quantitative real-time PCR
| Primer . | Sequence, 5′ → 3′ . |
|---|---|
| Mouse HIF-1α forward | GAAATGGCCCAGTGAGAAAA |
| Mouse HIF-1α-exon2 forward | GGCGAAGCAAAGAGTCTGAAG |
| Mouse HIF-1α reverse | CTTCCACGTTGCTGACTTGA |
| Mouse HIF-2α forward | CCTGCAGCCTCAGTGTATCA |
| Mouse HIF-2α reverse | GTGTGGCTTGAACAGGGATT |
| Mouse VEGF forward | TGTCACCACCACGCCATCA |
| Mouse VEGF reverse | GGAATCCCAGAAACAACCCTAATC |
| Mouse Glut-1 forward | GAGGAGCTCTTCCACCCTCT |
| Mouse Glut-1 reverse | TCCTCCTGGACTTCACTGCT |
| Mouse ribosomal 16S protein forward | AGATGATCGAGCCGCGC |
| Mouse ribosomal 16S protein reverse | GCTACCAGGGCCTTTGAGATGGA |
| Human HIF-1α forward | CTCAAAGTCGGACAGCCTCA |
| Human HIF-1α reverse | CCCTGCAGTAGGTTTCTGCT |
| Human actin forward | TGACGGGGTCACCCACACTGTGCCCATCTA |
| Human actin reverse | CTAGAAGCATTTGCGGTCGACGATGGAGGG |
| Primer . | Sequence, 5′ → 3′ . |
|---|---|
| Mouse HIF-1α forward | GAAATGGCCCAGTGAGAAAA |
| Mouse HIF-1α-exon2 forward | GGCGAAGCAAAGAGTCTGAAG |
| Mouse HIF-1α reverse | CTTCCACGTTGCTGACTTGA |
| Mouse HIF-2α forward | CCTGCAGCCTCAGTGTATCA |
| Mouse HIF-2α reverse | GTGTGGCTTGAACAGGGATT |
| Mouse VEGF forward | TGTCACCACCACGCCATCA |
| Mouse VEGF reverse | GGAATCCCAGAAACAACCCTAATC |
| Mouse Glut-1 forward | GAGGAGCTCTTCCACCCTCT |
| Mouse Glut-1 reverse | TCCTCCTGGACTTCACTGCT |
| Mouse ribosomal 16S protein forward | AGATGATCGAGCCGCGC |
| Mouse ribosomal 16S protein reverse | GCTACCAGGGCCTTTGAGATGGA |
| Human HIF-1α forward | CTCAAAGTCGGACAGCCTCA |
| Human HIF-1α reverse | CCCTGCAGTAGGTTTCTGCT |
| Human actin forward | TGACGGGGTCACCCACACTGTGCCCATCTA |
| Human actin reverse | CTAGAAGCATTTGCGGTCGACGATGGAGGG |
Reporter analysis
The HIF-1 luciferase reporter (pGL3-EPO-HRE) was previously described.25 A luciferase reporter plasmid driven by a 800-bp fragment of the HIF-1α promoter was provided by Dr Joachim Fandrey (University of Duisburg-Essen).22 A NFAT-dependent myocyte-enriched calcineurin-interacting protein (MCIP)-luciferase reporter plasmid and the NFAT expression plasmid were provided by Dr Beverly Rothermel (University of Texas Southwestern Medical Center).31 As a control plasmid, pcDNA3.1 (Invitrogen) was used. Cells were transfected with 2 μg of individual plasmids (0.02 μg of Renilla luciferase [Promega]), using jetPEI-transfection reagent. For cotransfection experiments, 1 μg of each plasmid was used. After 8 hours with transfection reagent, medium was changed and cells were treated as indicated for an additional 16-hour period. Cells were lysed, and firefly luciferase/Renilla luciferase activity was measured using a dual luciferase reporter assay kit. Otherwise, luciferase activity was normalized to protein. Relative values of treated cells were calculated by normalizing values to those of control cells (set to 1).
Electrophoretic mobility shift assay
Cells were harvested, washed with cold PBS, centrifuged (5 minutes, 500g, 4°C), resuspended in cell lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA [ethylenediaminetetraacetic acid], 0.5% Nonidet P-40, 1% Triton X-100), incubated on ice for 20 minutes, and centrifuged (1 minute, 12 000g, 4°C). Pellets were resuspended in nuclear protein extraction buffer (50 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]/KOH, 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 10% glycerol, pH 7.9) and incubated while shaking for 30 minutes at 4°C. A total of 1 mM dithiothreitol, 1 × protease inhibitor mixture, and 10 mM NaF was added freshly to both buffers. Lysates were centrifuged (20 minutes, 12 000g, 4°C), and protein amount was determined. Electrophoretic mobility shift assay (EMSA) analysis using 10 μg of nuclear protein was carried out basically as described.13 Oligonucleotides, including the sequence of the putative NFAT binding site (bold letters) of the murine HIF-1α promoter, were used, as follows: 5′-TTTTCAACT[b]GGAAACTCGGGCGGG-3′; 5′-CCCGCCCGAGTTTCCAGTTGAAA-3′. A specific NFATc1 AB (2 ng/mL) was added for supershift experiments.
Embryonic body assay
To generate embryonic bodies (EB), media without LIF was used. A total of 1000 cells/35 μl was seeded in hanging drops for 3 days. EB were then plated on Lumox dishes (Greiner Bio-one) for 2 days before starting incubations. CMPM was diluted 1/3 with culture medium and added to the EB for 24 hours. Phase-contrast pictures of plated EB were recorded using an inverted fluorescence microscope (Axiovert 200 M) with an A-plan × 10/0.25 Ph1 objective lens, equipped with an AxioCam MRm camera (Zeiss) and the SlideBook software (Intelligent Imaging Innovation).
