Abstract
Acute myelogenous leukemia is driven by leukemic stem cells (LSCs) generated by mutations that confer (or maintain) self-renewal potential coupled to an aberrant differentiation program. Using retroviral mutagenesis, we identified genes that generate LSCs in collaboration with genetic disruption of the gene encoding interferon response factor 8 (Irf8), which induces a myeloproliferation in vivo. Among the targeted genes, we identified Mef2c, encoding a MCM1-agamous-deficiens-serum response factor transcription factor, and confirmed that overexpression induced a myelomonocytic leukemia in cooperation with Irf8 deficiency. Strikingly, several of the genes identified in our screen have been reported to be up-regulated in the mixed-lineage leukemia (MLL) subtype. High MEF2C expression levels were confirmed in acute myelogenous leukemia patient samples with MLL gene disruptions, prompting an investigation of the causal interplay. Using a conditional mouse strain, we demonstrated that Mef2c deficiency does not impair the establishment or maintenance of LSCs generated in vitro by MLL/ENL fusion proteins; however, its loss led to compromised homing and invasiveness of the tumor cells. Mef2c-dependent targets included several genes encoding matrix metalloproteinases and chemokine ligands and receptors, providing a mechanistic link to increased homing and motility. Thus, MEF2C up-regulation may be responsible for the aggressive nature of this leukemia subtype.
Introduction
Acute myelogenous leukemia (AML) is maintained by a small population of leukemic stem cells (LSCs), which share 2 main characteristics with hematopoietic stem cells (HSCs): (1) self-renewal ability, thereby creating daughter cells with the capacity to replicate almost indefinitely, and (2) differentiation capacity, by which progressively differentiating cells are generated and which is responsible for the cellular heterogeneity found in leukemia.1,2 Fusion proteins generated from translocations involving the MLL gene on chromosome 11q23, which are associated with infant mixed-lineage leukemia (MLL) and adult AML with poor prognosis, have been shown to generate LSCs from committed myeloid progenitors.3,4 MLL regulates gene transcription at the chromatin level and MLL leukemias are invariably associated with aberrant expression of selected HOX and TALE homeobox genes, which have been implicated in the establishment of the LSCs in a cooperative manner.5-7 More recently, the Mef2c transcription factor, encoded by a member of the MCM1-agamous-deficiens-serum response factor (MADS) family of homeotic genes, has also been implicated in the establishment of MLL-induced LSCs.3 Nevertheless, the role of these proteins or other downstream effectors in the conversion of a progenitor cell into an LSCs capable of inducing an aggressive clinical leukemia is not well defined.
We initiated this study to identify novel mechanisms in the generation of AML LSCs using a genetic approach, in which mice deficient for the interferon regulatory factor-8 (Irf8) were infected with murine leukemia virus (MuLV) to induce acute leukemia. Irf8−/− mice spontaneously develop a chronic myeloproliferative syndrome,8 most probably resulting from the combined effects of impaired apoptosis and increased sensitivity to proliferation stimuli in the myeloid compartment.9-11 Infection with MuLV would be expected to accelerate the progression to an acute phase by activating cooperating oncogenes (or inactivating tumor suppressors) through random integration into the cellular DNA, resulting in the selective outgrowth of LSCs that feed an overt AML. Using this approach, we identified integrations in several genes that have been implicated in the generation of LSCs by an MLL-fusion protein, such as HoxA9, Meis1, and Myb, but also Mef2c. This result prompted us not only to confirm the cooperative action of Mef2c overexpression and Irf8 deficiency, but also to explore the correlation between Mef2c and MLL.
Mef2c is 1 of 4 myocyte enhancer factor 2 (MEF2) proteins, which make up 1 of 3 distinct classes of MADS-box transcription factors. Mef2 proteins are central regulators of diverse developmental programs,12 although a role in hematopoiesis was unappreciated until recently. We have previously demonstrated that Mef2c is an important determinant of myeloid cell fate,13 a result supported by the demonstration that Mef2c is regulated by the microRNA-223 during granulocytic differentiation.14 In addition, a critical role of Mef2c in mature B-cell proliferation, survival, and homeostasis has recently been demonstrated.15-17
We show here that Mef2c induces myelomonocytic leukemia in cooperation with Irf8 deficiency, and also confirm that high Mef2c expression levels are found in patient samples with disrupted MLL. However, taking advantage of mice in which Mef2c can be inducibly inactivated, we show that Mef2c is dispensable for both establishment and maintenance of LSCs generated by MLL fusion proteins. Instead, we provide evidence that demonstrates the importance of Mef2c for modulating the homing and invasive properties of MLL-AML.
Methods
Mouse strains
The generation of conditional (Mef2cfl) and nonconditional (Mef2c−) inactivating alleles of Mef2c has been previously described.18,19 Mef2cfl/+ and Mef2c−/+ mice were backcrossed into the C57BL/6J PtprccPepca (here called B6) mouse strain (for > 10 generations) and then crossed to generate Mef2cfl/− and Mef2c+/+ B6 littermates, which were used for these studies. To induce Mef2c excision, mice were crossed to MxCre B6 mice20 and newborns were injected with a double dose of 500 μg polyinosinic-polycytidylic (pIpC) acid (Sigma-Aldrich) dissolved in phosphate-buffered saline (PBS). Alternatively, bone marrow (BM) cells were infected with a retroviral vector coexpressing the Cre recombinase and enhanced green fluorescent protein (eGFP) (pSF91-CRE-iGFP; R860).21 In transplanted mice, pIpC (300 μg) was injected 3 times at 2-day intervals, as indicated. To validate Mef2c excision, DNA from blood or harvested cells was subjected to polymerase chain reaction (PCR), as previously described.13 B6.SJL-PtprcaPep3b/BoyJ (here called B6-Ptprca) mice were used as hosts for all BM transplantations. Irf8−/− B6 mice8 (also known as Icsbp−/−) and NOD.CB17-Prkdcscid/J mice (here called NOD/scid) were originally obtained from Ivan Horak (Leibniz Institute of Molecular Pharmacology, Berlin, Germany) and Charles River Laboratories, respectively, and were maintained in the Heinrich-Pette-Institut animal facilities. All animal experiments were approved by the Commission for Animal Experiments at the Heinrich-Pette-Institute together with Hamburg Office of Health. Survival curves of experimental animals were calculated by the method of Kaplan-Meier, and statistical analysis was performed using the SPSS Version 15.0 software.
MuLV infections and isolation of integration sites
Newborn Irf8−/− B6 mice were infected intraperitoneally with approximately 5 × 105 infectious units of Moloney (Mo)–MuLV harvested from SC1 fibroblasts as previously described.22 Retroviral integration sites were isolated from genomic DNA of tumors using a modified protocol of the ligase-mediated extension-primer tag selection PCR reaction.23
Retroviral gene transduction and BM transplantation
Retroviral vectors were used to transduce the murine Mef2c-α1 isoform (SF91-iGFP)24 or the MLL/ENL fusion protein (MSCVneo,25 MYs-GFP,26 or an MYs-Venus variant) into murine BM progenitors. Retroviral pseudotypes were generated in 293T cells transiently transfected with the retroviral vector plasmid plus plasmids expressing MoMuLV env and gagpol genes. For BM infections, virus particles were fixed onto 35-mm plates coated with Retronectin (Cambrex) before the addition of BM progenitors suspended in StemSpan SFEM medium (StemCell Technologies) supplemented with 1% glutamine, 50 ng/mL mSCF (PeproTech), 100 ng/mL each hFlt3-L and hIL-11 (StemCell Technologies), and 10 ng/mL mIL-3 (Strathmann). Infections were repeated after 24 hours. For Mef2c transductions, BM was isolated from mice that received intraperitoneal injections of 5-fluorouracil (150 mg/kg) 4 days before isolation. For MLL/ENL transductions, BM was isolated from untreated mice and progenitors (Linneg) were enriched using the Midi MAX Lineage Depletion Kit (Miltenyi Biotec).
Transduced BM cells (106) supplemented with untreated whole BM cells (105) were transplantated into lethally irradiated (9Gy) B6-Ptprca mice. For retransplantation experiments, 5 × 106 cells isolated from spleens of diseased mice were injected intravenously into either nontreated recipients. Mice were monitored regularly for disease symptoms. Fluorescence-activated cell sorter (FACS) and histologic analysis of hematopoietic cells and organs were performed as previously described.24,27 The following antibody clones were used: mouse anti-CD44 allophycocyanin (APC)–conjugated (clone IM7), mouse anti-CD184 phycoerythrin (PE)–conjugated (clone CXCR4), mouse anti-CD11b APC-conjugated (clone M1/70), mouse anti-Gr1 PE-conjugated (clone RB6-8C5), and mouse anti-F4/80 PE-conjugated (clone MCA497PEB). Unconjugated anti-CD16/32 (clone 93) was used to block unspecific binding. All micrographs were acquired with an Axioplan 2 microscope (Zeiss) with objectives indicated in figure legends using a ProgRes C12Plus digital camera (Jenoptik) and Image Access5 (release 221) software (Image Bildverarbeitung). Images were processed using Adobe Photoshop CS (Version 8.0.1; Adobe Systems Incorporated).