Immunofluorescence staining
After incubations, EB were washed with cold PBS and fixed with cold methanol/acetone (7:3) at −20°C for 1 day. Specimens were washed twice with 0.1% Triton-PBS, and blocking of unspecific binding was done for 1 hour at room temperature with 10% FCS in 0.01% Triton-PBS. Anti-CD31 AB was incubated overnight at 4°C. Cells were then washed 4 times with 0.01% Triton-PBS and incubated with anti–rat-Cy3 AB for 1 hour at room temperature. DAPI was added the last 30 minutes to stain nuclei. After washing 3 times with 0.01% Triton-PBS, specimens were mounted on glass slides and fluorescence was analyzed with the Laser Scanning System LSM 510 META (Configuration 30 Meta Zen 2007) from Zeiss equipped with the ZEN software (Zeiss). Objective lens EC Plan-Neofluar 40×/1.30 Oil DIC M27, a 1 mW helium/neon laser, excitation 543 nm (excitation of Cy3) and a 25 mW diode laser, excitation 405 nm (excitation of DAPI), was used. For statistical analysis of the CD31 fluorescence, the mean intensity of the monolayer areas was analyzed for DAPI and CD31 fluorescence with the histogram function of the ZEN software. Background intensity was subtracted, and the ratio of the mean intensity of CD31 versus DAPI fluorescence under control conditions was set to 1.
Statistical analysis
Each experiment was performed at least 3 times. Representative data are shown. Data in bar graphs are given as mean values plus or minus standard deviation (SD). Means were checked for statistical significance using 1-way analysis of variance, followed by Tukey tests.
Differences having a P value less than .05 were considered statistically significant and refer to controls if not otherwise indicated.
Results
AC supernatants induce HIF-1α and activate HIF-1 in macrophages
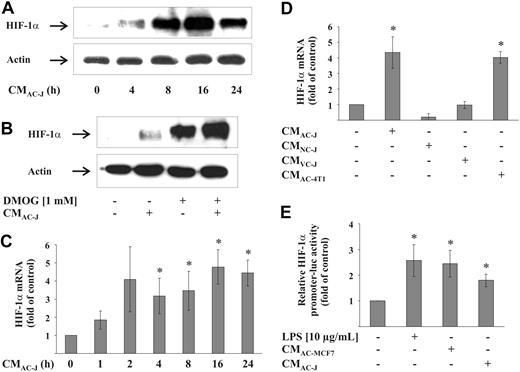
In a first approach, RAW264.7 macrophages were incubated with the supernatant of apoptotic T-lymphoma Jurkat cells (CMAC-J), which provoked a time-dependent HIF-1α protein accumulation with maximal protein expression at 16 hours (Figure 1A). Considering that CMAC-J might attenuate degradation of HIF-1α via the 26S proteasome, we used dimethyloxaloylglycine (DMOG) to block prolylhydroxylase (PHD) activity, which stabilized HIF-1α because of impaired proteasomal degradation. Cotreatment with CMAC-J and DMOG elevated HIF-1α protein accumulation in RAW264.7 cells compared with treatments with DMOG alone (Figure 1B), indicating enhanced protein production in the presence of CMAC-J. Interestingly, besides protein accumulation, CMAC-J caused a time-dependent accumulation of HIF-1α mRNA, with RNA levels reaching an approximately 4-fold increase over controls at 16 hours (Figure 1C). HIF-2α mRNA was not induced (data not shown). Up-regulation of HIF-1α mRNA was also observed in primary human (data not shown) and mouse (Figure 2D) macrophages treated with CMAC-J. Treating RAW264.7 cells with supernatants of viable or necrotic Jurkat cells failed to induce HIF-1α mRNA (Figure 1D). Furthermore, CMAC generated from mouse mammary 4T1 cells up-regulated HIF-1α mRNA in RAW264.7 comparable with CMAC-J (Figure 1D). To emphasize that HIF-1α mRNA accumulation resulted from transcriptional regulation rather than mRNA stabilization, we used a HIF-1α promoter reporter fragment. Incubating RAW264.7 macrophages with CMAC-J or the supernatant of apoptotic MCF-7 breast carcinoma cells (CMAC-MCF7) significantly elevated reporter activity compared with controls (Figure 1E). The known effect of LPS in activating the HIF-1α promoter was reproduced as an internal control. These data suggest that the supernatant of AC, irrespective of whether generated from Jurkat, 4T1, or MCF-7 cells, increased the mRNA of HIF-1α via transcriptional regulation.
HIF-1α mRNA and protein accumulation in macrophages by CM. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 4 to 24 hours. (B) RAW264.7 cells were treated with CMAC-J, 1 mM DMOG, or the combination of both for 16 hours. Relative expression of HIF-1α and actin was followed by Western blot analysis. Results are representative for 3 individual experiments. (C) RAW264.7 cells were treated with CMAC-J for 1 to 24 hours. (D) RAW264.7 cells were treated with CMAC-J, the supernatant of necrotic (CMNC-J) or viable (CMVC-J) Jurkat cells as well as CMAC-4T1 for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of HIF-1α versus ribosomal 16S protein mRNA under control conditions was set to 1. (E) RAW264.7 cells were transfected with the HIF-1α–luciferase promoter plasmid and treated with 10 μg/mL LPS, CMAC-J, or CMAC-MCF7 for 16 hours. Luciferase activity was measured and normalized to protein. Control conditions were set to 1. Data (C-E) are the mean ± SD (n ≥ 3). *Significant alterations compared with controls.
HIF-1α mRNA and protein accumulation in macrophages by CM. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 4 to 24 hours. (B) RAW264.7 cells were treated with CMAC-J, 1 mM DMOG, or the combination of both for 16 hours. Relative expression of HIF-1α and actin was followed by Western blot analysis. Results are representative for 3 individual experiments. (C) RAW264.7 cells were treated with CMAC-J for 1 to 24 hours. (D) RAW264.7 cells were treated with CMAC-J, the supernatant of necrotic (CMNC-J) or viable (CMVC-J) Jurkat cells as well as CMAC-4T1 for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of HIF-1α versus ribosomal 16S protein mRNA under control conditions was set to 1. (E) RAW264.7 cells were transfected with the HIF-1α–luciferase promoter plasmid and treated with 10 μg/mL LPS, CMAC-J, or CMAC-MCF7 for 16 hours. Luciferase activity was measured and normalized to protein. Control conditions were set to 1. Data (C-E) are the mean ± SD (n ≥ 3). *Significant alterations compared with controls.