Establishment of cell M/E cell lines and colony-forming assay
Two independent sets of MLL/ENL expressing cell lines (M/E cells) were generated. In the first set, BM from Mef2cfl/− or Mef2c+/+ littermates was transformed with MSCVneo-MLL/ENL, as previously described.24,28 These were subsequently infected with either with SF91-iGFP or SF91-CREiGFP. The second set was generated from BM from Mef2cfl/−MxCRE and Mef2c+/+MxCRE littermates (treated with pIpC as newborns) and infected with MYs-MLL/ENL-GFP. Transformed cells were isolated by serial replating (7-day intervals) in Methocult M3434 (StemCell Technologies). Single colonies from the quaternary cloning assay were picked at culture in RPMI 1640 medium, supplemented with 10% fetal calf serum, 1% glutamine, 10 ng/mL each mIL3 (Strathmann), mGM-CSF, hIL-6 (PeproTech), and 100 ng/mL mSCF (generously supplied by Ursula Just, University Kiel). Several independent clones were analyzed, but no difference in phenotype or proliferation was observed. For subsequent studies, Mef2cΔ/−M/E cells were infected with MYs-iPac vectors, expressing a puromycin resistance marker alone, or with wild-type Mef2c or a previously characterized histone deacetylase (HDAC)-binding Mef2c mutant29 (Mef2c-VLL65-67ASR, here called Mef2cASR), which retains the ability of Mef2c to induce differentiation (A.S., C.S., unpublished results, January 2009). Both sets of cell lines were analyzed extensively in vitro (differentiation markers, proliferation, and migration assays). The first set (M/E-Mef2cΔ/−-CRE/GFP and M/E-Mef2c+/+-CRE/GFP cells) was used to test tumorigenicity in vivo by intraperitoneal injection of 106 cells into nonirradiated NOD/scid mice.
Gene expression analysis of AML samples and MLL/ENL-transformed cells
A total of 285 AML cases have been analyzed using Affymetrix HGU133A GeneChips (Affymetrix).30 Relative expression values for IRF8 and MEF2C for each patient were used to determine mean values for patient clusters. For gene expression profiling of M/E cells, RNA was isolated from M/E-Mef2cΔ/− cells infected with MYs-iPAC or MYs-pMef2cASR after puromycin selection at 3 different time points, pooled, and hybridized against the Agilent Whole Mouse Genome Microarray 4 × 44K using the one-color service of Miltenyi. Transcript levels were verified by real-time reverse-transcribed (RT)-PCR using the SYBRGreen Reaction Mix (Roche Mannheim) in a Roche Light-Cycler. cDNA levels were normalized against Hprt transcript levels. Primers and amplification conditions are available on request. The microarray data are available in the Gene Expression Omnibus (GEO) public database under accession number GSE17231.
In vivo and in vitro homing and proliferation assays
In vivo hematopoietic cell homing assay was performed as described.31 Briefly, freshly isolated BM cells were labeled with the fluorescent dye carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes), washed, and then 107 cells were injected per recipient mouse irradiated (9 Gy) 1 hour before transplantation. Mice were killed 4 hours after the transplantation, and the percentage of CFSE+ cells in the BM and spleen was determined by FACS. To obtain a statistical reproducibility, at least 7 × 106 events were analyzed for each sample. Transmigration assays were performed using Transwells (5-μm pore size; Costar) in 24-well tissue culture plates. Briefly, 1 to 2 × 105 M/E cells were plated in the upper chamber of each Transwell in medium without chemoattractant. The medium in the lower chambers was supplemented with either 100 ng/mL SDF1α or 1× PBS. Cell numbers were determined after 3 to 5 hours. Cobblestone formation assays were performed by plating 106 M/E-Mef2c/GFP or M/E-GFP cells onto a monolayer of stromal MS-5 cells in 6-well plates (Falcon) for 24 hours.
To determine the proliferation rate in vivo, Lin− BM progenitors isolated from pIpC treated Mef2c+/+MxCre and Mef2c−/flMxCre littermates were infected with MYs-MLL/ENL-Venus. Infected BM cells (1.2 × 106) were transplanted into lethally irradiated (9 Gy) B6-Ptprca mice. Thirteen days after transplantation, mice were injected intraperitoneally with 2 mg bromodeoxyuridine (BrdU) and analyzed 2 hours later. BM cells were sorted for Venus+ cells, and these cells were subsequently stained with the APC BrdU Flow Kit (BD Biosciences) following the manufacturer's instructions.
Results
Identification and confirmation of Mef2c oncogenic activity in a mouse model
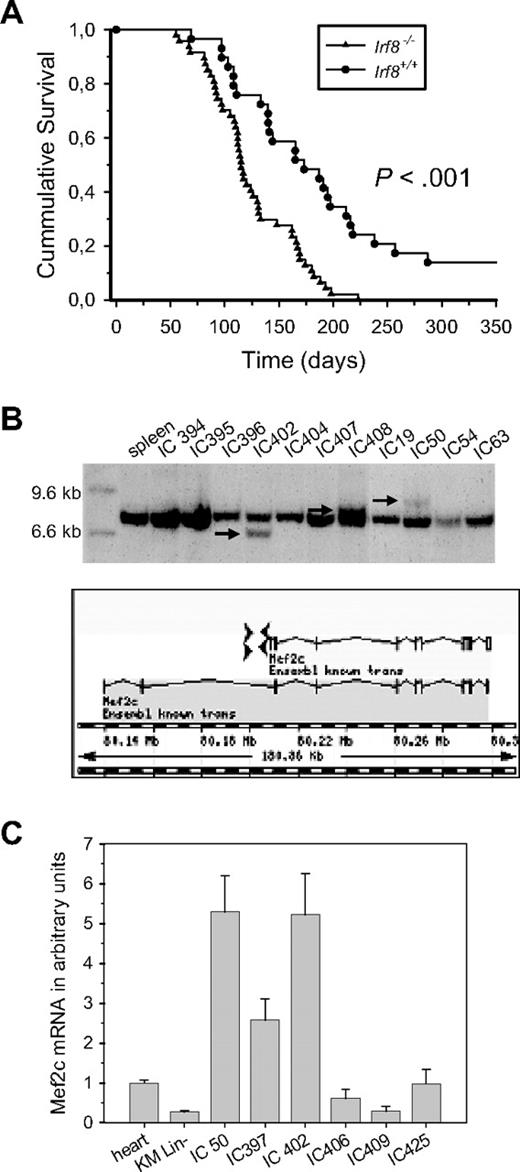
To identify novel genes that cooperate with other genetic mutations to generate AML, we performed retroviral insertional mutagenesis on Irf8−/− mice, which spontaneously develop a mild chronic myeloproliferative disease resulting from Irf8 deficiency.8 Newborn Irf8+/+ and Irf8−/− B6 mice were infected with Mo-MuLV and monitored for leukemia development. MuLV-induced leukemia was accelerated in Irf8−/− compared with Irf8+/+ B6 mice (Figure 1A). Furthermore, although Mo-MuLV induced a T-cell leukemia (CD3+) in 100% of the control mice, myeloid leukemia (CD11b+) was observed in 29% of Irf8−/− mice. These results confirm a cooperative effect between Irf8 deficiency and MuLV insertional mutagenesis. Retroviral integration sites mapping to the HoxA7, HoxA9, Meis1, and Myb gene loci were found in several independent myeloid tumors, similar to other mouse models.32 In addition, integrations in the Mef2c gene locus, mapping within 4 kb upstream of the first coding exon, were found in approximately 20% of the tumors analyzed (Figure 1B). A 5-fold increase in Mef2c mRNA was observed in tumors carrying proviral integrations in the Mef2c locus, compared with tumors with the same phenotype but without apparent disruption of the Mef2c gene (Figure 1C).
Identification of Mef2c as a common integration site in MuLV-induced myeloid tumors of Irf8−/− mice. (A) Kaplan-Meier survival curves of Irf8−/− (n = 47) or Irf8+/+ (n = 28) B6 mice infected with Mo-MuLV. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .001) increase in disease latency in the Irf8-deficient background. The mean survival was 125 (± 6) days and 184 (± 15) days, respectively. (B) Cloning and analysis of sequences flanking retroviral integration sites revealed integrations upstream of the first coding exon of Mef2c, in both 5′ and 3′ orientation to the gene, as indicated. Southern blot analysis of HindIII digested genomic DNA isolated from myeloid tumors originating from MuLV-infected Irf8−/− mice confirmed disruption of the Mef2c locus, as indicated by arrows. (C) Tumor samples with Mef2c integrations have high levels of Mef2c transcripts. The mean levels of Mef2c transcripts in different tumors (IC) relative to the level in heart tissue were determined by quantitative RT-PCR in 2 independent experiments performed in duplicate. Sequence analysis of RT-PCR fragments demonstrated coding sequences for both α1 and α1-β-γ isoforms in normal BM and tumors, the former being the more prominent form.
Identification of Mef2c as a common integration site in MuLV-induced myeloid tumors of Irf8−/− mice. (A) Kaplan-Meier survival curves of Irf8−/− (n = 47) or Irf8+/+ (n = 28) B6 mice infected with Mo-MuLV. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .001) increase in disease latency in the Irf8-deficient background. The mean survival was 125 (± 6) days and 184 (± 15) days, respectively. (B) Cloning and analysis of sequences flanking retroviral integration sites revealed integrations upstream of the first coding exon of Mef2c, in both 5′ and 3′ orientation to the gene, as indicated. Southern blot analysis of HindIII digested genomic DNA isolated from myeloid tumors originating from MuLV-infected Irf8−/− mice confirmed disruption of the Mef2c locus, as indicated by arrows. (C) Tumor samples with Mef2c integrations have high levels of Mef2c transcripts. The mean levels of Mef2c transcripts in different tumors (IC) relative to the level in heart tissue were determined by quantitative RT-PCR in 2 independent experiments performed in duplicate. Sequence analysis of RT-PCR fragments demonstrated coding sequences for both α1 and α1-β-γ isoforms in normal BM and tumors, the former being the more prominent form.
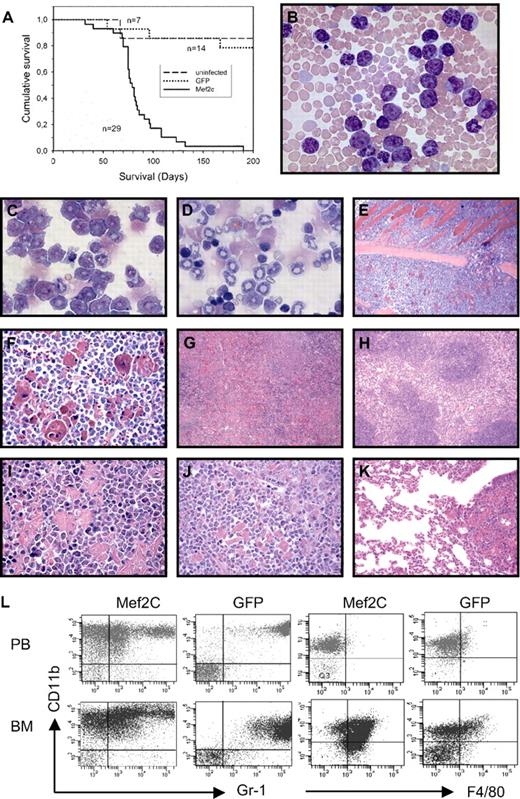
To determine whether constitutive and high expression levels of Mef2c were causative in leukemia development, BM cells from Irf8−/− and Irf8+/+ were infected with retroviral vectors expressing Mef2c and transplanted into syngenic hosts. With a median latency of 79 days after transplantation, all mice receiving Irf8−/− BM expressing Mef2c died of AML (Figure 2A). Moribund mice were characterized by cachexia and dyspnea and had hepatosplenomegaly. Consistent with a diagnosis of acute leukemia, most mice were anemic and peripheral blood (PB) and BM contained a high proportion of blasts and immature monocytic and granulocytic forms (Figure 2B-D). Peripheral white blood counts were highly elevated in 90% of the animals (mean value, 170 × 106 cells/mL; range, 5 × 106-800 × 106 cells/mL) compared with the GFP controls (range, 3.5 × 106-9 × 106). The leukemia was highly invasive, and a striking accumulation of crystal-storing macrophages was observed (Figure 2E-K). Flow cytometric analyses of PB and BM confirmed that 80% to 95% of the organs were positive for both GFP (as a surrogate marker for Mef2c), and the majority of the tumor cells were CD11bhiGr1lo and expressed the monocytic marker F4/80 in the BM compartment, consistent with a high incidence of immature monocytes and blasts (Figure 2L). Notably, no disease was observed over a 9-month period in mice receiving Irf8+/+ BM cells that had been transduced with Mef2c vectors with similar efficiencies (18% and 40% GFP+ cells, 2 independent experiments). These results demonstrate a striking synergy between high Mef2c expression and Irf8 deficiency in the induction of invasive acute myelomonocytic leukemia.
Ectopic expression of Mef2c into Irf8−/− BM progenitors induces acute myelomonocytic leukemia in recipient mice. (A) Kaplan-Meier survival curves of mice receiving Irf8−/− BM cells after infection with retroviral vectors carrying Mef2c/GFP or GFP alone, or uninfected controls, are shown. (B) Blood smear of moribund mouse receiving Mef2c-Irf−/− BM cells shows leukocytosis composed predominately of immature blastlike monocytic and granulocytic cells. The heterochromatic erythrocytes reflect a moderate anemia, confirmed by hematocrit values between 27 and 42 (normal values, 42-48; Pappenheim stain; 63×/1.4 oil). (C-D) Cytospin of BM cells from mice transplanted with either Mef2c/GFP (C) or GFP (D) Irf8−/− BM. The accumulation of blastlike cells and immature monocytes is observed in BM with ectopic Mef2c expression, in contrast to controls, which show the typical increase in granulopoiesis and immature erythropoiesis characteristic of Irf8−/− mice. In addition to juvenile forms, mature banded granulocytes are the prominent form (Pappenheim stain; 63×/1.4 oil). (E-F) Sternal BM of mouse transplanted with Mef2c-Irf8−/− BM showing considerable hypercellularity associated with left-shifted monocytosis and granulocytosis and reduced numbers of erythroid cells. The sinusoidal vascular system is compressed and obscured and an eruption of leukemic cells through the nutrient foramina, which normally allows vascularization of the BM cavity, is observed, leading to the invasion of leukemic BM cells into the neighboring thoracic musculature (top half of microphotograph; 10×/0.45). Higher magnification clearly shows myeloid blast and immature forms of the invasive cells and the numerous phagocytic cells with an eosinophilic storage phenomenon in their cytoplasm (hematoxylin and eosin stain; 40×/1.3 oil). (G-I) Splenic sections of mice receiving either Mef2c (G,I) or control GFP (H) Irf8−/− BM. The regular splenic architecture observed in control mice (H) is completely effaced in Mef2c mice because of the myeloid hyperplasia of the red pulp. As in the BM, an abundance of pseudo pseudo-Gaucher cells with intracellular accumulation of para-crystalline substance at different stages of development can be observed. Large storage cells showing hemophagocytosis are also present (I). Pockets of erythroid cells are observed, which compensates for the loss of erythropoiesis in the BM (hematoxylin and eosin stain; 10×/0.45 and 63×/1.4 oil). (J-K) The highly invasive behavior of the leukemic cells is evidenced by infiltration into nonhematopoietic tissue. (J) Similar to the spleen, the normal liver lobular architecture is completely destroyed and periportal and perivenous liver parenchyma is replaced by infiltrating leukemic myeloblasts and immature forms, as well as phagocytic histiocytes (hematoxylin and eosin stain; 40×/1.3 oil). (K). Pulmonary infiltration with hematopoietic cells was observed in all animals analyzed. Focal peribronchial and perivascular accumulation of immature myeloid and phagocytic histocytes is seen, as well as the diffuse thickening of the avelolar septa by leukemic infiltration (hematoxylin and eosin stain; 20×/0.75). (L) Flow-cytometric analysis of cells isolated from BM and PB from Mef2C and control GFP-transduced mice. Expression analysis of CD11b, Gr1, and F4/80 confirmed the prominent monocytic component of the AML induced by Mef2c. Only GFP+ cells are shown. The results are representative of all mice analyzed.
Ectopic expression of Mef2c into Irf8−/− BM progenitors induces acute myelomonocytic leukemia in recipient mice. (A) Kaplan-Meier survival curves of mice receiving Irf8−/− BM cells after infection with retroviral vectors carrying Mef2c/GFP or GFP alone, or uninfected controls, are shown. (B) Blood smear of moribund mouse receiving Mef2c-Irf−/− BM cells shows leukocytosis composed predominately of immature blastlike monocytic and granulocytic cells. The heterochromatic erythrocytes reflect a moderate anemia, confirmed by hematocrit values between 27 and 42 (normal values, 42-48; Pappenheim stain; 63×/1.4 oil). (C-D) Cytospin of BM cells from mice transplanted with either Mef2c/GFP (C) or GFP (D) Irf8−/− BM. The accumulation of blastlike cells and immature monocytes is observed in BM with ectopic Mef2c expression, in contrast to controls, which show the typical increase in granulopoiesis and immature erythropoiesis characteristic of Irf8−/− mice. In addition to juvenile forms, mature banded granulocytes are the prominent form (Pappenheim stain; 63×/1.4 oil). (E-F) Sternal BM of mouse transplanted with Mef2c-Irf8−/− BM showing considerable hypercellularity associated with left-shifted monocytosis and granulocytosis and reduced numbers of erythroid cells. The sinusoidal vascular system is compressed and obscured and an eruption of leukemic cells through the nutrient foramina, which normally allows vascularization of the BM cavity, is observed, leading to the invasion of leukemic BM cells into the neighboring thoracic musculature (top half of microphotograph; 10×/0.45). Higher magnification clearly shows myeloid blast and immature forms of the invasive cells and the numerous phagocytic cells with an eosinophilic storage phenomenon in their cytoplasm (hematoxylin and eosin stain; 40×/1.3 oil). (G-I) Splenic sections of mice receiving either Mef2c (G,I) or control GFP (H) Irf8−/− BM. The regular splenic architecture observed in control mice (H) is completely effaced in Mef2c mice because of the myeloid hyperplasia of the red pulp. As in the BM, an abundance of pseudo pseudo-Gaucher cells with intracellular accumulation of para-crystalline substance at different stages of development can be observed. Large storage cells showing hemophagocytosis are also present (I). Pockets of erythroid cells are observed, which compensates for the loss of erythropoiesis in the BM (hematoxylin and eosin stain; 10×/0.45 and 63×/1.4 oil). (J-K) The highly invasive behavior of the leukemic cells is evidenced by infiltration into nonhematopoietic tissue. (J) Similar to the spleen, the normal liver lobular architecture is completely destroyed and periportal and perivenous liver parenchyma is replaced by infiltrating leukemic myeloblasts and immature forms, as well as phagocytic histiocytes (hematoxylin and eosin stain; 40×/1.3 oil). (K). Pulmonary infiltration with hematopoietic cells was observed in all animals analyzed. Focal peribronchial and perivascular accumulation of immature myeloid and phagocytic histocytes is seen, as well as the diffuse thickening of the avelolar septa by leukemic infiltration (hematoxylin and eosin stain; 20×/0.75). (L) Flow-cytometric analysis of cells isolated from BM and PB from Mef2C and control GFP-transduced mice. Expression analysis of CD11b, Gr1, and F4/80 confirmed the prominent monocytic component of the AML induced by Mef2c. Only GFP+ cells are shown. The results are representative of all mice analyzed.