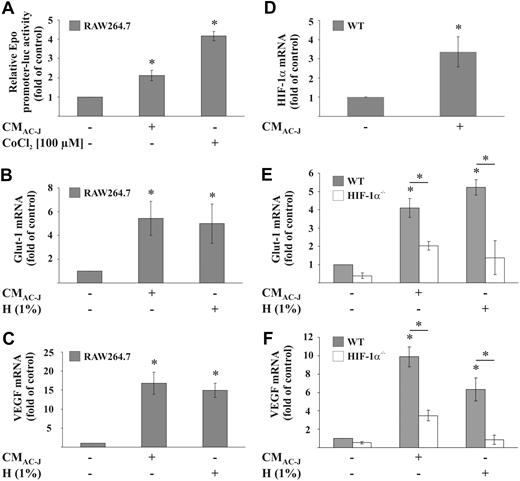
CM induces HIF-1 activity in RAW264.7 and primary mouse macrophages. (A) RAW264.7 cells were transfected with the pGL-3xEPO-HRE plasmid and treated with 100 μM CoCl2 or the supernatant of apoptotic Jurkat cells (CMAC-J) for 16 hours. Control conditions were set to 1. (B-C) RAW264.7 cells or (E-F) primary mouse macrophages were incubated under 1% hypoxia (H) or treated with CMAC-J for 16 hours. Glut-1 (B,E) and VEGF (C,F) as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of ribosomal 16S protein versus Glut-1 or VEGF mRNA under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
CM induces HIF-1 activity in RAW264.7 and primary mouse macrophages. (A) RAW264.7 cells were transfected with the pGL-3xEPO-HRE plasmid and treated with 100 μM CoCl2 or the supernatant of apoptotic Jurkat cells (CMAC-J) for 16 hours. Control conditions were set to 1. (B-C) RAW264.7 cells or (E-F) primary mouse macrophages were incubated under 1% hypoxia (H) or treated with CMAC-J for 16 hours. Glut-1 (B,E) and VEGF (C,F) as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of ribosomal 16S protein versus Glut-1 or VEGF mRNA under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
To gain information as to whether the increase in HIF-1α protein stimulates HIF-1 transcriptional activity, we first used an Epo-promoter-luciferase construct, which contains multiple HRE sites. Whereas CMAC-J up-regulated the promoter activity 2-fold, the hypoxic mimetic CoCl2 induced promoter activity approximately 4-fold (Figure 2A). In a second approach, we demonstrated up-regulation of 2 well-described HIF-1 targets, glucose transporter (Glut)–1 and vascular endothelial growth factor (VEGF), after 16 hours of incubation (Figure 2B-C). CMAC-J and hypoxia (1% oxygen) were equally effective, up-regulating Glut-1 6-fold, whereas VEGF was induced approximately 17-fold. To unequivocally prove that CMAC-J causes HIF-1 to up-regulate VEGF and Glut-1, we used primary macrophages from HIF-1α−/− mice compared with wild-type (wt) littermates. Primary wt macrophages showed a 3-fold up-regulation of HIF-1α mRNA after CMAC-J (Figure 2D). Whereas hypoxia and CMAC-J induced mRNA expression of Glut-1 (4-fold induction) and VEGF (10-fold induction) in primary wt macrophages, up-regulation was largely attenuated in primary HIF-1α−/− macrophages (Figure 2E-F). Residual induction in HIF-1α knockout cells might be explained by additional transcription factors regulating either Glut-1 and/or VEGF. Whereas STAT3 may regulate VEGF synergistically with HIF-1α,32 c-Myc and AKT have been shown to regulate Glut-1 (for review, see Ganapathy et al33 ).
CMAC-activated macrophages caused differentiation of embryonic stem cells
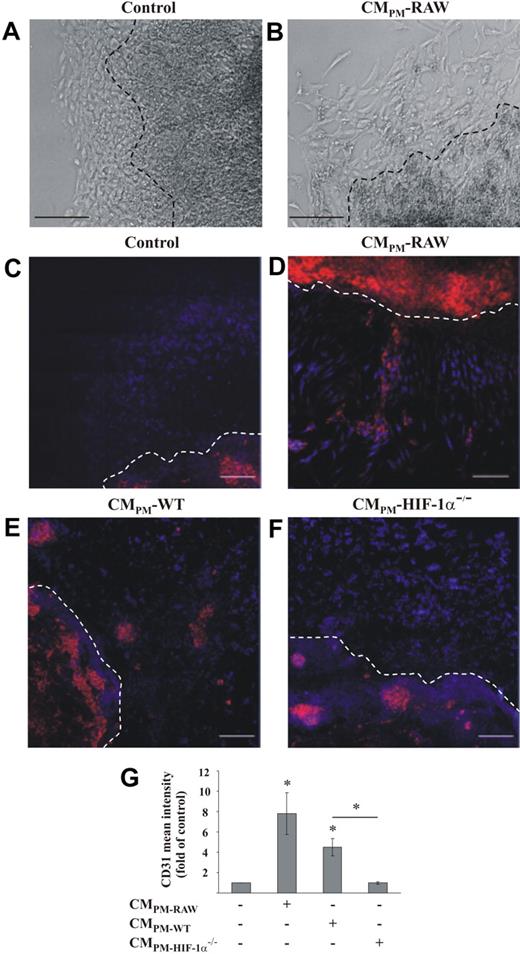
Having established that conditioned medium of AC activated HIF-1 in macrophages, we were interested to determine functional consequences. Therefore, we exposed macrophages for 16 hours to CMAC-J, removed the media, and continued incubations with fresh media for 5 hours. The supernatant from these polarized macrophages, termed (CMPM), was harvested either from RAW264.7 macrophages, primary HIF-1α+/+, or HIF-1α−/− macrophages, and EB were incubated for 24 hours with these supernatants. Plating EB on Lumox dishes initiated growth of a monolayer rim of cells around the multilayer body of stem cells (Figure 3A-B). Hannig et al34 showed that in differentiating embryonic stem cells, CD31 expression peaked at day 6–8 after retrieval of LIF. We monitored cell differentiation at day 6. Using CMPM from RAW264.7 cells, we noticed morphologic alterations of stem cells in the monolayer zone compared with standard culture medium (Figure 3A-B). More defined than differences in the phase-contrast pictures, we observed an increase in CD31-positive cells after adding CMPM from RAW264.7 macrophages compared with controls (Figure 3C-D). Under control conditions, autodifferentiation into CD31-positive cells in the multilayer body of EB was noticed (data not shown). Differentiation toward CD31-positive cells was strongly induced not only in the multilayer body, but also in the monolayer zone surrounding the EB when incubated with CMPM from RAW264.7 cells (Figure 3D). Interestingly, we observed no differentiation toward CD31-positive cells in the monolayer zone under control conditions (Figure 3C). Therefore, we only focused on CD31 differentiation in the monolayer zone. Treatment of stem cells with the supernatant of RAW264.7 cells, not previously exposed to CMAC, revealed no differences compared with the exposure with standard culture medium (data not shown). When primary mouse macrophages instead of RAW264.7 were treated with CMAC-J to generate CMPM, formation of CD31-positive cells became apparent in the monolayer zone (Figure 3E). Stem cells incubated with CMPM derived from HIF-1α−/− macrophages did not differentiate to CD31-positive cells in the monolayer zone (Figure 3F).