Relatively high levels of MEF2C transcripts in specific AML subtypes
To address the potential relevance of our findings to human disease, we analyzed MEF2C transcript levels in a dataset of leukemic BM and PB cells of 285 persons with AML.30 Previous analysis of this dataset identified 16 groups of AML with distinct gene expression profiles. Surprisingly, we found a strong positive correlation (r = 0.40) between IRF8 and MEF2C gene expression. These results contrast to our experimental system, in which Irf8 deficiency synergized with Mef2c overexpression and thus suggest alternative mechanisms or cooperating partners in human AML. A probable cooperating mechanism is the deregulation of the MLL gene, as suggested by the high mean expression levels of MEF2C in patients carrying MLL gene disruptions, which are found in both patient clusters 1 and 16 (Figure 3). MLL-associated AML is predominately associated with a monocytic phenotype; however, high levels of MEF2C expression were not found in clusters 5 or 9, which also contain high levels of monocytic and myelomonocytic cells, refuting the theory that high MEF2C levels correlate strictly with the differentiation phenotype. A relatively high mean expression level was also found in cluster 10 (with a predominantly M1 phenotype), but this was significantly less than that found in MLL-associated AML. Interestingly, other common integration sites found in our screen (Meis1, HoxA7, HoxA9, and Myb) have also been shown to be important players in MLL-associated leukemogenesis.6,33 Furthermore, the high levels of MEF2C expression in MLL-leukemia samples agree with recent findings in which MEF2C was identified as a direct target of the MLL/AF9 fusion protein within the LSC compartment.3
High levels of MEF2C gene expression found in clusters of AML patients with high incidence of MLL. Shown are pairwise correlations of gene expression profiles between AML patients calculated by OmiViz software (Version 3.6) and viewed by HeatMapper, as previously described.30 The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors indicating higher positive (red) or negative (blue) correlations. Histograms next to each tumor represent expression levels for the IRF8 or MEF2C probe sets (bars are proportional to the level of expression). Six of 16 clusters identified in the cohort of 285 AML patients on the basis of the unsupervised clustering results, and other parameters, such as clinical and molecular characteristics of the AML patients, are indicated.30 These correspond to clusters with a high incidence of t(11q23) (percentage is indicated), French-American-British classifications for myelomonocytic (M4) or monocytic (M5) AML, or aberrant high EVI1 expression. Bar diagram represents the mean expression levels (± SE) of MEF2C for patient samples with known MLL gene rearrangements (MLL) compared with all other patient samples (other) or with that in the patients samples within the indicated clusters. Patient numbers are indicated. P values were determined by the Student t test: **P < .001, *P = .02 compared with patients with 11q23 [MLL] disruptions.
High levels of MEF2C gene expression found in clusters of AML patients with high incidence of MLL. Shown are pairwise correlations of gene expression profiles between AML patients calculated by OmiViz software (Version 3.6) and viewed by HeatMapper, as previously described.30 The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors indicating higher positive (red) or negative (blue) correlations. Histograms next to each tumor represent expression levels for the IRF8 or MEF2C probe sets (bars are proportional to the level of expression). Six of 16 clusters identified in the cohort of 285 AML patients on the basis of the unsupervised clustering results, and other parameters, such as clinical and molecular characteristics of the AML patients, are indicated.30 These correspond to clusters with a high incidence of t(11q23) (percentage is indicated), French-American-British classifications for myelomonocytic (M4) or monocytic (M5) AML, or aberrant high EVI1 expression. Bar diagram represents the mean expression levels (± SE) of MEF2C for patient samples with known MLL gene rearrangements (MLL) compared with all other patient samples (other) or with that in the patients samples within the indicated clusters. Patient numbers are indicated. P values were determined by the Student t test: **P < .001, *P = .02 compared with patients with 11q23 [MLL] disruptions.
Mef2c expression is not necessary for MLL/ENL transformation in vitro
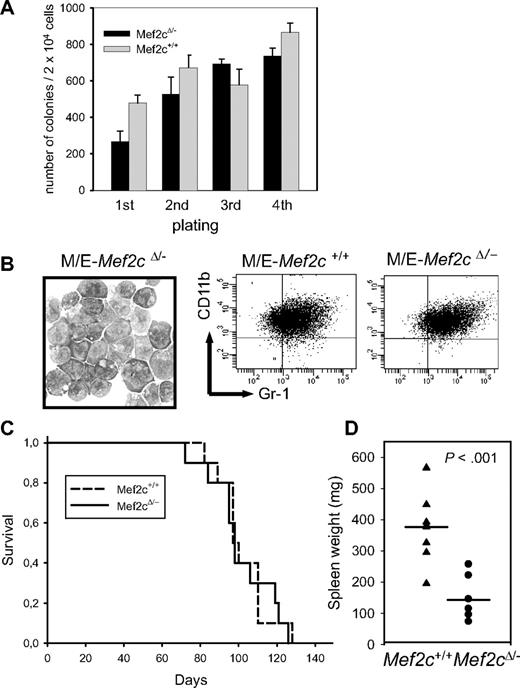
To test the relevance of high MEF2C expression levels in MLL-deregulated AML, we used littermates from a cross between mice carrying a conditional Mef2c null allele (Mef2cfl),18 which is flanked by loxP sites for deletion by Cre recombinase, and mice carrying a constitutive Mef2c null allele (Mef2c−; homozygous Mef2c null mice are embryonic lethal).19 In a first set of experiments, BM cells from Mef2c+/+ or Mef2cfl/− littermates was infected with MLL/ENL retroviral vectors to establish transformed cell lines (M/E cells). The MLL/ENL fusion protein, which is the product of a common translocation involving the MLL gene, readily immortalizes myelomonocytic progenitors in vitro, which are tumorigenic in vivo.28 The M/E cells were then infected with a CRE/GFP expression vector to determine whether Mef2c was necessary for the maintenance of the transformed cell lines. Cells were sorted for GFP expression, and excision was confirmed by PCR analysis. Notably, cells lacking functional Mef2c continued to proliferate at the same rate as Mef2c+/+ transformed cells, also receiving the CRE vector, as measured by incorporation of thymidine over a 48-hour period and cell counts over 6 days (data not shown). In a second set of experiments, BM was isolated from Mef2cfl/−-Mx1CRE mice, in which the floxed allele was deleted by activating CRE expression by pIpC, generating Mef2cΔ/−. Transformed M/E cell lines could be readily established for both genotypes, as demonstrated in a replating assay (Figure 4A). The morphology and the immunophenotype of the cells were similar for all genotypes, although a slight but reproducible increase in Gr1 expression levels was consistently observed in all cells lacking Mef2c (Figure 4B). Incubation in different myeloid cytokines (singly or as cocktails) did not change either their morphology or surface antigen expression. In all cases, the excised allele was confirmed by PCR analysis of cellular DNA (data not shown). Thus, Mef2c is not necessary for either the establishment or maintenance of the immortalized state or for the block in terminal differentiation in MLL/ENL-transformed cultures.
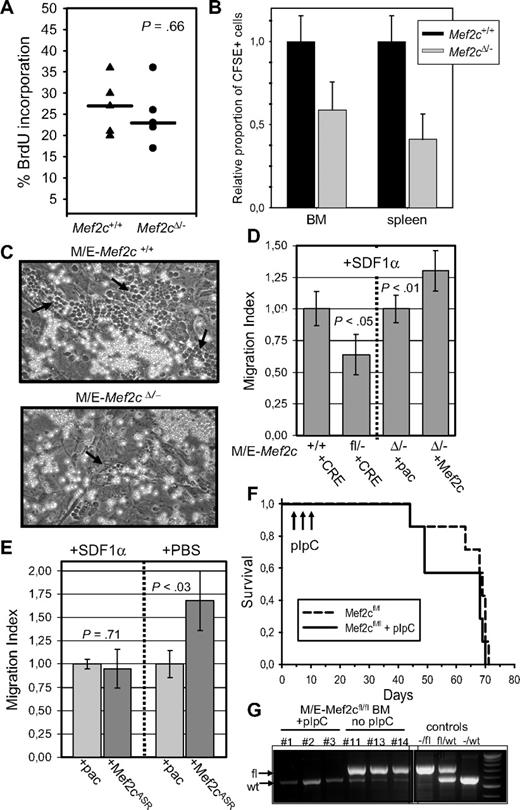
Mef2c is not necessary for MLL/ENL-induced transformation or maintenance in culture, but for homing and/or spread of the MLL/ENL leukemic cells in vivo. (A) Establishment of transformed cell lines with MLL/ENL is not impaired after deletion of the floxed Mef2c allele by CRE recombinase. Results show the mean colony number (± SD) of serially replated cultures seeded with 2 × 104 cells in methylcellulose each week. The first plating was done immediately after MLL/ENL transduction, so that variation in the colony numbers reflects also infection efficiencies. Similar results were obtained for 2 independent M/E cell lines. (B) Inactivation of Mef2c in MLL/ENL-transformed Mef2cfl/− cells did not alter their morphology (Pappenheim stain; 100×/1.3 oil) or gross immunophenotype, as assessed by FACS analysis. (C) Survival curves of NOD/scid mice injected intraperitoneally with 106 M/E cells with indicated genotype and infected with a CRE vector. Mice were killed when large tumors in abdominal cavity were clearly visible. (D) Spleen weights of animals receiving M/E cells with the indicated genetic background. Vertical line indicates median value. P value was calculated by the Student t test.
Mef2c is not necessary for MLL/ENL-induced transformation or maintenance in culture, but for homing and/or spread of the MLL/ENL leukemic cells in vivo. (A) Establishment of transformed cell lines with MLL/ENL is not impaired after deletion of the floxed Mef2c allele by CRE recombinase. Results show the mean colony number (± SD) of serially replated cultures seeded with 2 × 104 cells in methylcellulose each week. The first plating was done immediately after MLL/ENL transduction, so that variation in the colony numbers reflects also infection efficiencies. Similar results were obtained for 2 independent M/E cell lines. (B) Inactivation of Mef2c in MLL/ENL-transformed Mef2cfl/− cells did not alter their morphology (Pappenheim stain; 100×/1.3 oil) or gross immunophenotype, as assessed by FACS analysis. (C) Survival curves of NOD/scid mice injected intraperitoneally with 106 M/E cells with indicated genotype and infected with a CRE vector. Mice were killed when large tumors in abdominal cavity were clearly visible. (D) Spleen weights of animals receiving M/E cells with the indicated genetic background. Vertical line indicates median value. P value was calculated by the Student t test.