CM enhances differentiation of EB into CD31-positive cells. Plated EB were treated either with (A,C) control medium or the supernatant of macrophages, which were prestimulated for 16 hours with the supernatant of apoptotic Jurkat cells and then incubated for 5 hours with fresh medium (CMPM). CMPM was derived from RAW264.7 (B,D), primary mouse wt macrophages (E), or primary mouse HIF-1α−/− macrophages (F), and added to EB for 24 hours. The appearance of CD31-positive cells was monitored by immunofluorescence staining (red). DAPI is stained in blue. Scale bar represents 200 μm (A-B) and 100 μm (C-F). Results are representative for 3 individual experiments. The dotted line indicates the border between the multilayer body and the surrounding monolayer rim of the EB. Original magnifications, ×10 (A-B) and ×40 (C-E). (G) Mean CD31 fluorescence intensity in the monolayer rim of EB was measured, and the ratio of CD31 versus DAPI under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
CM enhances differentiation of EB into CD31-positive cells. Plated EB were treated either with (A,C) control medium or the supernatant of macrophages, which were prestimulated for 16 hours with the supernatant of apoptotic Jurkat cells and then incubated for 5 hours with fresh medium (CMPM). CMPM was derived from RAW264.7 (B,D), primary mouse wt macrophages (E), or primary mouse HIF-1α−/− macrophages (F), and added to EB for 24 hours. The appearance of CD31-positive cells was monitored by immunofluorescence staining (red). DAPI is stained in blue. Scale bar represents 200 μm (A-B) and 100 μm (C-F). Results are representative for 3 individual experiments. The dotted line indicates the border between the multilayer body and the surrounding monolayer rim of the EB. Original magnifications, ×10 (A-B) and ×40 (C-E). (G) Mean CD31 fluorescence intensity in the monolayer rim of EB was measured, and the ratio of CD31 versus DAPI under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
Quantification of the CD31 intensity showed an 8-fold increase with CMPM-RAW and a 4.5-fold increase with CMPM-WT in the monolayer zone compared with controls. Using CMPM-HIF-1α−/− revealed background staining only (Figure 3G). Results to date suggest that up-regulation of HIF-1α by CMAC activated HIF-1 target gene expression in macrophages, which subsequently initiated embryonic stem cell differentiation, as followed by CD31 expression.
Role of S1P in provoking HIF-1α responses
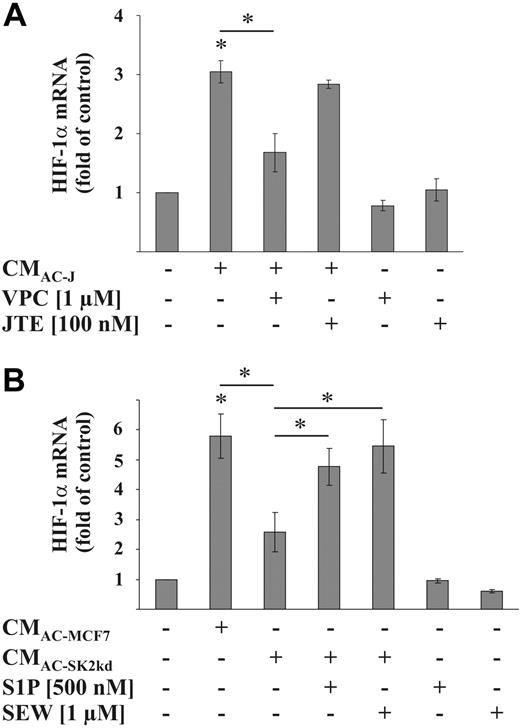
The following set of experiments elucidated molecular details as to how CMAC induced HIF-1α mRNA. Because it is known that S1P is an important mediator released from AC with the ability to regulate macrophage responses, we explored its potential impact on HIF-1 expression. Using VPC23019, a S1P receptor-1 and -3 (S1P1/S1P3) antagonist, up-regulation of HIF-1α mRNA by CMAC-J was attenuated, whereas JTE-013, a S1P receptor-2 (S1P2) antagonist, showed no effect (Figure 4A). To corroborate a role of S1P in up-regulating HIF-1α mRNA, we also used conditioned medium of apoptotic MCF-7 cells (CMAC-MCF7) compared with medium derived from MCF-7 cells with a knockdown of SK2 (CMAC-SK2kd), which is known to contain a reduced amount of S1P (4 ng/mL S1P vs 0.65 ng/mL S1P in the supernatants of 2 × 105 apoptotic MCF-7 respective MCF-7-SK2kd cells/mL13 ; 45 ng/mL S1P in the supernatants from 2.5 × 106 apoptotic Jurkat cells/mL, whereas S1P in the supernatant of viable cells was not detectable9 ). Compared with CMAC-MCF7, which caused a 6-fold increase in HIF-1α mRNA, CMAC-SK2kd only showed a 2- to 3-fold HIF-1α mRNA increase in RAW264.7 macrophages (Figure 4B). The addition of either authentic S1P or SEW2871, a S1P1 agonist, to CMAC-SK2kd restored induction of HIF-1α mRNA compared with values seen with CMAC-MCF7 (Figure 4B). Taking into consideration that CMAC-SK2kd did not lower HIF-1α mRNA to control levels with the further notion that neither S1P nor SEW2871 alone induced HIF-1α mRNA in RAW264.7 cells (Figure 4B), we concluded that additional mediators participated in up-regulating HIF-1α.
Role of S1P in HIF-1α mRNA induction. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 16 hours in the presence or absence of 1 μM VPC23019 or 100 nM JTE-013. VPC23019 and JTE-013 were preincubated for 1 hour. (B) RAW264.7 cells were treated with the supernatant of apoptotic MCF-7 cells (CMAC-MCF7) or the supernatant of apoptotic MCF-7 cells with a knockdown of SK2 (CMAC-SK2kd), with or without the further addition of 500 nM S1P or 1 μM SEW2871 for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of HIF-1α versus ribosomal 16S protein mRNA under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
Role of S1P in HIF-1α mRNA induction. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 16 hours in the presence or absence of 1 μM VPC23019 or 100 nM JTE-013. VPC23019 and JTE-013 were preincubated for 1 hour. (B) RAW264.7 cells were treated with the supernatant of apoptotic MCF-7 cells (CMAC-MCF7) or the supernatant of apoptotic MCF-7 cells with a knockdown of SK2 (CMAC-SK2kd), with or without the further addition of 500 nM S1P or 1 μM SEW2871 for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined by qRT-PCR. The ratio of HIF-1α versus ribosomal 16S protein mRNA under control conditions was set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
TGF-β participated in regulating HIF-1α mRNA expression
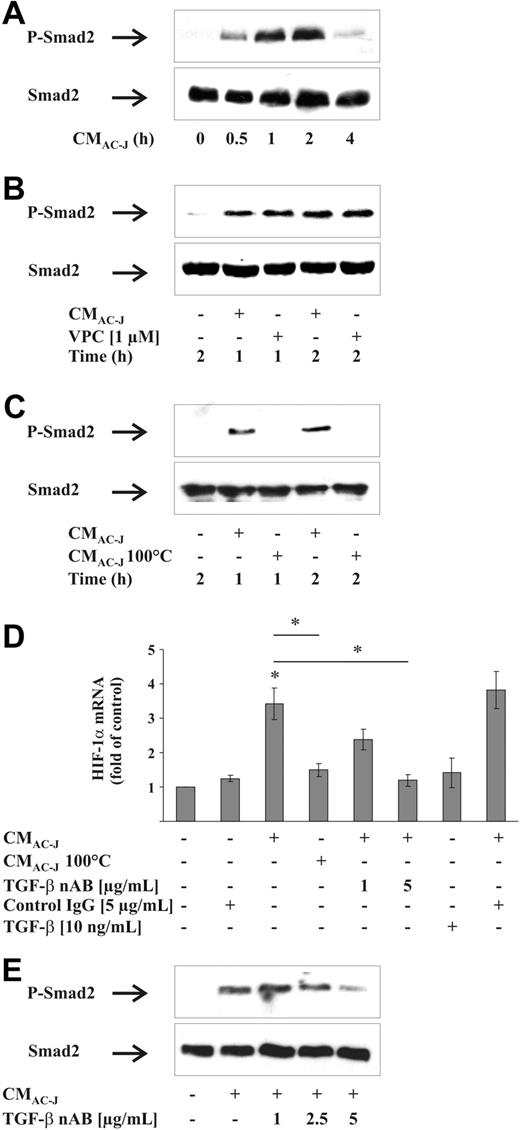
It is established that AC release TGF-β as an immuoregulatory component, but it was also shown that activation of S1P receptors by S1P induces cross-signaling via the TGF-β receptor.35 Therefore, we examined whether TGF-β contributed to increase HIF-1α mRNA content. To prove TGF-β receptor activation, we followed phosphorylation of Smad2, an established TGF-β downstream signaling event. CMAC-J time dependently increased Smad2 phosphorylation in RAW264.7 cells, with most pronounced activation at 1 to 2 hours, whereas total Smad2 remained unchanged (Figure 5A). To exclude activation of the TGF-β receptor due to cross-signaling by the S1P1/S1P3 receptor, we coincubated cells with CMAC-J and VPC23019 for 1 and 2 hours and followed Smad2 phosphorylation, which was not affected by S1P1/S1P3 inhibition (Figure 5B). Furthermore, we incubated cells with CMAC-J or denatured CMAC-J for 1 or 2 hours. Heating CMAC-J at 100°C for 1 hour (CMAC-J 100°C) inactivated proteins, presumably TGF-β, whereas lipids such as S1P should remain active. Measurements of S1P showed equal amounts of S1P in heated versus control CMAC-J (data not shown). Denaturated CMAC-J failed to phosphorylate Smad2 (Figure 5C), suggesting the involvement of a heat-labile factor. To check whether in our system TGF-β receptor and S1P receptor signaling interfere, we treated RAW264.7 cells with 10 ng/mL TGF-β in the absence/presence of VPC23019 for 1 or 2 hours. Blocking S1P receptors did not interfere with TGF-β receptor-mediated Smad2 phosphorylation (data not shown). The use of VPC23019 (Figure 5B) and denaturated CMAC-J (Figure 5C) excluded cross-activation of the TGF-β receptor by S1P. Along this line, denaturated CMAC-J did not induce HIF-1α mRNA (Figure 5D). Supporting experiments revealed that the TGF-β neutralizing AB (nAB) lowered the expression of HIF-1α mRNA in response to CMAC-J, whereas an isotype IgG control AB had no effect on HIF1-α mRNA (Figure 5D). However, as shown for S1P, TGF-β added alone failed to induce the HIF-1α mRNA content (Figure 5D). To verify the involvement of TGF-β, the decrease in Smad2 phosphorylation without altering total Smad2 expression due to the use of the TGF-β nAB with CMAC-J was monitored (Figure 5E). Apparently, besides S1P, also TGF-β regulates HIF-1α mRNA expression in response to the supernatant of AC.