Mef2c is not required for MLL/ENL-induced leukemia but shortens the disease latency and increases the dissemination of leukemic cells in vivo
To determine whether Mef2c expression was necessary for tumorigenesis in vivo, mice were injected intraperitoneally with cells from the established Mef2cΔ/− or Mef2c+/+ M/E cell lines (both carrying a CRE retroviral vector) and monitored for disease induction. Mice of each cohort developed large tumor masses within the peritoneal cavity with similar size and kinetics (Figure 4C and data not shown). These results demonstrated that loss of functional Mef2c did not impair tumor/leukemia formation in vivo, even if the M/E cells were initially transformed in the presence of Mef2c. However, postmortem inspection of hematopoietic organs within these mice revealed a striking difference between mice receiving Mef2cΔ/− or Mef2c+/+ M/E cells. In contrast to mice receiving M/E-Mef2c+/+ cells, in which infiltrating tumor cells were readily observed in the spleen, as monitored by GFP positivity (median, 89%; range, 66%-100%) and size (Figure 4D), mice receiving M/E-Mef2cΔ/− cells had smaller spleens with lower GFP levels (median, 29%; range, 8%-85%; Figure 4D). A similar reduction in the spread to BM was also observed, as measured by proportion of GFP+ cells (mean, 13.5% ± 8% for Mef2cΔ/− cells, n = 6, compared with 82% ± 16% for Mef2c+/+ cells, n = 7). Normal hematocrit levels (range, 34%-43%) and leukocyte counts (range, 2-12 × 106 cells/mL) were observed in both cohorts.
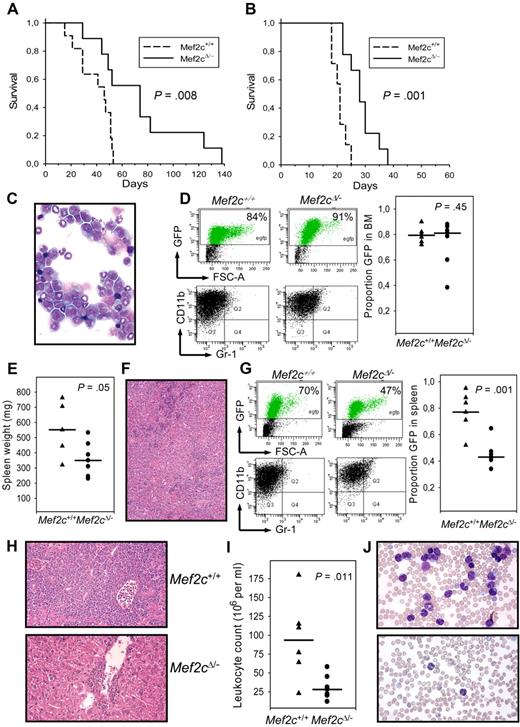
To verify that the impaired homing and dissemination were not the result of a specific characteristic of the immortalized cells, Mef2cΔ/−MxCRE or Mef2c+/+MxCRE BM cells (from mice previously treated with pIpC) were freshly infected with MLL/ENL-GFP viral vectors with similar efficiencies (29% and 25%, respectively) and transplanted intravenously into conditioned mice. All mice receiving Mef2c+/+ or Mef2cΔ/− BM cells infected with MLL/ENL vectors died of a rapid disease, characterized by expansion of myeloid CD11b+ cells in the BM and spleen, but with different latencies (median latency of 52 and 75 days, respectively, for Mef2c+/+ or Mef2cΔ/− BM recipients; Figure 5A). Retransplantation of tumorigenic cells into secondary (nonirradiated) recipients led to a rapid disease, regardless of the genotype, although a slight but significant increase in disease latency was observed in mice receiving Mef2cΔ/− leukemic cells (Figure 5B).
Mef2c is not required for in the induction of leukemia in vivo. (A) Kaplan-Meier survival curves of B6 mice receiving BM from mice with indicated genotype after transduction with retroviral vectors coexpressing MLL/ENL and GFP. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .008) increase in disease latency in mice receiving Mef2-deficient BM. (B) Survival curves of B6 mice receiving 5 × 106 tumor cells (intravenously) from 1 of 3 independent mice (for each genotype) from experiment shown in panel A. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .001) increase in disease latency in mice retransplaned with Mef2-deficient tumors. (C) Cytospin of BM cells from mice transplanted with MLL/ENL infected Mef2cΔ/− BM (5% efficiency) and killed 50 days later. An accumulation of blastlike cells and immature myeloid forms is clearly seen (Pappenheim stain; 63×/1.4 oil). (D) Flow cytometric analysis of cells isolated from BM of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). High proportion of GFP+ cells was observed in all mice. Each symbol in the dot diagram to the right represents an independent mouse. Horizontal line indicates median value. P value was calculated by the Student t test. Gating on the GFP+ population of the BM (bottom panels) demonstrates that the MLL/ENL/GFP+ cells are CD11b+Gr1− /lo. (E) Dot diagram showing the spleen weights of mice analyzed 50 days after BM transplantation. Horizontal line indicates median value. P value was calculated by the Student t test. (F) Splenic section of mice receiving Mef2cΔ/− BM infected with MLL/ENL vectors. The regular splenic architecture is completely effaced in mice receiving MLL/ENL-infected BM because of the myeloid hyperplasia of the red pulp. Pockets of erythroid cells are observed, which compensates for the loss of erythropoiesis in the BM (hematoxylin and eosin stain; 10×/0.45). (G) Flow cytometric analysis of cells isolated from spleens of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). A higher proportion of GFP+ cells was observed in mice receiving Mef2c+/+ BM compared with Mef2cΔ/− BM. Each symbol in the dot diagram to the right represents an independent mouse. P value was calculated by the Student t test. Gating on the GFP population of the spleen (bottom panels) demonstrates that the MLL/ENL/GFP+ cells are CD11b+Gr1neg /lo. (H) The highly invasive behavior of the MLL/ENL Mef2c+/+ leukemic cells is evidenced by infiltration into the liver. The normal liver lobular architecture is completely destroyed and periportal and perivenous liver parenchyma is replaced by infiltrating leukemic myeloblasts and immature forms (top panel). In contrast, invading cells are limited to periportal regions in mice receiving MLL/ENL Mef2cΔ/− BM (hematoxylin and eosin stain; 20×/0.75). (I) Leukocyte counts of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). (J) Blood smears of mice receiving MLL/ENL Mef2c+/+ (top panel) or Mef2cΔ/− BM (bottom panel). The leukocytosis is predominantly composed of immature blastlike monocytic and granulocytic cells (Pappenheim stain; 40×/1.3 oil).
Mef2c is not required for in the induction of leukemia in vivo. (A) Kaplan-Meier survival curves of B6 mice receiving BM from mice with indicated genotype after transduction with retroviral vectors coexpressing MLL/ENL and GFP. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .008) increase in disease latency in mice receiving Mef2-deficient BM. (B) Survival curves of B6 mice receiving 5 × 106 tumor cells (intravenously) from 1 of 3 independent mice (for each genotype) from experiment shown in panel A. The log-rank test for comparison of cumulative incidence curves confirmed a significant (P < .001) increase in disease latency in mice retransplaned with Mef2-deficient tumors. (C) Cytospin of BM cells from mice transplanted with MLL/ENL infected Mef2cΔ/− BM (5% efficiency) and killed 50 days later. An accumulation of blastlike cells and immature myeloid forms is clearly seen (Pappenheim stain; 63×/1.4 oil). (D) Flow cytometric analysis of cells isolated from BM of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). High proportion of GFP+ cells was observed in all mice. Each symbol in the dot diagram to the right represents an independent mouse. Horizontal line indicates median value. P value was calculated by the Student t test. Gating on the GFP+ population of the BM (bottom panels) demonstrates that the MLL/ENL/GFP+ cells are CD11b+Gr1− /lo. (E) Dot diagram showing the spleen weights of mice analyzed 50 days after BM transplantation. Horizontal line indicates median value. P value was calculated by the Student t test. (F) Splenic section of mice receiving Mef2cΔ/− BM infected with MLL/ENL vectors. The regular splenic architecture is completely effaced in mice receiving MLL/ENL-infected BM because of the myeloid hyperplasia of the red pulp. Pockets of erythroid cells are observed, which compensates for the loss of erythropoiesis in the BM (hematoxylin and eosin stain; 10×/0.45). (G) Flow cytometric analysis of cells isolated from spleens of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). A higher proportion of GFP+ cells was observed in mice receiving Mef2c+/+ BM compared with Mef2cΔ/− BM. Each symbol in the dot diagram to the right represents an independent mouse. P value was calculated by the Student t test. Gating on the GFP population of the spleen (bottom panels) demonstrates that the MLL/ENL/GFP+ cells are CD11b+Gr1neg /lo. (H) The highly invasive behavior of the MLL/ENL Mef2c+/+ leukemic cells is evidenced by infiltration into the liver. The normal liver lobular architecture is completely destroyed and periportal and perivenous liver parenchyma is replaced by infiltrating leukemic myeloblasts and immature forms (top panel). In contrast, invading cells are limited to periportal regions in mice receiving MLL/ENL Mef2cΔ/− BM (hematoxylin and eosin stain; 20×/0.75). (I) Leukocyte counts of mice 50 days after receiving BM (with the indicated genotype) infected with MLL/ENL vector (5% efficiency). (J) Blood smears of mice receiving MLL/ENL Mef2c+/+ (top panel) or Mef2cΔ/− BM (bottom panel). The leukocytosis is predominantly composed of immature blastlike monocytic and granulocytic cells (Pappenheim stain; 40×/1.3 oil).