Role of TGF-β in HIF-1α mRNA induction. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 30 minutes to 4 hours. (B) RAW264.7 cells were treated with CMAC-J with or without 1 μM VPC23019 for 1 or 2 hours. VPC23019 was preincubated for 1 hour. (C) RAW264.7 cells were treated with CMAC-J or with denatured CMAC-J (CMAC-J 100°C) for 1 or 2 hours. Relative expression of Smad2 and the phosphorylation of Smad2 (P-Smad2) were followed by Western analysis. Results are representative for 3 individual experiments. (D) RAW264.7 cells were treated with CMAC-J in the presence/absence of 1 or 5 μg/mL TGF-β nAB, with 5 μg/mL control IgG, with CMAC-J 100°C, or with 10 ng/mL TGF-β for 16 hours. The nAB and the control IgG were preincubated with CMAC-J for 1 hour at 37°C. HIF-1α as well as ribosomal 16S protein mRNAs were determined by qRT-PCR. For details, see Figure 4. (E) RAW264.7 cells were treated with CMAC-J with or without increasing concentrations of a TGF-β nAB for 2 hours. The nAB was preincubated with CMAC-J for 1 hour at 37°C. Relative expression of Smad2 and P-Smad2 was followed by Western analysis. Results are representative for 3 individual experiments.
Role of TGF-β in HIF-1α mRNA induction. (A) RAW264.7 cells were treated with the supernatant of apoptotic Jurkat cells (CMAC-J) for 30 minutes to 4 hours. (B) RAW264.7 cells were treated with CMAC-J with or without 1 μM VPC23019 for 1 or 2 hours. VPC23019 was preincubated for 1 hour. (C) RAW264.7 cells were treated with CMAC-J or with denatured CMAC-J (CMAC-J 100°C) for 1 or 2 hours. Relative expression of Smad2 and the phosphorylation of Smad2 (P-Smad2) were followed by Western analysis. Results are representative for 3 individual experiments. (D) RAW264.7 cells were treated with CMAC-J in the presence/absence of 1 or 5 μg/mL TGF-β nAB, with 5 μg/mL control IgG, with CMAC-J 100°C, or with 10 ng/mL TGF-β for 16 hours. The nAB and the control IgG were preincubated with CMAC-J for 1 hour at 37°C. HIF-1α as well as ribosomal 16S protein mRNAs were determined by qRT-PCR. For details, see Figure 4. (E) RAW264.7 cells were treated with CMAC-J with or without increasing concentrations of a TGF-β nAB for 2 hours. The nAB was preincubated with CMAC-J for 1 hour at 37°C. Relative expression of Smad2 and P-Smad2 was followed by Western analysis. Results are representative for 3 individual experiments.
NFAT induced up-regulation of HIF-1α mRNA
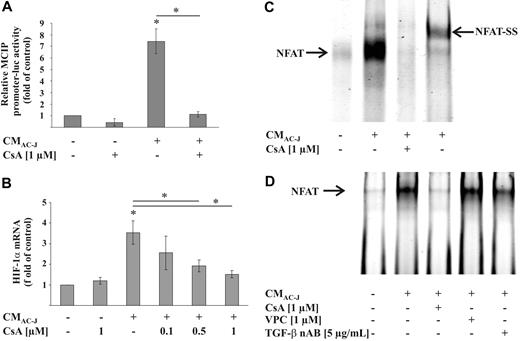
Although TGF-β activated Smad2, no repeats of AGAC or GTCT sequences, described as Smad-binding elements,36 were located in the HIF-1α promoter. Therefore, a direct impact of Smad2 downstream of TGF-β signaling in affecting HIF-1α mRNA was rather unlikely. Notably, NF-κB is established to induce HIF-1α transcription. We used the NF-κB inhibitors Bay11-7082 as well as SN50. Although blocking NF-κB in our system, these inhibitors did not interfere with CMAC-J in elevating the mRNA of HIF-1α (data not shown). Because NFATc1 up-regulated HIF-1α mRNA in mast cells, we examined whether CMAC-J induced NFAT in macrophages. A NFAT-dependent MCIP-luciferase construct was approximately 8-fold up-regulated by CMAC-J (Figure 6A). Considering that NFAT activation requires dephosphorylation by calcineurin, we used CsA to interfere with the phosphatase reaction. CsA reversed CMAC-J-dependent induction of MCIP reporter activity (Figure 6A). To prove the importance of NFAT in up-regulating HIF-1α mRNA in response to CMAC-J, we added increasing concentrations of CsA in combination with CMAC-J and noticed a gradual decrease in HIF-1α mRNA expression (Figure 6B). To show direct binding of NFAT to the HIF-1α promoter, we used the described NFAT-binding sequence of the HIF-1α promoter for EMSA analysis.23 Treatment of RAW264.7 cells with CMAC-J enhanced binding of NFAT to the promoter sequence, which again could be reversed when cells were exposed to CsA (Figure 6C). Supershift experiments with the NFATc1 AB proved the specificity of the oligonucleotide-protein complex. Using a NF-κB p50 AB as a negative control during supershift analysis did not affect NFAT binding, thus substantiating specificity (data not shown). Unfortunately, neither adding S1P or TGF-β alone or in combination, nor blocking the S1P1/S1P3 receptor with VPC23019 or neutralizing TGF-β affected the NFAT activity of the MCIP reporter (data not shown). EMSA analysis revealed a slight reduction of CMAC-J-induced NFAT binding when TGF-β was neutralized, whereas binding was not affected by VPC23019 (Figure 6D). Further experiments need to analyze the cross-talk between multiple signaling pathways using NFAT to up-regulate HIF-1α mRNA.
Role of NFAT in HIF-1α mRNA expression. (A) RAW264.7 cells were transfected with a NFAT-dependent reporter and prl-cmv-renilla plasmid and treated with the supernatant of apoptotic Jurkat cells (CMAC-J) with our without 1 μM CsA for 16 hours or with 1 μM CsA alone. Luciferase activity, normalized to Renilla activity, was measured. (B) RAW264.7 cells were treated with CMAC-J with or without increasing concentrations of CsA for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined, as described in Figure 4. (C) RAW264.7 cells were treated with CMAC-J with or without 1 μM CsA for 4 hours. Supershift analysis was performed with the addition of the NFATc1 AB. (D) RAW264.7 cells were treated with CMAC-J with or without 1 μM CsA, 1 μM VPC23019, or 5 μg/mL TGF-β nAB for 4 hours. The TGF-β nAB was preincubated with CMAC-J for 1 hour at 37°C. Binding of NFAT to a HIF-1α promoter sequence was analyzed by EMSA using a specific 5′-IRD700-labeled oligonucleotide. CsA and VPC23019 were preincubated for 1 hour. Results are representative for 3 individual experiments.