To better compare the disease with and without Mef2c expression, another MLL/ENL transduction/BM transplantation was performed, in which the infection frequency was adjusted to obtain 5% GFP BM cells. After the first signs of mortality (∼ day 50), mice from the 2 cohorts (mice receiving MLL/ENL transduced BM from either Mef2c+/+ or Mef2cΔ/− mice; n = 8 per genotype) were analyzed. Inspection of sternal BM sections of all animals revealed a rather homogeneous picture resulting from the predominance of transformed myeloid cells and striking hypercellularity, leading to almost complete suppression of the sinusoidal vascular system (data not shown). Morphologic and surface expression analysis confirmed that the BM was composed almost exclusively of CD11b+ myeloblasts and immature forms (Figure 5C). No significant difference between Mef2c+/+ or Mef2cΔ/− BM recipients was observed, as quantified by determining the proportion of GFP+ (ie, MLL/ENL expressing) cells by FACS analysis (Figure 5D). Indeed, the very high proportion of transformed cells in this organ suggests that this is the origin of the leukemic cell transformation. However, the spread of the transformed cells to the periphery and to other lymphoid or nonhematopoietic organs was strikingly different between the 2 cohorts, manifesting in 2 distinct clinical diseases in the 2 genotypic cohorts. First, whereas all mice showed splenomegaly, the median spleen weight of Mef2c+/+ recipients was significantly higher than mice receiving Mef2cΔ/− (Figure 5E), although, in both cohorts, histologic analysis of the splenic sections showed complete effacement of the normal architecture resulting from myeloid hyperplasia of the red pulp (Figure 5F). FACS analysis demonstrated a high incidence of MLL/ENL (GFP+) transformed myeloblasts (CD11b+) in both cohorts, although a significantly lower proportion was observed in Mef2cΔ/− BM recipients (Figure 5G). Furthermore, whereas hepatomegaly was observed in 6 of 7 mice receiving MLL/ENL Mef2c+/+ BM, this was observed in only 2 of 8 Mef2cΔ/− recipients. Histologic inspection showed that the normal liver lobular architecture was destroyed in all mice with enlarged livers, the periportal and perivenous liver parenchyma being replaced by infiltrating leukemic myeloblasts and immature forms (Figure 5H). In contrast, the nonenlarged livers of Mef2cΔ/− BM recipients showed only limited myeloblasts infiltration (Figure 5H). Similarly, lymphadenopathy was only observed in 2 of 8 mice of the MLL/ENL Mef2cΔ/− cohort, but in 5 of 7 Mef2c+/+. Finally, analysis of the blood also revealed a significant difference in the spread of the transformed cells, as evidenced by the extremely high leukocyte counts in the Mef2c+/+ but not the Mef2cΔ/− cohort (Figure 5I). Blood smears and FACS analysis confirmed that this was the result of abnormal egression of MLL/ENL-infected Mef2c+/+ myeloblasts and abnormal myeloid forms from the BM (Figure 5J; and data not shown). Taken together, these results support the hypothesis that Mef2c is important for either the engraftment/mobilization/or proliferation of leukemic cells into permissive environments.
Mef2c expression correlates with increased homing and motility properties and with increased expression levels of chemokines and receptors
The importance of Mef2c in proliferation, homing, engraftment, and/or mobilization was investigated in several different assays. First, although in vitro proliferation assays of M/E cells did not reveal a difference between Mef2c+/+ and Mef2cΔ/− cells, we sought to investigate the proliferation of MLL/ENL-transformed cells in the BM. Two weeks after transplantation of freshly infected MLL/ENL-Venus Mef2c+/+ and Mef2cΔ/− cell BM progenitors, mice were injected with BrdU and the levels of incorporation were assayed in Venus+ cells isolated from the BM 2 hours later. As shown in Figure 6A, no significant difference in BrdU uptake and thus proliferation between Mef2c+/+ and Mef2cΔ/− cells was observed.
Mef2c-deficient cells proliferate in vivo but show reduced homing and motility. (A) Percentage of BrdU incorporation after a 2-hour pulse in MLL/ENL-YFP-infected Mef2cΔ/− MxCRE or Mef2c+/+ MxCRE BM cells transplanted into irradiated mice. Each square represents an independent mouse receiving BM with the indicated phenotype. Cells were sorted for YFP expression (19%-34% of total BM) before staining for BrdU. (B) BM cells from Mef2cΔ/− MxCRE or Mef2c+/+ MxCRE mice (treated with pIpC) were labeled in vitro with CFSE and then injected via the tail vein into lethally irradiated syngeneic recipient mice. Shown is the relative mean proportion (± SE) of dye-positive cells from BM or spleen isolated 4 hours after transplantation from 4 mice from 2 independent experiments. An average of 68 (± 9.5) and 78 (± 32) CFSE+Mef2c+/+ cells per 105 total spleen or BM cells, respectively, were detected. (C) Visualization of MS-5 stroma cell cultures 24 hours after 106 M/E Mef2c+/+ or Mef2cΔ/− cells were seeded onto monolayer. Cells transcending the monolayer are clearly seen as phase-dark cells (indicated by ↙). White translucent cells are above the stroma layer. The experiment was repeated 3 times with identical results. (D) Migration studies were compared between M/E cells in culture in which Mef2c was either deleted (by infection with CRE vector) or in which Mef2c was reintroduced with a Mef2c vector. Migration index was calculated 3 to 5 hours after cells from freshly infected and selected cultures were seeded into the upper well of a Transwell with SDF1α in the bottom chamber. Shown is the mean (± SE) of 2 independent experiments, each performed in duplicate. P values were calculated by the Student t test for each pair. (E) Migration index of M/E cells deficient for Mef2c that were either infected with an empty (pac) vector or a vector expressing the HDAC-biding mutant Mef2cASR. Migration index was calculated 3 to 5 hours after cells from freshly infected and selected cultures were seeded into the upper well of a Transwell with either PBS or SDF1α in the bottom chamber, as indicated. Shown is the mean (± SE) of 2 independent experiments, each performed in duplicate. P values were calculated by the Student t test for each pair. (F) Survival curves of mice receiving MLL/ENL-transduced Mef2fl/fl BM, half of which received pIpC injections at the indicated times. (G) Verification of complete excision of the Mef2c floxed alleles after pIpC injection, PCR analysis was performed on DNA isolated from BM cells of diseased mice (3 from each cohort). Primers were designed to detect either the floxed (fl) or wild-type (wt) allele. Faint wt alleles can be detected in all animals resulting from residual host cells. Tail DNA from different genotypes was used as positive controls.
Mef2c-deficient cells proliferate in vivo but show reduced homing and motility. (A) Percentage of BrdU incorporation after a 2-hour pulse in MLL/ENL-YFP-infected Mef2cΔ/− MxCRE or Mef2c+/+ MxCRE BM cells transplanted into irradiated mice. Each square represents an independent mouse receiving BM with the indicated phenotype. Cells were sorted for YFP expression (19%-34% of total BM) before staining for BrdU. (B) BM cells from Mef2cΔ/− MxCRE or Mef2c+/+ MxCRE mice (treated with pIpC) were labeled in vitro with CFSE and then injected via the tail vein into lethally irradiated syngeneic recipient mice. Shown is the relative mean proportion (± SE) of dye-positive cells from BM or spleen isolated 4 hours after transplantation from 4 mice from 2 independent experiments. An average of 68 (± 9.5) and 78 (± 32) CFSE+Mef2c+/+ cells per 105 total spleen or BM cells, respectively, were detected. (C) Visualization of MS-5 stroma cell cultures 24 hours after 106 M/E Mef2c+/+ or Mef2cΔ/− cells were seeded onto monolayer. Cells transcending the monolayer are clearly seen as phase-dark cells (indicated by ↙). White translucent cells are above the stroma layer. The experiment was repeated 3 times with identical results. (D) Migration studies were compared between M/E cells in culture in which Mef2c was either deleted (by infection with CRE vector) or in which Mef2c was reintroduced with a Mef2c vector. Migration index was calculated 3 to 5 hours after cells from freshly infected and selected cultures were seeded into the upper well of a Transwell with SDF1α in the bottom chamber. Shown is the mean (± SE) of 2 independent experiments, each performed in duplicate. P values were calculated by the Student t test for each pair. (E) Migration index of M/E cells deficient for Mef2c that were either infected with an empty (pac) vector or a vector expressing the HDAC-biding mutant Mef2cASR. Migration index was calculated 3 to 5 hours after cells from freshly infected and selected cultures were seeded into the upper well of a Transwell with either PBS or SDF1α in the bottom chamber, as indicated. Shown is the mean (± SE) of 2 independent experiments, each performed in duplicate. P values were calculated by the Student t test for each pair. (F) Survival curves of mice receiving MLL/ENL-transduced Mef2fl/fl BM, half of which received pIpC injections at the indicated times. (G) Verification of complete excision of the Mef2c floxed alleles after pIpC injection, PCR analysis was performed on DNA isolated from BM cells of diseased mice (3 from each cohort). Primers were designed to detect either the floxed (fl) or wild-type (wt) allele. Faint wt alleles can be detected in all animals resulting from residual host cells. Tail DNA from different genotypes was used as positive controls.
In a second set of experiments, the homing potential of Mef2cΔ/− or Mef2c+/+ BM cells was directly assayed by tracking transplanted cells labeled with a fluorescent dye (CFSE). Four hours after transplantation into conditioned mice, significantly lower numbers of transplanted Mef2cΔ/− compared with Mef2c+/+ cells could be detected in both BM and spleen (Figure 6B), suggesting impaired homing in the absence of Mef2c. Similarly, a strikingly reduced ability of Mef2c-deficient MLL/ENL-transformed cells to form cobblestone clusters underneath a stroma cell layer in vitro was observed (Figure 6C), a characteristic ascribed to the interaction of the homing factor SDF1α (CXCL12) secreted by stroma and its receptor CXCR4.34 To quantitate this observation and to ensure that the difference was the result of Mef2c itself, a migration assay in the presence of the SDF1α chemoattractant was performed on M/E Mef2c+/+, M/E Mef2cΔ/−, as well as M/E Mef2cΔ/−+Pac (conferring resistance to puromycin) and M/E Mef2cΔ/−+Mef2c/Pac (in which Mef2c was reintroduced into the Mef2c-deficient cells). These experiments demonstrated that loss of Mef2c lead to decreased migration capacity, whereas reintroduction of Mef2c could rescue this effect (Figure 6D). As Mef2c protein activity is highly dependent on type IIa HDAC binding, which is controlled by extracellular cues, we sought to determine whether relief of HDAC inhibition may reveal a stronger migration activity. To test this, a Mef2c mutant (Mef2cASR), which retains transcription activity but can no longer be repressed by type IIa HDACs,29 was tested in our assay. Expression of this HDAC-binding mutant in Mef2c-deficient cells resulted in a high migration index relative to control cells in the absence of SDF1α (Figure 6E), possibly reflecting an autocrine mechanism. However, this activity was inhibited by addition of SDF1α (Figure 6E), suggesting reciprocal inhibition with other chemokines, as previously described.35
To determine whether loss of effective homing to various hematopoietic organs was the key mechanism by which loss of Mef2c function impaired MLL/ENL induced disease, a transduction/ BM transplantation assay was performed on Mef2cfl/fl MxCRE BM cells not exposed to pIpC. One week after transplantation of MLL/ENL-transduced BM cells, half of the mice (n = 7) were injected with pIpC to induce Mef2c excision. The disease latency of these mice were comparable with that of mice expressing functional Mef2c (Figure 6F). PCR analysis confirmed the efficient recombination of the floxed alleles in the developing tumors (Figure 6G). No significant difference in spleen weights or GFP percentage was observed in these 2 mouse cohorts (data not shown), although a higher incidence of hepatomegaly was observed in mice with tumors expressing functional versus inactivated Mef2c (+pIpC; 0.8 vs 0.3). These results demonstrate that defective homing capacity of MLL/ENL BM cells lacking functional Mef2c is a key factor in the increased disease latency of MLL/ENL-induced leukemia in these cells.
Microarray gene expression profiling reveals up-regulation of several chemokine ligands and receptors as well as matrix metalloproteinase genes in Mef2c+ cells
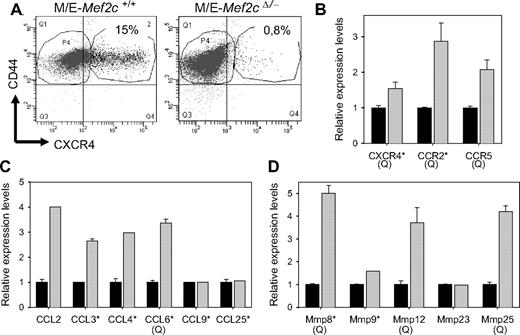
To determine the mechanism behind the defective homing capacity of Mef2c-deficient cells, the surface expression of CXCR4 and CD44, both known to be important in homing and engraftment of hematopoietic cells, was determined (Figure 7A). CD44 was detected at high levels on all MLL/ENL-transformed cells, regardless of Mef2c status. In contrast, a small but consistent subpopulation (up to 15%) of MLL/ENL-transformed Mef2c+/+ cells expressed high levels of CXCR4, whereas CXCR4 could not be detected on transformed cells lacking Mef2c (Figure 7A). Somewhat surprisingly, although CXCR4 was not detected on the surface of Mef2c−/− cells, high levels of CXCR4 transcripts were detected (Figure 7B), suggesting a more complex regulation of CXCR4 surface expression.
Mef2c expression in MLL/ENL Mef2cΔ/−-transformed cells results in increased expression of several genes implicated in homing and invasive growth. (A) FACS analysis detected increase levels of CXCR4 expression on a subset of M/E cells of the indicated genotype immediately after recloning in methylcellulose cultures, which enriches for clonogenic or engrafting cells. Shown is one representative experiment of 3. (B-D) Microarray analysis was performed on RNA isolated from M/E Mef2cΔ/− cells infected with Mef2cASR/pac or pac vectors and subjected to puromycin selection. The relative expression levels (± SD) of the indicated gene for cultures expressing either Mef2cASR/pac ( ) or pac (■) vectors are indicated. For some genes (marked with a Q), the results shown are that from quantitative RT-PCR, which in all cases confirmed the results obtained from the microarray. Results are shown for only genes that showed signals more than 4-fold above background. (B) Several chemokine receptors are up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater than 50-fold over background. (C) Several chemokines are highly expressed in MLL/ENL cultures and show up-regulation in Mef2c+ cultures. *Genes with mean signal intensities greater than 100-fold above background. (D) The expression of several MMP genes is up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater that 20-fold above background.
) or pac (■) vectors are indicated. For some genes (marked with a Q), the results shown are that from quantitative RT-PCR, which in all cases confirmed the results obtained from the microarray. Results are shown for only genes that showed signals more than 4-fold above background. (B) Several chemokine receptors are up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater than 50-fold over background. (C) Several chemokines are highly expressed in MLL/ENL cultures and show up-regulation in Mef2c+ cultures. *Genes with mean signal intensities greater than 100-fold above background. (D) The expression of several MMP genes is up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater that 20-fold above background.
Mef2c expression in MLL/ENL Mef2cΔ/−-transformed cells results in increased expression of several genes implicated in homing and invasive growth. (A) FACS analysis detected increase levels of CXCR4 expression on a subset of M/E cells of the indicated genotype immediately after recloning in methylcellulose cultures, which enriches for clonogenic or engrafting cells. Shown is one representative experiment of 3. (B-D) Microarray analysis was performed on RNA isolated from M/E Mef2cΔ/− cells infected with Mef2cASR/pac or pac vectors and subjected to puromycin selection. The relative expression levels (± SD) of the indicated gene for cultures expressing either Mef2cASR/pac ( ) or pac (■) vectors are indicated. For some genes (marked with a Q), the results shown are that from quantitative RT-PCR, which in all cases confirmed the results obtained from the microarray. Results are shown for only genes that showed signals more than 4-fold above background. (B) Several chemokine receptors are up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater than 50-fold over background. (C) Several chemokines are highly expressed in MLL/ENL cultures and show up-regulation in Mef2c+ cultures. *Genes with mean signal intensities greater than 100-fold above background. (D) The expression of several MMP genes is up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater that 20-fold above background.
) or pac (■) vectors are indicated. For some genes (marked with a Q), the results shown are that from quantitative RT-PCR, which in all cases confirmed the results obtained from the microarray. Results are shown for only genes that showed signals more than 4-fold above background. (B) Several chemokine receptors are up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater than 50-fold over background. (C) Several chemokines are highly expressed in MLL/ENL cultures and show up-regulation in Mef2c+ cultures. *Genes with mean signal intensities greater than 100-fold above background. (D) The expression of several MMP genes is up-regulated in Mef2c+ cultures. *Genes with mean signal intensities greater that 20-fold above background.
To gain insight into the possible mechanism by which Mef2c may impart increased homing and invasion properties to the MLL/ENL-transformed cells, microarray analysis was used to compare gene expression profiles. For this assay, we used the Mef2cASR mutant, in which the protein sites known to interact with type IIa HDACs were mutated, to ensure that Mef2c transcriptional activity was not repressed.29 A striking up-regulation of transcripts encoding chemokine ligands and receptors was observed in cells engineered to express Mef2cASR, which was confirmed by RT-PCR in most cases (Figure 7B-C). Most prominent was the close to 3-fold up-regulation of CCR2, a shared receptor for several ligands of the CC family, including CCL2. The closely related CCR5 receptor was also up-regulated, but overall expression levels were one-tenth of that of CCR2, as judged by signal intensity on microarrays. Indeed, CCR2 and CXCR4 were the only 2 chemokine-receptor genes expressed at high levels in the MLL/ENL-transformed cells, giving signals that were more than 70-fold higher than background levels on the arrays. In agreement with our analysis of M/E Mef2cΔ/− and Mef2c+/+ cells, only a minor difference in CXCR4 transcript levels was observed when Mef2c was expressed in the null background (Figure 7A-B).
In addition to chemokine receptors, we observed increased expression of several CC ligands (Figure 7B). In addition to CCL2, which belongs to the monocytic chemotactic protein family of CC ligands, several members (CCL3, CCL4, and CCL6) of the macrophage inflammatory protein of CC ligands were highly expressed and up-regulated in Mef2c+ cells. No significant signals for CXCL12 (SDF1α) transcript levels, or for other CXC ligands, were observed in these cells.
Finally, analysis of the microarray data also revealed significant increases in transcripts encoding several matrix metalloproteinases (MMP; Figure 7D), which have also been repeatedly implicated in cell motility, tissue invasion, and metastasis.36 Highest expression levels were observed for Mmp8 and Mmp9 (> 30-fold above background signals). Although only slightly higher levels of Mmp9 were observed in Mef2c+ cells, a more than 5-fold increase of Mmp8 transcripts was observed in these cells. In addition, up to 4-fold higher levels of Mmp12 and Mmp25 were also observed.
Discussion
The study presented here provides new insight into the role of the MADS Mef2 transcription factors in leukemia induction. Two observations were made in our study: (1) In the absence of the tumor suppressor Irf8, but not in its presence, deregulation of the Mef2c gene induces a rapid and aggressive myelomonocytic leukemia with close to 100% penetrance, demonstrating the Mef2c can act as a cooperating oncogene; and (2) high levels of Mef2c are found in the myelomonocytic leukemia associated with MLL transformation, but Mef2c was not necessary for either the establishment or maintenance of the LSCs that drives MLL leukemia induction in an experimental model, but rather modulated the clinical manifestations of the disease by imparting motility and invasive properties. It is of interest to note that many of the downstream oncogenes of MLL fusion proteins characterized to date have been identified by retroviral insertional mutagenesis in Irf8-deficient mice, either in the mouse strain we used or in BXH-2 mice.37 We propose that this primarily reflects the relatively large number of genes that are deregulated by MLL disruption and that alone cannot induce leukemia, however, either in concert with other MLL targets or in cooperation with the Irf8-deficient “preleukemic” phenotype are potent disease inducers. A key component of the Irf8 system is most probably related to its decrease sensitivity to apoptotic stimuli, providing a cellular environment where secondary events leading to acute leukemia can occur.9-11
By what mechanism does Mef2c act as an oncogene in leukemia induction?