Role of NFAT in HIF-1α mRNA expression. (A) RAW264.7 cells were transfected with a NFAT-dependent reporter and prl-cmv-renilla plasmid and treated with the supernatant of apoptotic Jurkat cells (CMAC-J) with our without 1 μM CsA for 16 hours or with 1 μM CsA alone. Luciferase activity, normalized to Renilla activity, was measured. (B) RAW264.7 cells were treated with CMAC-J with or without increasing concentrations of CsA for 16 hours. HIF-1α as well as ribosomal 16S protein mRNA were determined, as described in Figure 4. (C) RAW264.7 cells were treated with CMAC-J with or without 1 μM CsA for 4 hours. Supershift analysis was performed with the addition of the NFATc1 AB. (D) RAW264.7 cells were treated with CMAC-J with or without 1 μM CsA, 1 μM VPC23019, or 5 μg/mL TGF-β nAB for 4 hours. The TGF-β nAB was preincubated with CMAC-J for 1 hour at 37°C. Binding of NFAT to a HIF-1α promoter sequence was analyzed by EMSA using a specific 5′-IRD700-labeled oligonucleotide. CsA and VPC23019 were preincubated for 1 hour. Results are representative for 3 individual experiments.
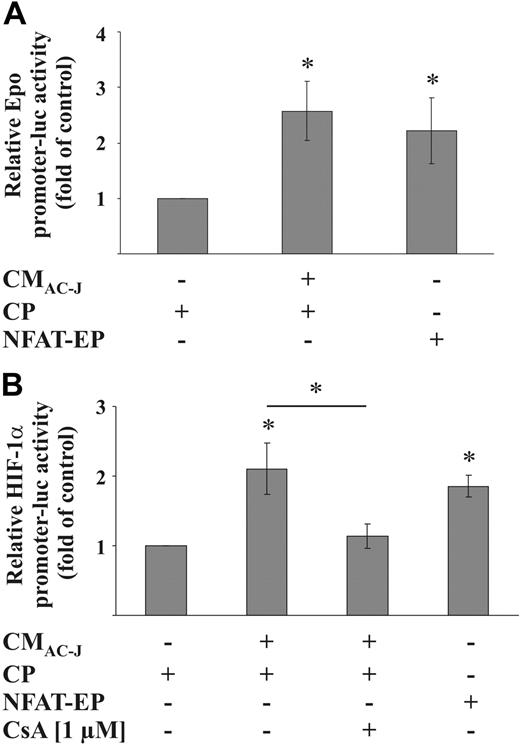
To strengthen the role of NFAT in up-regulating HIF-1α mRNA and activating HIF-1, RAW264.7 cells were cotransfected with either the Epo-promoter-luciferase construct or the HIF-1α-promoter-luciferase construct and a NFAT expression plasmid. NFAT expression was controlled with the MCIP reporter plasmid (data not shown). Overexpression of NFAT increased Epo-reporter activity (Figure 7A) and activated the HIF-1α promoter construct (Figure 7B). The CMAC-J-induced HIF-1α promoter activity was sensitive to CsA treatment and, thus, NFAT dependent (Figure 7B).
NFAT induces HIF-1 activity and the HIF-1α promoter. (A) RAW264.7 cells were transfected with the pGL-3xEPO-HRE plasmid, cotransfected with a control plasmid (CP) or an NFAT expression plasmid (NFAT-EP), treated with CMAC-J, or remained as controls for 16 hours. (B) RAW264.7 cells were transfected with the HIF-1α-luciferase promoter plasmid, cotransfected with a control plasmid (CP) or the NFAT-EP, and treated in the absence or presence of CsA (preincubated for 1 hour) with CMAC-J, or remained as controls for 16 hours. Luciferase activity was normalized to protein. Control conditions were set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
NFAT induces HIF-1 activity and the HIF-1α promoter. (A) RAW264.7 cells were transfected with the pGL-3xEPO-HRE plasmid, cotransfected with a control plasmid (CP) or an NFAT expression plasmid (NFAT-EP), treated with CMAC-J, or remained as controls for 16 hours. (B) RAW264.7 cells were transfected with the HIF-1α-luciferase promoter plasmid, cotransfected with a control plasmid (CP) or the NFAT-EP, and treated in the absence or presence of CsA (preincubated for 1 hour) with CMAC-J, or remained as controls for 16 hours. Luciferase activity was normalized to protein. Control conditions were set to 1. Data are the mean ± SD (n ≥ 3). *Significant alterations compared with controls (or otherwise as indicated).
Conclusively, CMAC-J activated NFAT, which induced the HIF-1α promoter to up-regulate HIF-1α mRNA expression. Consequently, HIF-1 released proangiogenic factors from macrophages.
Discussion
Macrophages display a remarkable plasticity and alter their phenotype in response to environmental signals. In the tumor microenvironment, macrophages known as TAMs are critical players stimulating angiogenesis and exhibiting an anti-inflammatory and antiapoptotic regulatory phenotype. As TAMs often reside in hypoxic tumor environments, it seems rational to allude this to their HIF-1 activity,1 although TAMs are not necessarily localized in hypoxic regions.37 We provide evidence that supernatants of AC activate NFAT to induce transcriptional regulation of HIF-1α, and that S1P in combination with TGF-β participates in the regulation of HIF-1α mRNA. Activation of HIF-1 even under normoxic conditions contributes to TAM polarization by inducing VEGF and stimulating differentiation of embryonic stem cells to become CD31 positive.
Hypoxia with the activation of HIF-1 is crucial for tumor growth, but its distinct role in TAMs is not fully explored. Recently, we noticed that the supernatant of AC (CMAC) stimulated STAT1-dependent VEGF production within 1 hour in human macrophages.38 In this study, we used CMAC to polarize mouse macrophages for 16 hours and found an increase in HIF-1α mRNA, protein, and HIF-1 activity. Up-regulation of HIF-1α mRNA was also evident in primary mouse as well as primary human macrophages. Experiments in macrophages from leukocyte conditional HIF-1α−/− mice established a causative role of HIF-1 in transmitting CMAC responses. It appears that VEGF is regulated by distinct pathways in response to CMAC. Whereas early signals were transmitted by STAT1, late signals used established circuits via HIF-1.
The function of TAMs in the tumor microenvironment determines tumor progression by inducing the angiogenic switch. To investigate consequences of HIF-1 activation in macrophages in response to CMAC, the supernatant of polarized macrophages (CMPM) was used in a functional readout. We incubated EB with CMPM to follow their differentiation toward CD31-positive cells. CD31 is an established marker for endothelial cell differentiation and is used in similar assays with stem cells as a readout for vasculogenesis and angiogenesis.34,39 As reported by others,39 we noticed autodifferentiation into CD31-positive cells in the multilayer body of the EB. Interestingly, we observed no differentiation toward CD31-positive cells in the monolayer zone surrounding the EB center, unless EB were treated with CMPM. With the knockout of HIF-1α in macrophages, differentiation to CD31-positive cells by CMPM in the monolayer zone was completely blocked. This implies functional consequences of HIF-1 activation in macrophages toward the production of potential proangiogenic factors such as VEGF. These results add to our knowledge as to how macrophages respond to AC and how regulatory macrophage subsets such as TAMs may use HIF-1 to deliver differentiation signals. Early in tumor development, macrophages attack tumor cells to kill them. In turn, apoptotic tumor cells as well as tumor-derived cytokines reprogram macrophages to support rather than to oppose tumor progression.1,2,7-10,12,13,30,40,41 As shown in our study, the release of tumor-promoting molecules such as VEGF is not restricted to a hypoxic compartment, but tightly linked to HIF-1α expression. This may point to a crucial role of HIF-1 in macrophages in delivering a molecular signature characterizing TAMs.
In this study, we identified S1P and TGF-β as relevant mediators released from AC that up-regulated HIF-1α mRNA and protein. The impact of S1P and S1P1 in tumor angiogenesis was illustrated in an in vivo model, in which an anti-S1P1 AB attenuated S1P-induced angiogenesis.18 Furthermore, it has been reported that the S1P1 homozygous knockout is lethal because of incomplete vessel development.19 Our experiments with S1P receptor antagonists suggested a role of S1P1 and/or S1P3 activation in triggering HIF-1α expression. Considering that mouse macrophages only express S1P1 and S1P214 points to a crucial role of S1P1 in our system. S1P has been linked to increased VEGF expression, with uncertainties concerning signal transduction pathways.42 CMAC-SK2kd, which contains substantially less S1P,13 did not initiate HIF-1α transcription, pointing to HIF-1 as the missing link between S1P and VEGF up-regulation. However, the notion that neither a specific S1P1 agonist nor S1P alone enhanced HIF1-α mRNA, and the further indication that both stimuli were active in combination with CMAC-SK2kd, suggests that additional factors besides S1P are needed.
After Smad2 phosphorylation by CMAC, an established TGF-β downstream signaling event, we corroborated the release of TGF-β from AC.4 Whereas TGF-β often restricts tumor growth and angiogenesis in the early phase of tumor development, there is evidence for its proangiogenic and tumor-promoting activities during late-stage tumor progression.43 TGF-β–induced angiogenesis was linked to urokinase-type plasminogen activator receptor and its type 1 inhibitor up-regulation.43 Furthermore, TGF-β was shown to up-regulate VEGF-C.44 Our results suggest that both S1P and TGF-β are present in the CMAC and are needed for HIF-α mRNA expression regulation in macrophages. As neither TGF-β nor S1P alone or in combination was sufficient in inducing HIF-1α, we assume that combinatory and thus more complex signaling circuits are operating.
More recent studies identified NF-κB, STAT3, and NFAT as transcriptional regulators of HIF-1α.22-24 Having excluded a contribution of NF-κB in our system, we concentrated on STAT3 and NFAT. Whereas STAT3 binding to the HIF-1α promoter sequence in EMSA analysis was not enhanced by CMAC (data not shown), NFAT binding as well as NFAT activity were strongly induced. Both S1P receptor activation and TGF-β have been shown to activate calcineurin and thereby to stimulate NFAT.45,46 NFAT is known for its important role in T-cell development, T helper–cell differentiation, and T-cell activation. However, recently, NFAT has also been shown to induce cytokines or genes associated with cancer development and angiogenesis (reviewed by Lu and Huan47 ). Our results add to these findings and define a molecular pathway showing that NFAT might use HIF-1 to promote angiogenesis. Unfortunately, we were unable to directly link either S1P or TGF-β to NFAT activation. We assume additional factors to be released from apoptotic cells, which contribute to HIF-1α mRNA regulation. Adenosine might be released from apoptotic cells, and it has been shown that HIF-1α mRNA can be up-regulated via activation of adenosine A2a receptor.48 Recently, we showed that CMAC-J induced the adenosine A2a receptor in a S1P/heme oxygenase-1–dependent manner.38 Furthermore, PGE2 induced HIF-1α mRNA,49 and we reported that PGE2 is released from macrophages after treatment with CMAC-J.30 These results underline the complexity of signals delivered by AC, and extensive new sets of experiments will be needed to analyze details of the signaling cross-talk initiated during CMAC-induced macrophage polarization.
In this study, we could show that the interaction of macrophages with dying tumor cells provokes macrophage activation with features that characterize TAM, such as providing proangiogenic signals. Transcriptional regulation of HIF-1α mRNA by AC-derived mediators such as S1P and TGF-β offers an explanation to understand macrophage polarization in the tumor microenvironment even under normoxic conditions.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Deutsche Forschungsgemeinschaft (SFB 815, FOG 784, Excellence Cluster Cardiopulmonary System), Deutsche Krebshilfe, Sander Foundation (2007.070.1), LOEWE (LiFF) program, and European Community (PROLIGEN).
Authorship
Contribution: C.W. performed qRT-PCR experiments; D.N. and J.Z. carried out NFAT and Epo-luc reporter analysis; H.M. contributed to the EB assay and assisted in data analysis; A.W. performed the SK2 knockdown in MCF-7 cells and supplied CMAC-MCF7 and CMAC-SK2kd; B.H. designed and performed experiments and analyzed data; N.D. assisted with the design of experiments and data analysis; B.B. supervised the project; and B.H. and B.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernhard Brüne, Goethe-University, Faculty of Medicine, Biochemistry I, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany; e-mail: bruene@pathobiochemie1.de.