The strong oncogenic potential of Mef2c was clearly manifested in the Irf8-deficient background. Earlier work has also demonstrated cooperation between ectopic Mef2c expression and Sox4 activation in leukemia induction.38 Somewhat unexpectedly, our study showed that Mef2c expression is not required for MLL/ENL transformation in vitro or in vivo, although disruption of the MLL locus is associated with high levels of Mef2c expression. Even if transformation in vitro initially occurred in Mef2c+ cells, subsequent excision of functional Mef2c did not impact on its ability to induce tumors in vivo. The most probable explanation is that Mef2c oncogenic function is redundant with other downstream targets of MLL fusion proteins, such as HoxA9, Meis1, and Myb. Notably, many of the downstream targets of MLL fusion proteins have complementary but also overlapping transforming properties.5,6,39 Alternatively, loss of Mef2c may be compensated by up-regulation of other Mef2 proteins in these cells. We did not observe up-regulation of Mef2d expression in these tumors (M.S., S. Roscher, C.S., unpublished results, June 2007), but we cannot rule out that expression of Mef2a or Mef2b is affected.
What is the mechanism behind the transforming capacity of Mef2c observed in the Ir8-deficient BM?
Recent studies of both AML and ALL have shown that transcription factors playing pivotal roles in differentiation are frequently deregulated in acute leukemia.40,41 We thus hypothesize that up-regulated Mef2c expression also contributes to the leukemogenic process by deregulating the normal controls of myelomonocytic differentiation. This hypothesis is primarily based on our recent findings showing that Mef2c is a critical modulator of monocytic differentiation in response to external stimuli13 ; in the absence of Mef2c, a reduction in macrophage/monocytic progenitors is observed in the presence of various cytokines, whereas ectopic Mef2c expression leads to increased levels of monopoiesis at the expense of granulopoiesis. The ability of Mef2c to modulate monocytic differentiation is also observed in the striking monocytic phenotype of the leukemia induced by enforced Mef2c expression in Irf8−/− BM cells. In light of the fact that the Irf8−/− background is associated with increased granulopoiesis and defective monopoiesis,8,42,43 this was clearly unexpected. It may be these contradicting or incomplete signaling pathways, stipulating decisions between monocytic and granulocytic differentiation, that led to the observed block in myeloid differentiation in our mouse model and is the hallmark of acute myelogenous leukemia. An important downstream target of Mef2c for modulating monocytic differentiation is Jun,13 which has also been identified as a critical target in the differentiation block observed in leukemic mice with hypomorphic mutations affecting the PU.1 transcription factor.44 However, other genes that are normally up-regulated in activated macrophages were also shown to be up-regulated in Mef2c+ transformed cells, which may also be key to the aggressive nature of these tumors.
Finally, it is important to note that high levels of Mef2c transcripts are found in the HSC compartment3,13 ; thus, Mef2c may impart specific features found in HSCs to the LSCs. Competitive repopulation assays have revealed a small but consistent disadvantage to Mef2cΔ/− hematopoietic stem cells, which is at least partly attributable to the reduced homing capacity of these cells (A.E., M.S., C.S., unpublished results, December 2008). Although studies by Krivtsov et al3 suggested the importance of Mef2c in imparting “self-renewal,” as demonstrated in a single plating assay of MLL/AF9 transformed BM progenitors, our studies do not support this hypothesis, but we cannot rule out that this discrepancy is the result of the different transforming fusion proteins or perhaps different target cells.
How may Mef2c regulate homing and invasiveness?
Although functional Mef2c was not found to be necessary for either initiating or maintaining MLL/ENL-induced transformation, its expression did impact on the clinical symptoms (eg, increased dissemination to other organs) and disease latency of the leukemia. The earlier study of Krivtsov et al also observed an increased latency of leukemia induction by MLL/AF9 in the absence of Mef2c, using siRNA technology.3 Although they predicted that Mef2c up-regulation is necessary for establishment of MLL-induced LSCs, our cumulative results suggest that a more probable interpretation is that Mef2c expression modulates the clinical disease by impinging on homing, motility, and invasive properties of the leukemic cells. This hypothesis is supported by the highly aggressive and metastatic nature of the AML induced by Mef2c expression in Irf8-deficient cells, but also by various in vitro assays and animal experiments demonstrating impaired homing and motility of cells deficient for Mef2c. Homing and engraftment of HSCs to the BM and extramedullary tissue, and presumably also the homing and spread of LSCs, involves common mechanisms of adhesion, migration signaling through cytokines and chemoattractants, and MMP activation.45-47 Although we observed no difference in surface expression levels of CD44 on MLL/ENL-transformed cells, we consistently observed up-regulated surface expression of CXCR4, the receptor for CXCL12 (SDF1α), in a subset of Mef2c+ cells, which was not observed in Mef2cΔ/− cells. However, this regulation was not at the transcriptional level, as similar high levels of CXCR4 transcripts were observed in Mef2c+/+ or Mef2cΔ/−, with or without enforced expression of Mef2c. Surface expression levels of CXCR4 have been shown to be regulated by several different mechanisms, including ubiquitination,48 cytokine stimulation,49,50 and by the formation of heterodimers.48 Notably, both CCR2 and CCR5, which were down-regulated in the absence of Mef2c, have been demonstrated to form dimers with CXCR4, leading to increased cell surface expression and/or increased activity,51,52 suggesting a mechanism by which surface levels of CXCR4 may be differentially regulated in cells expressing Mef2c.
We cannot rule out, however, the contribution of other Mef2c target genes. For instance, the increased expression levels of several CC chemokines observed in our analysis may contribute to the increased motility and invasiveness of the Mef2c+ tumors by either autocrine or paracrine mechanism by increasing chemotaxis, stimulating proliferation and/or survival, or altering the stroma environment.53-55 Notably, increased levels of CCR2 and its ligands have been observed in AML with a myelomonocytic or monocytic phenotype, and correlated with increased extramedullary involvement.54 Furthermore, CCL3, CCL4, CCL6, and CCL9 are all highly expressed in the MLL/ENL-transformed cells and also in activated macrophages, providing an additional link between the leukemic phenotype and the importance of Mef2c during normal macrophage differentiation. Similarly, our microarray analysis also revealed several MMPs that were up-regulated in Mef2c+ cells, which have often been linked to tumor metastasis and leukemic survival rates.56,57 In analogy to the highly related MMP10 gene, which has been shown to be directly activated by Mef2 transcription factors,58 a conserved Mef2c-binding site within the first intron of MMP8 suggests that it may also be targeted by Mef2c.
Implications for deregulated Mef2 transcription factors and human leukemia
Taken together, our study has clearly demonstrated the oncogenic role of Mef2c in AML, which is most probably attributable to its important role in modulating myeloid differentiation, but also regulating migration and homing of hematopoietic progenitors to the BM and spleen. Although high expression levels of MEF2C in AML patients samples were limited to a few patient clusters, it is well established that Mef2c is tightly regulated at a posttranscriptional level by several different mechanisms.14,59 This includes regulation by the microRNA-223, a newly recognized target of AML1-ETO, the fusion product of the relatively common t(8;21) AML.60 Thus, deregulation of Mef2 proteins may be critically involved in a wide range of acute leukemias.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the Stocking laboratory for help in this study, with special thanks to Ulla Bergholz, Arne Düsedau, Gundula Pilnitz-Stolze, Ulla Müller, Susanne Roscher, Sylvia Wegerich, and Marion Ziegler for invaluable assistance in many of the experiments, as well as Daniel Bayer for expert assistance in the statistical analysis.
This work was supported by the Deutsche José Carreras Leukämie Stiftung and the Japanese Human Science Foundation (in collaboration with T. Hara). The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit und soziale Sicherung.
Authorship
Contribution: M.S., A.S., M.F., and A.E. designed and performed the majority of the experiments, analyzed data, and helped write the manuscript; R.D. and P.J.V. performed analysis of AML patient material; M.A.A. and E.N.O. provided the Mef2c conditional mouse strain; J.L. provided pathology and histology expertise; R.K.S. designed and performed experiments and analyzed data; and C.S. designed, performed, and supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carol Stocking, Heinrich-Pette-Institut, PO Box 201652, D-20206 Hamburg, Germany; e-mail: stocking@hpi.uni-hamburg.de.
References
Author notes
M.S. and A.S. contributed equally to this study.

![Figure 3. High levels of MEF2C gene expression found in clusters of AML patients with high incidence of MLL. Shown are pairwise correlations of gene expression profiles between AML patients calculated by OmiViz software (Version 3.6) and viewed by HeatMapper, as previously described.30 The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors indicating higher positive (red) or negative (blue) correlations. Histograms next to each tumor represent expression levels for the IRF8 or MEF2C probe sets (bars are proportional to the level of expression). Six of 16 clusters identified in the cohort of 285 AML patients on the basis of the unsupervised clustering results, and other parameters, such as clinical and molecular characteristics of the AML patients, are indicated.30 These correspond to clusters with a high incidence of t(11q23) (percentage is indicated), French-American-British classifications for myelomonocytic (M4) or monocytic (M5) AML, or aberrant high EVI1 expression. Bar diagram represents the mean expression levels (± SE) of MEF2C for patient samples with known MLL gene rearrangements (MLL) compared with all other patient samples (other) or with that in the patients samples within the indicated clusters. Patient numbers are indicated. P values were determined by the Student t test: **P < .001, *P = .02 compared with patients with 11q23 [MLL] disruptions.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/12/10.1182_blood-2008-05-158196/4/m_zh89990941110003.jpeg?Expires=1769106769&Signature=afiX18E6EHgk939AYrpDxfg9XKGX~H2RK8F-gN7qmn2vypzGW4j8E4gvwdsHfAmn9G0EEuT6Lqntczb6FJJOIX~7luIKCzr~vyaQ2FoMg5OcI~Es0IXz9I37JN5pp-4OxOeVIyHlVV6c1up3wYLtL051vpk3dlnGWDO8wfbwgczS~c0lZGyBpdkmM9DiQNMq16pXzASmz1YHWZurBwsA6K3gP4ggAcyc5Tr0eBGAAPq0AXjZlvs9WvhWd1kcje5qIygRepplgTKXflKIlv0DrdkatF2K6KQMo5ux6zdz5wxvBKdWOmo0Hi0mviy-Li~cPQ7w7JWVso55lYR4A7QGLQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal