Abstract
Translocations involving chromosome 11q23 frequently occur in pediatric acute myeloid leukemia (AML) and are associated with poor prognosis. In most cases, the MLL gene is involved, and more than 50 translocation partners have been described. Clinical outcome data of the 11q23-rearranged subgroups are scarce because most 11q23 series are too small for meaningful analysis of subgroups, although some studies suggest that patients with t(9;11)(p22;q23) have a more favorable prognosis. We retrospectively collected outcome data of 756 children with 11q23- or MLL-rearranged AML from 11 collaborative groups to identify differences in outcome based on translocation partners. All karyotypes were centrally reviewed before assigning patients to subgroups. The event-free survival of 11q23/MLL-rearranged pediatric AML at 5 years from diagnosis was 44% (± 5%), with large differences across subgroups (11% ± 5% to 92% ± 5%). Multivariate analysis identified the following subgroups as independent prognostic predictors: t(1;11)(q21;q23) (hazard ratio [HR] = 0.1, P = .004); t(6;11)(q27;q23) (HR = 2.2, P < .001); t(10;11)(p12;q23) (HR = 1.5, P = .005); and t(10;11)(p11.2;q23) (HR = 2.5, P = .005). We could not confirm the favorable prognosis of the t(9;11)(p22;q23) subgroup. We identified large differences in outcome within 11q23/MLL-rearranged pediatric AML and novel subgroups based on translocation partners that independently predict clinical outcome. Screening for these translocation partners is needed for accurate treatment stratification at diagnosis.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease. Currently, response to therapy and cytogenetic abnormalities, including abnormalities such as t(8;21), inv(16), and t(15;17), which together are defined as good-risk AML, are the main prognostic factors.1 Translocations involving chromosome 11q23 is composed of 15% to 20% of all pediatric AML cases and are, in general, associated with a poor outcome.2-4 In more than 95% of cases involving 11q23 rearrangements, the mixed lineage leukemia (MLL) gene is involved, which requires confirmation with molecular analyses such as fluorescence in situ hybridization (FISH).5 The heterogeneity of MLL-rearranged AML is further reflected by the identification of more than 50 different fusion partners of this gene.6
In AML, the most common 11q23 rearrangements are t(9;11)(p22;q23) (in ∼ 50% of cases), t(11;19)(q23;p13.1), t(11;19)(q23;p13.3), t(6;11)(q27;q23), and t(10;11)(p12;q23).2,3 Most AML samples with 11q23 rearrangements are morphologically classified as French-American-British morphology classification (FAB)-M4 or FAB-M5.7
In some studies, the t(9;11)(p22;q23) subgroup has been associated with a better prognosis, which may at least partially be the result of enhanced sensitivity to different classes of drugs, as demonstrated for this subgroup.4,8-10 In the current treatment protocols of St Jude Children's Research Hospital and the Nordic Society for Pediatric Hematology and Oncology (NOPHO), this has resulted in low-risk stratification of these patients. Studies on the clinical outcome of various 11q23 rearrangements in pediatric acute lymphoblastic leukemia showed age-specific outcome differences. For instance, in children older than 1 year, t(4;11)(q21;q23) showed an adverse outcome compared with that of children who carried other 11q23 rearrangements,11 which is not the case in infants.12 However, clinical outcome data of pediatric patients with AML expressing other 11q23 rearrangements are extremely scarce because most collaborative group series are too small for meaningful outcome analysis of these rare subgroups.
Therefore, a very large international study was undertaken to collected clinical outcome data of 756 children with 11q23 or MLL-rearranged pediatric AML from 11 collaborative study groups from 15 countries. Our aim was to identify differences in clinical outcome based on the various 11q23 or MLL rearrangements, which may result in better risk-group stratification and risk-group–directed therapy for these patients.
Methods
Patients
Data on 756 patients with 11q23- or MLL-rearranged pediatric AML were collected from 11 collaborative study groups, including the Berlin-Frankfurt-Münster Study Group (Germany and Austria; n = 160), the Japanese Pediatric Leukemia/Lymphoma Study Group (Japan; n = 75), the Leucémies Aiguës Myéloblastiques de l'Enfant Cooperative Group (France; n = 61), the Czech Pediatric Hematology Working Group (Czech Republic; n = 18), the St Jude Children's Research Hospital (n = 44), the Associazione Italiana Ematologia Oncologia Pediatrica (Italy; n = 34); Belarus (n = 25), the Children's Oncology Group (both including the Children's Cancer Group and the Pediatric Oncology Group studies; n = 215), the NOPHO (Denmark, Finland, Iceland, Norway, and Sweden; n = 59), the Dutch Children's Oncology Group (The Netherlands; n = 34), and 2 centers of the Medical Research Council (United Kingdom; n = 31).
The patients were identified by the study groups reviewing their karyotypic records and including those with 11q23- or MLL-rearranged AML, as determined by G-, Q-, or R-banded karyotyping, FISH, or reverse-transcribed polymerase chain reaction (RT-PCR). For the purpose of this study, these data were grouped as 11q23/MLL-rearranged AML. A predefined set of data were collected for each case and checked for consistency and completeness. This set of data consisted of clinical data obtained at initial diagnosis, including sex, age, white blood cell count (WBC), hemoglobin, platelets, extramedullary disease, and FAB morphology. In addition, we collected data on treatment, such as therapy protocol, including hematopoietic stem cell transplantation (HSCT), and all events (including nonresponders, relapse, second malignancy, or death) during follow-up. Only patients younger than 18 years at diagnosis were included in the analysis. Patients were eligible when diagnosed between January 1, 1993 and January 1, 2005. The Children's Cancer Group, however, included patients diagnosed between January 1, 1995 and January 1, 2003; the Pediatric Oncology Group included patients diagnosed between January 1, 1993 and January 1, 2000 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Exclusion criteria consisted of secondary AML after congenital bone marrow failure disorders (eg, Fanconi anemia, severe congenital neutropenia, and Shwachman syndrome), aplastic anemia, prior chemotherapy or radiotherapy for other diseases, and prior myelodysplastic syndrome.
Patients were treated on national/collaborative group AML trials.4,13-22 The treatment protocols were approved according to local law and guidelines, by the Institutional Review Boards of each participating center with informed consent obtained in accordance with the Declaration of Helsinki. Patients received anthracycline- and cytarabine-based induction courses, followed by consolidation courses that included high-dose cytarabine (supplemental Table 2). Different anthracyclines were used. To calculate the cumulative anthracycline dose prescribed in the various protocols, we used a ratio of 1:5 (in mg/m2) for daunorubicin/idarubicin or mitoxantrone and a ratio of 1:1 for daunorubicin/doxorubicin.23 A total of 108 patients received allogeneic HSCT in first complete remission, whereas 59 patients underwent autologous HSCT; these patients were included in the “chemotherapy only” group.24
Categories of 11q23 or MLL rearrangements
Most patients were entered into the study based on available karyotypic reports showing 11q23/MLL rearrangements. Assignment to 11q23/MLL-rearranged subgroups based on translocation partners took place after independent central review by the coauthors (S.C.R., J.H.) of the breakpoint in the corresponding chromosome and followed the International System for Human Cytogenetic Nomenclature (ISCN2005).25 Molecular confirmation of an MLL rearrangement with FISH and/or RT-PCR was not required because conventional cytogenetics methods, RT-PCR, and FISH are usually complementary and allow reliable identification of 11q23/MLL gene rearrangements in leukemic clones. Patients with an MLL rearrangement detected by FISH and an unknown translocation partner were classified in the “11q23/MLL-other” group. At least 10 patients had to be included per subgroup; otherwise, the cases were allocated to the 11q23/MLL-other group.
There were 328 (43%) patients with t(9;11)(p22;q23), 98 (13%) with t(10;11)(p12;q23), 35 (5%) with t(6;11)(q27;q23), 34 (5%) with t(11;19)(q23;p13.1), 25 (3%) with t(11;19)(q23;p13.3), 31 (4%) with t(11;19)(q23;p13)-without ascertained subband, 25 (3%) with t(1;11)(q21;q23), 13 (2%) with t(4;11)(q21;q23), 12 (2%) with t(10;11)(p11.2;q23), and 12 (2%) with t(11;17)(q23;q21). The remaining 143 (19%) patients, including 4 patients with t(9;11) and another breakpoint on chromosome 9, were assigned to the 11q23/MLL-other subgroup (Table 1).
Characteristics of pediatric patients with 11q23/MLL-rearranged AML grouped on the basis of translocation partners
| . | Total . | t(9;11)(p22;q23) . | t(10;11)(p12;q23) . | t(6;11)(q27;q23) . | t(11;19)(q23;p13) . | t(11;19)(q23;p13.1) . | t(11;19)(q23;p13.3) . | t(1;11)(q21;q23) . | t(4;11)(q21;q23) . | t(10;11)(p11.2;q23) . | t(11;17)(q23;q21) . | Other . | P* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 756 | 328 | 98 | 35 | 31 | 34 | 25 | 25 | 13 | 12 | 12 | 143 | |
| Male sex, n (%) | 367 (48.5) | 157 (47.9) | 55 (56.1) | 19 (54.3) | 13 (41.9) | 15 (44.1) | 11 (44.0) | 9 (36.0) | 3 (23.1) | 6 (50) | 7 (58.3) | 72 (50.3) | .495 |
| Median age (N = 756) | 2.2 | 2.6 | 1.3 | 12.4 | 1.6 | 4.6 | 7.1 | 1.3 | 1.3 | 1.0 | 9.0 | 1.9 | |
| Less than 2 y, n (%) | 358 (47.4) | 137 (41.8) | 61 (62.2) | 3 (8.6) | 18 (58.1) | 14 (41.2) | 9 (36.0) | 19 (76.0) | 8 (61.5) | 9 (75.0) | 4 (33.3) | 72 (50.3) | < .001 |
| 2 to 9 y, n (%) | 227 (30.0) | 117 (35.7) | 22 (22.4) | 12 (34.3) | 8 (25.8) | 11 (32.4) | 5 (20.0) | 6 (24.0) | 2 (15.4) | 3 (25.0) | 3 (25.0) | 41 (28.6) | |
| 10 or more y, n (%) | 171 (22.6) | 74 (22.6) | 15 (15.3) | 20 (57.1) | 5 (16.1) | 9 (26.5) | 11 (44.0) | — | 3 (23.1) | — | 5 (41.7) | 30 (21.0) | |
| Median WBC (N = 754) | 20.9 | 13.7 | 10.9 | 59.8 | 76.0 | 27.0 | 17.5 | 33.4 | 20.0 | 17.2 | 87.8 | 17.2 | |
| Less than 20 × 109/L, n (%) | 371 (49.1) | 180 (54.9) | 58 (59.2) | 8 (22.9) | 10 (32.3) | 15 (44.1) | 14 (56.09) | 9 (36.0) | 6 (46.2) | 6 (50.0) | 4 (33.3) | 61 (42.7) | |
| 20 to less than 100 × 109/L, n (%) | 206 (27.2) | 82 (25.0) | 22 (22.4) | 12 (34.3) | 9 (29.0) | 13 (38.2) | 3 (12.0) | 11 (44.0) | 3 (23.1) | 2 (16.7) | 3 (25.0) | 46 (32.2) | < .001 |
| 100 or more × 109/L, n (%) | 177 (23.4) | 65 (19.8) | 18 (18.4) | 15 (42.9) | 12 (38.7) | 6 (17.6) | 8 (32.0) | 5 (20.0) | 4 (30.8) | 4 (33.3) | 5 (41.7) | 35 (24.5) | |
| FAB type, n (%) (N = 722) | < .001 | ||||||||||||
| FAB-M0 | 23 (3.2) | 3 (1.0) | 4 (4.3) | — | 1 (3.2) | 1 (3.0) | 3 (12.0) | — | 3 (25.0) | 1 (9.1) | 1 (8.3) | 6 (4.5) | |
| FAB-M1 | 40 (5.5) | 9 (2.9) | 3 (3.2) | 5 (14.7) | 1 (3.2) | 6 (18.2) | 4 (16.0) | 3 (12.0) | 1 (8.3) | 1 (9.1) | — | 7 (5.3) | |
| FAB-M2 | 33 (4.6) | 10 (3.2) | 2 (2.2) | 2 (5.9) | 2 (6.5) | 5 (15.2) | — | 2 (8.0) | — | — | 1 (8.3) | 9 (6.8) | |
| FAB-M3 | — | — | — | — | — | — | — | — | — | — | — | — | |
| FAB-M4 | 137 (19.0) | 23 (7.3) | 13 (14.0) | 12 (35.3) | 13 (41.9) | 10 (30.3) | 5 (20.0) | 14 (56.0) | 2 (16.7) | 3 (27.3) | 4 (33.3) | 38 (28.6) | |
| FAB-M5 | 462 (64.0) | 254 (81.2) | 67 (72.0) | 14 (41.2) | 14 (45.2) | 11 (33.3) | 12 (48.0) | 6 (24.0) | 5 (41.7) | 6 (54.5) | 6 (50.0) | 67 (50.4) | |
| FAB-M6 | — | — | — | — | — | — | — | — | — | — | — | — | |
| FAB-M7 | 20 (2.8) | 14 (4.5) | 3 (3.2) | — | — | — | 1 (4.0) | — | 1 (8.3) | — | — | 1 (0.8) | |
| FAB unspecified | 7 (1.0) | — | 1 (1.1) | 1 (2.9) | — | — | — | — | — | — | — | 5 (3.8) | |
| Median blast in BM (%) (N = 653) | 80.0 | 82.0 | 82.0 | 88.5 | 72.0 | 70.0 | 86.0 | 69.0 | 72.0 | 45.0 | 85.0 | 79.0 | |
| CNS involvement, n (%) (N = 693) | 100 (14.4) | 33 (11.0) | 16 (17.6) | 5 (15.2) | 8 (27.6) | 6 (19.4) | 2 (9.1) | 2 (8.0) | 1 (9.1) | 2 (22.2) | 3 (25.0) | 22 (17.1) | .272 |
| Hepatomegaly, n (%) (N = 634) | 371 (58.8) | 163 (57.0) | 55 (63.2) | 17 (63.0) | 21 (67.7) | 11 (40.7) | 10 (62.5) | 12 (57.1) | 6 (60.0) | 4 (50.0) | 7 (63.6) | 65 (59.1) | .782 |
| Splenomegaly, n (%) (N = 641) | 278 (43.4) | 123 (43.2) | 44 (50.0) | 15 (55.6) | 15 (48.4) | 9 (34.6) | 7 (43.8) | 4 (20.0) | 4 (20.0) | 5 (62.5) | 3 (27.3) | 49 (45.0) | .347 |
| Additional cytogenetic aberrations, n (%) (N = 736) | 375 (51.0) | 164 (51.4) | 42 (43.8) | 19 (54.3) | 21 (66.7) | 21 (61.8) | 12 (48.0) | 18 (75.0) | 5 (38.5) | 5 (41.7) | 8 (66.7) | 60 (44.4) | .062 |
| . | Total . | t(9;11)(p22;q23) . | t(10;11)(p12;q23) . | t(6;11)(q27;q23) . | t(11;19)(q23;p13) . | t(11;19)(q23;p13.1) . | t(11;19)(q23;p13.3) . | t(1;11)(q21;q23) . | t(4;11)(q21;q23) . | t(10;11)(p11.2;q23) . | t(11;17)(q23;q21) . | Other . | P* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 756 | 328 | 98 | 35 | 31 | 34 | 25 | 25 | 13 | 12 | 12 | 143 | |
| Male sex, n (%) | 367 (48.5) | 157 (47.9) | 55 (56.1) | 19 (54.3) | 13 (41.9) | 15 (44.1) | 11 (44.0) | 9 (36.0) | 3 (23.1) | 6 (50) | 7 (58.3) | 72 (50.3) | .495 |
| Median age (N = 756) | 2.2 | 2.6 | 1.3 | 12.4 | 1.6 | 4.6 | 7.1 | 1.3 | 1.3 | 1.0 | 9.0 | 1.9 | |
| Less than 2 y, n (%) | 358 (47.4) | 137 (41.8) | 61 (62.2) | 3 (8.6) | 18 (58.1) | 14 (41.2) | 9 (36.0) | 19 (76.0) | 8 (61.5) | 9 (75.0) | 4 (33.3) | 72 (50.3) | < .001 |
| 2 to 9 y, n (%) | 227 (30.0) | 117 (35.7) | 22 (22.4) | 12 (34.3) | 8 (25.8) | 11 (32.4) | 5 (20.0) | 6 (24.0) | 2 (15.4) | 3 (25.0) | 3 (25.0) | 41 (28.6) | |
| 10 or more y, n (%) | 171 (22.6) | 74 (22.6) | 15 (15.3) | 20 (57.1) | 5 (16.1) | 9 (26.5) | 11 (44.0) | — | 3 (23.1) | — | 5 (41.7) | 30 (21.0) | |
| Median WBC (N = 754) | 20.9 | 13.7 | 10.9 | 59.8 | 76.0 | 27.0 | 17.5 | 33.4 | 20.0 | 17.2 | 87.8 | 17.2 | |
| Less than 20 × 109/L, n (%) | 371 (49.1) | 180 (54.9) | 58 (59.2) | 8 (22.9) | 10 (32.3) | 15 (44.1) | 14 (56.09) | 9 (36.0) | 6 (46.2) | 6 (50.0) | 4 (33.3) | 61 (42.7) | |
| 20 to less than 100 × 109/L, n (%) | 206 (27.2) | 82 (25.0) | 22 (22.4) | 12 (34.3) | 9 (29.0) | 13 (38.2) | 3 (12.0) | 11 (44.0) | 3 (23.1) | 2 (16.7) | 3 (25.0) | 46 (32.2) | < .001 |
| 100 or more × 109/L, n (%) | 177 (23.4) | 65 (19.8) | 18 (18.4) | 15 (42.9) | 12 (38.7) | 6 (17.6) | 8 (32.0) | 5 (20.0) | 4 (30.8) | 4 (33.3) | 5 (41.7) | 35 (24.5) | |
| FAB type, n (%) (N = 722) | < .001 | ||||||||||||
| FAB-M0 | 23 (3.2) | 3 (1.0) | 4 (4.3) | — | 1 (3.2) | 1 (3.0) | 3 (12.0) | — | 3 (25.0) | 1 (9.1) | 1 (8.3) | 6 (4.5) | |
| FAB-M1 | 40 (5.5) | 9 (2.9) | 3 (3.2) | 5 (14.7) | 1 (3.2) | 6 (18.2) | 4 (16.0) | 3 (12.0) | 1 (8.3) | 1 (9.1) | — | 7 (5.3) | |
| FAB-M2 | 33 (4.6) | 10 (3.2) | 2 (2.2) | 2 (5.9) | 2 (6.5) | 5 (15.2) | — | 2 (8.0) | — | — | 1 (8.3) | 9 (6.8) | |
| FAB-M3 | — | — | — | — | — | — | — | — | — | — | — | — | |
| FAB-M4 | 137 (19.0) | 23 (7.3) | 13 (14.0) | 12 (35.3) | 13 (41.9) | 10 (30.3) | 5 (20.0) | 14 (56.0) | 2 (16.7) | 3 (27.3) | 4 (33.3) | 38 (28.6) | |
| FAB-M5 | 462 (64.0) | 254 (81.2) | 67 (72.0) | 14 (41.2) | 14 (45.2) | 11 (33.3) | 12 (48.0) | 6 (24.0) | 5 (41.7) | 6 (54.5) | 6 (50.0) | 67 (50.4) | |
| FAB-M6 | — | — | — | — | — | — | — | — | — | — | — | — | |
| FAB-M7 | 20 (2.8) | 14 (4.5) | 3 (3.2) | — | — | — | 1 (4.0) | — | 1 (8.3) | — | — | 1 (0.8) | |
| FAB unspecified | 7 (1.0) | — | 1 (1.1) | 1 (2.9) | — | — | — | — | — | — | — | 5 (3.8) | |
| Median blast in BM (%) (N = 653) | 80.0 | 82.0 | 82.0 | 88.5 | 72.0 | 70.0 | 86.0 | 69.0 | 72.0 | 45.0 | 85.0 | 79.0 | |
| CNS involvement, n (%) (N = 693) | 100 (14.4) | 33 (11.0) | 16 (17.6) | 5 (15.2) | 8 (27.6) | 6 (19.4) | 2 (9.1) | 2 (8.0) | 1 (9.1) | 2 (22.2) | 3 (25.0) | 22 (17.1) | .272 |
| Hepatomegaly, n (%) (N = 634) | 371 (58.8) | 163 (57.0) | 55 (63.2) | 17 (63.0) | 21 (67.7) | 11 (40.7) | 10 (62.5) | 12 (57.1) | 6 (60.0) | 4 (50.0) | 7 (63.6) | 65 (59.1) | .782 |
| Splenomegaly, n (%) (N = 641) | 278 (43.4) | 123 (43.2) | 44 (50.0) | 15 (55.6) | 15 (48.4) | 9 (34.6) | 7 (43.8) | 4 (20.0) | 4 (20.0) | 5 (62.5) | 3 (27.3) | 49 (45.0) | .347 |
| Additional cytogenetic aberrations, n (%) (N = 736) | 375 (51.0) | 164 (51.4) | 42 (43.8) | 19 (54.3) | 21 (66.7) | 21 (61.8) | 12 (48.0) | 18 (75.0) | 5 (38.5) | 5 (41.7) | 8 (66.7) | 60 (44.4) | .062 |
WBC indicates white blood cell count; FAB, French-American-British morphology classification; BM, bone marrow; CNS, central nervous system; and —, zero.
P values indicate whether the differences are significant among the subgroups.
Statistical analyses
CR was defined as less than 5% blasts in the bone marrow, with regeneration of trilineage hematopoiesis plus absence of leukemic cells in the cerebrospinal fluid or elsewhere.26 Early death was defined as any death within the first 6 weeks of treatment. Treatment of patients who did not obtain CR was considered a failure on day 0. Overall survival (OS) was measured from the date of diagnosis to the date of last follow-up or death from any cause. Event-free survival (EFS) was calculated from the date of diagnosis to the first event or to the date of last follow-up. For OS and EFS analyses, patients who did not experience an event were censored at the time of last follow-up. The Kaplan-Meier method was used to estimate the 5-year probabilities of OS (pOS) and EFS (pEFS), and survival estimates were compared using the log-rank test. Cumulative incidence functions of relapse (with other events and death while in CR as competing events) were constructed by the method of Kalbfleisch and Prentice and compared by the Gray test. For multivariate analysis, the Cox proportional-hazard regression model was used; allogeneic HSCT was included as a time-dependent covariate. Continuous variables known to be of prognostic value in AML were categorized according to cutoff points (eg, age older than 1, 2, 5, or 10 years, WBC < 20 × 109/L or > 100 × 109/L). The χ2 or Fisher exact test was used to compare differences in percentages of variables among subgroups; the Mann-Whitney U test was used for continuous variables. All P values are descriptive and explorative and were considered significant if less than or equal to .01. Statistical analyses were conducted using SAS software (SAS-PC, Version 9.1).
Results
11q23 or MLL rearrangements
Of the 756 cases, 376 (50%) were evaluated by conventional cytogenetics only. A total of 380 (50%) cases were evaluated by conventional cytogenetics; in addition, an MLL rearrangement was confirmed with either FISH or RT-PCR. We did not detect differences in sex distribution, age at diagnosis, and diagnostic WBC between these 2 groups. No differences were detected for outcome between the cases for whom only conventional cytogenetics were available and the cases that were molecularly evaluated (pEFS 42.9% ± 2.6% vs 44.3% ± 2.6%, P = .70). Moreover, because conventional cytogenetic methods are usually complementary to RT-PCR and FISH and allow reliable identification of MLL gene rearrangements in leukemic clones, we addressed the total group as 11q23/MLL rearrangements.
Clinical characteristics
The patient characteristics of the various 11q23/MLL subgroups are described in detail in Table 1. There were no differences in sex distribution among the 11q23/MLL subgroups; however, significant differences were seen in age at diagnosis, diagnostic WBC, and FAB types. The median age at diagnosis for the entire cohort was 2.2 years (range, 0-17.9 years). In most 11q23/MLL subgroups, the median age at diagnosis was younger than 5 years; however, in the t(6;11)(q27;q23) and t(11;17)(q23;q21) subgroups, the median ages were 12.4 and 9.0 years, respectively (Table 1). Patients with a t(6;11)(q27;q23), t(11;17)(q23;q21), or t(11;19)(q23;p13)-without ascertained subband had a higher median WBC than other subgroups (59.8 × 109/L, 87.8 × 109/L, and 76.0 × 109/L, respectively; Table 1). Most of the AML cases were classified as FAB-M5 (64%) or FAB-M4 (19%); however, in some subgroups, another FAB-type distribution was found. For instance, in the t(1;11)(q21;q23) subgroup, only 25% of patients presented with FAB-M5 morphology and 56% presented with FAB-M4 (Table 1).
11q23/MLL rearrangements and survival
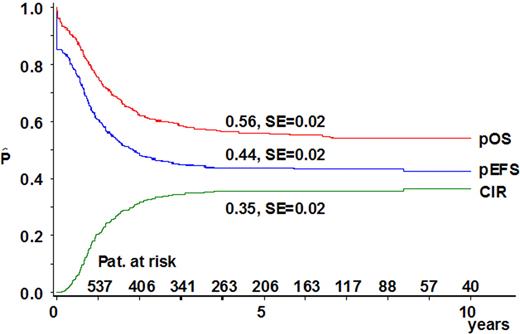
The median follow-up time of survivors was 4.8 years. In 99% of the patients, therapy was administered. The overall CR rate was 85% with no significant differences in CR rates among the subgroups (P = .64). For the cohort of 11q23/MLL-rearranged patients, 5-year EFS was 44% (± 2%), 5-year OS was 56% (± 2%), and the cumulative incidence of relapse at 5 years from diagnosis was 35% (± 2%) (Figure 1).
Survival curves of the 756 pediatric patients with 11q23/MLL-rearranged AML included in this study. The probability of overall survival (pOS) at 5 years from diagnosis was 56% ± 2% (312 events); the probability of event-free survival (pEFS) was 44% ± 2% (417 events); and the probability of cumulative incidence of relapse (pCIR) was 35% ± 2% (257 events).
Survival curves of the 756 pediatric patients with 11q23/MLL-rearranged AML included in this study. The probability of overall survival (pOS) at 5 years from diagnosis was 56% ± 2% (312 events); the probability of event-free survival (pEFS) was 44% ± 2% (417 events); and the probability of cumulative incidence of relapse (pCIR) was 35% ± 2% (257 events).
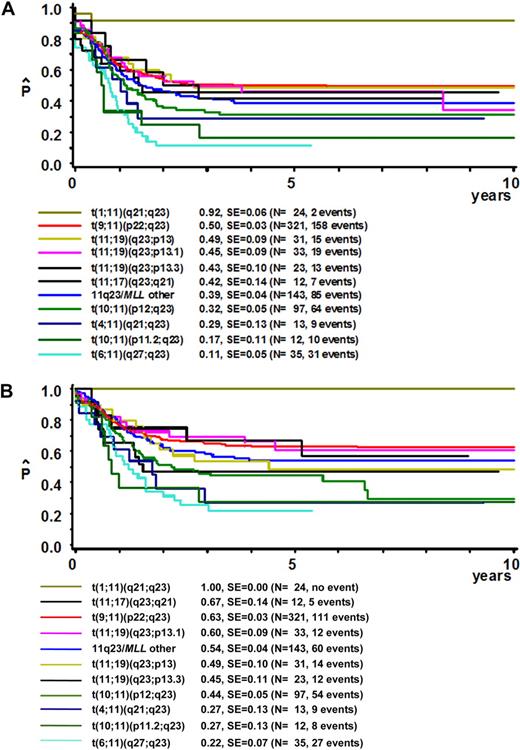
Interestingly, the outcome of patients with different 11q23/MLL rearrangements varied significantly (Table 2; Figure 2). The 24 patients with a t(1;11)(q21;q23) had an excellent outcome with a 5-year pEFS of 92% (± 5%) and an 5-year pOS of 100% (± 0%), as the 2 patients with an event were successfully rescued at relapse. On the other hand, the 35 patients with a t(6;11)(q27;q23) had the worst outcome: 5-year EFS was 11% (± 5%) and 5-year OS was 22% (± 7%). In addition, the 11q23 subgroups t(10;11)(p11.2;q23), t(4;11)(q21;q23), and t(10;11)(p12;q23) showed poor outcome (5-year pEFS was 17% ± 11%, 29% ± 13%, and 32% ± 5%, respectively); OS rates were in agreement with the 5-year pEFS data.
Survival estimates of prognostic factors in 11q23/MLL-rearranged AML, including subgroups based on translocation partners
| . | 5-year pEFS . | 5-year pCIR . | 5-year pOS . | |||
|---|---|---|---|---|---|---|
| Mean percentage (SE) . | P* (log rank) . | Mean percentage (SE) . | P* (Gray test) . | Mean percentage (SE) . | P* (log rank) . | |
| MLL rearrangement | ||||||
| All | 44 (2) | 35 (2) | 56 (2) | |||
| t(1;11)(q21;q23) | 92 (5) | < .001 | 4 (4) | < .001 | 100 (0) | < .001 |
| t(9;11)(p22;q23) | 50 (3) | 29 (3) | 63 (3) | |||
| t(11;19)(q23;p13) | 49 (9) | 24 (8) | 49 (10) | |||
| t(11;19)(q23;p13.1) | 46 (9) | 42 (9) | 61 (9) | |||
| t(11;19)(q23;p13.3) | 46 (10) | 21 (9) | 47 (11) | |||
| t(11;17)(q23;q21) | 42 (14) | 41 (15) | 67 (14) | |||
| t(10;11)(p12;q23) | 31 (5) | 52 (5) | 45 (5) | |||
| t(4;11)(q21;q23) | 29 (13) | 56 (16) | 27 (13) | |||
| t(10;11)(p11.2;q23) | 17 (11) | 50 (16) | 27 (13) | |||
| t(6;11)(q27;q23) | 11 (5) | 54 (9) | 22 (7) | |||
| Other | 39 (4) | 40 (5) | 54 (5) | |||
| Additional cytogenetic aberrations | ||||||
| No | 48 (3) | .01 | 31 (3) | .03 | 61 (3) | .002 |
| Yes | 39 (3) | 39 (3) | 48 (3) | |||
| WBC | ||||||
| Less than 20 × 109/L, n (%) | 48 (3) | .004 | 45 (3) | .6 | 59 (3) | .004 |
| 20 to less than 100 × 109/L, n (%) | 43 (4) | 42 (4) | 57 (4) | |||
| 100 × 109/L or more, n (%) | 36 (4) | 46 (4) | 47 (4) | |||
| Age | ||||||
| Less than 10 y | 46 (2) | .006 | 43 (3) | .048 | 60 (2) | .001 |
| 10 y or more | 34 (4) | 51 (4) | 42 (4) | |||
| . | 5-year pEFS . | 5-year pCIR . | 5-year pOS . | |||
|---|---|---|---|---|---|---|
| Mean percentage (SE) . | P* (log rank) . | Mean percentage (SE) . | P* (Gray test) . | Mean percentage (SE) . | P* (log rank) . | |
| MLL rearrangement | ||||||
| All | 44 (2) | 35 (2) | 56 (2) | |||
| t(1;11)(q21;q23) | 92 (5) | < .001 | 4 (4) | < .001 | 100 (0) | < .001 |
| t(9;11)(p22;q23) | 50 (3) | 29 (3) | 63 (3) | |||
| t(11;19)(q23;p13) | 49 (9) | 24 (8) | 49 (10) | |||
| t(11;19)(q23;p13.1) | 46 (9) | 42 (9) | 61 (9) | |||
| t(11;19)(q23;p13.3) | 46 (10) | 21 (9) | 47 (11) | |||
| t(11;17)(q23;q21) | 42 (14) | 41 (15) | 67 (14) | |||
| t(10;11)(p12;q23) | 31 (5) | 52 (5) | 45 (5) | |||
| t(4;11)(q21;q23) | 29 (13) | 56 (16) | 27 (13) | |||
| t(10;11)(p11.2;q23) | 17 (11) | 50 (16) | 27 (13) | |||
| t(6;11)(q27;q23) | 11 (5) | 54 (9) | 22 (7) | |||
| Other | 39 (4) | 40 (5) | 54 (5) | |||
| Additional cytogenetic aberrations | ||||||
| No | 48 (3) | .01 | 31 (3) | .03 | 61 (3) | .002 |
| Yes | 39 (3) | 39 (3) | 48 (3) | |||
| WBC | ||||||
| Less than 20 × 109/L, n (%) | 48 (3) | .004 | 45 (3) | .6 | 59 (3) | .004 |
| 20 to less than 100 × 109/L, n (%) | 43 (4) | 42 (4) | 57 (4) | |||
| 100 × 109/L or more, n (%) | 36 (4) | 46 (4) | 47 (4) | |||
| Age | ||||||
| Less than 10 y | 46 (2) | .006 | 43 (3) | .048 | 60 (2) | .001 |
| 10 y or more | 34 (4) | 51 (4) | 42 (4) | |||
pEFS indicates probability of event-free survival; pCIR, probability of cumulative incidence of relapse; pOS, probability of overall survival; WBC, white blood cell count.
P values indicate whether the differences are significant between the subgroups.
Survival curves for patients with 11q23/MLL-rearranged pediatric AML grouped on the basis of different translocation partners. (A) Event-free survival curves. (B) Overall survival curves. Assignment to 11q23-rearranged subgroups was based on translocation partners, as identified after central review of karyotyping. Some patients were assigned to 11q23 subgroups based on RT-PCR results only. If an MLL rearrangement was determined by FISH and the translocation partner was unknown, the patient was included in the “11q23/MLL-other” group. At least 10 patients had to be included to create a subgroup; otherwise, the cases were allocated to the 11q23/MLL-other group. Patients with a t(1;11)(q21;q23) showed independent favorable outcome with overall survival at 5 years of 100% ± 0%, and an event-free survival of 92% ± 5%. Several rearrangements were identified as predictors of poor clinical outcome, including t(6;11)(q27;q23), t(10;11)(p11.2;q23), t(4;11)(q21;q23), and t(10;11)(p12;q23).
Survival curves for patients with 11q23/MLL-rearranged pediatric AML grouped on the basis of different translocation partners. (A) Event-free survival curves. (B) Overall survival curves. Assignment to 11q23-rearranged subgroups was based on translocation partners, as identified after central review of karyotyping. Some patients were assigned to 11q23 subgroups based on RT-PCR results only. If an MLL rearrangement was determined by FISH and the translocation partner was unknown, the patient was included in the “11q23/MLL-other” group. At least 10 patients had to be included to create a subgroup; otherwise, the cases were allocated to the 11q23/MLL-other group. Patients with a t(1;11)(q21;q23) showed independent favorable outcome with overall survival at 5 years of 100% ± 0%, and an event-free survival of 92% ± 5%. Several rearrangements were identified as predictors of poor clinical outcome, including t(6;11)(q27;q23), t(10;11)(p11.2;q23), t(4;11)(q21;q23), and t(10;11)(p12;q23).
When analyzing the type of events, there were significant differences in the cumulative incidence of relapse between the various 11q23/MLL subgroups (pGray < .001; Table 2), whereas the 5-year cumulative incidence of death in CR (pGray = .468) did not differ significantly among the subgroups.
Other prognostic factors
Patients with a high WBC (> 100 × 109/L) had a significantly worse 5-year pEFS than did patients presenting with a low WBC (< 20 × 109/L; 36% ± 4% vs 48% ± 3%; P = .001). After evaluating various cutoff points for age, patients older than 10 years had significantly worse 5-year pEFS than patients younger than 10 years (34% ± 4% vs 46% ± 2%; P = .006). In addition, patients with additional cytogenetic aberrations had significantly worse 5-year pEFS than other patients (39% ± 3% vs 48% ± 3%; P = .01; Table 2). In addition, differences in outcome were seen across the study groups with 5-year pEFS ranging from 29% (± 10%) to 61% (± 7%; supplemental Table 3).
Prognostic factors in the t(9;11)(p22;q23) subgroup
The 5-year pEFS of patients with a t(9;11)(p22;q23) was 50% (± 3%). Within this subgroup, patients with FAB-M5 morphology had a 5-year pEFS of 56% (± 3%), and those with other FAB subtypes had 31% (± 7%) (P = .002; supplemental Table 4). No differences in outcome were detected according to age.
Comparing the outcome of the t(9;11)(p22;q23) subgroup with that of other 11q23 subgroups within the various collaborative study groups, we found, in the NOPHO protocols only, that patients with a t(9;11)(p22;q23) did better than those with other 11q23/MLL rearrangements (5-year pEFS, 77% ± 8% vs 38% ± 9%, P log-rank = .006).
Multivariate analysis of the total cohort of patients with 11q23/MLL rearrangements
Cox regression analysis of EFS from diagnosis revealed the subgroups t(6;11)(q27;q23) (hazard ratio [HR] = 2.3, P < .001), t(10;11)(p12;q23) (HR = 1.5, P = .004), and t(10;11)(p11.2;q23) (HR = 2.4, P = .007) as independent predictors of poor prognosis (Table 3). With an HR of 0.1 (P = .004), the t(1;11)(q21;q23) subgroup was associated with a favorable outcome. In addition, WBC greater than 100 × 109/L was an independent predictor of poor prognosis (HR = 1.4, P = .003). All of these factors also predicted OS from diagnosis. Allogeneic HSCT as a time-dependent variable or treatment according to study group was not an independent predictor of EFS or OS. When the analysis was restricted to the poor prognostic subgroups t(10;11)(p12;q23) or t(6;11)(q27;q23), allogeneic HSCT as time-dependent variable again did not predict for EFS or OS.
Multivariate analysis of survival parameters of pediatric patients with 11q23/MLL-rearranged AML
| . | pEFS . | pOS . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| MLL translocation | ||||||
| t(9;11)(p22;q23) | 1.0 | Reference | 1.0 | Reference | ||
| t(10;11)(p12;q23) | 1.5 | 1.1-2.1 | .004 | 1.9 | 1.3-2.6 | < .001 |
| t(6;11)(q27;q23) | 2.3 | 1.5-3.4 | < .001 | 2.5 | 1.6-4.0 | < .001 |
| t(11;19)(q23;p13) | 1.0 | 0.6-1.7 | .91 | 1.4 | 0.8-2.4 | .27 |
| t(11;19)(q23;p13.1) | 1.1 | 0.7-1.8 | .77 | 0.9 | 0.5-1.7 | .83 |
| t(11;19)(q23;p13.3) | 1.1 | 0.6-1.9 | .85 | 1.5 | 0.8-2.7 | .17 |
| t(1;11)(q21;q23) | 0.1 | 0.0-0.6 | .007 | — | — | — |
| t(4;11)(q21;q23) | 1.6 | 0.8-3.2 | .15 | 2.3 | 1.2-4.6 | .02 |
| t(10;11)(p11.2;q23) | 2.4 | 1.3-4.6 | .007 | 3 | 1.5-6.3 | .003 |
| t(11;17)(q23;q21) | 1.0 | 0.5-2.2 | .94 | 1 | 0.4-2.6 | .94 |
| Other | 1.3 | 1.0-1.7 | .05 | 1.3 | 0.9-1.8 | .11 |
| Additional cytogenetic aberrations | ||||||
| No | 1.0 | Reference | 1.0 | Reference | ||
| Yes | 1.2 | 1.0-1.5 | .05 | 1.3 | 1.1-1.7 | .02 |
| WBC | ||||||
| Less than 100 × 109/L | 1.0 | Reference | 1.0 | Reference | ||
| 100 × 109/L or more | 1.4 | 1.1-1.7 | .003 | 1.4 | 1.1-1.8 | .007 |
| Age | ||||||
| Less than 10 y | 1.0 | Reference | 1.0 | Reference | ||
| 10 y or more | 1.3 | 1.0-1.6 | .04 | 1.4 | 1.1-1.8 | .01 |
| Allogeneic BMT | ||||||
| No | 1.0 | Reference | 1.0 | Reference | ||
| Yes | 0.9 | 0.6-1.3 | .59 | 0.9 | 0.6-1.2 | .41 |
| . | pEFS . | pOS . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| MLL translocation | ||||||
| t(9;11)(p22;q23) | 1.0 | Reference | 1.0 | Reference | ||
| t(10;11)(p12;q23) | 1.5 | 1.1-2.1 | .004 | 1.9 | 1.3-2.6 | < .001 |
| t(6;11)(q27;q23) | 2.3 | 1.5-3.4 | < .001 | 2.5 | 1.6-4.0 | < .001 |
| t(11;19)(q23;p13) | 1.0 | 0.6-1.7 | .91 | 1.4 | 0.8-2.4 | .27 |
| t(11;19)(q23;p13.1) | 1.1 | 0.7-1.8 | .77 | 0.9 | 0.5-1.7 | .83 |
| t(11;19)(q23;p13.3) | 1.1 | 0.6-1.9 | .85 | 1.5 | 0.8-2.7 | .17 |
| t(1;11)(q21;q23) | 0.1 | 0.0-0.6 | .007 | — | — | — |
| t(4;11)(q21;q23) | 1.6 | 0.8-3.2 | .15 | 2.3 | 1.2-4.6 | .02 |
| t(10;11)(p11.2;q23) | 2.4 | 1.3-4.6 | .007 | 3 | 1.5-6.3 | .003 |
| t(11;17)(q23;q21) | 1.0 | 0.5-2.2 | .94 | 1 | 0.4-2.6 | .94 |
| Other | 1.3 | 1.0-1.7 | .05 | 1.3 | 0.9-1.8 | .11 |
| Additional cytogenetic aberrations | ||||||
| No | 1.0 | Reference | 1.0 | Reference | ||
| Yes | 1.2 | 1.0-1.5 | .05 | 1.3 | 1.1-1.7 | .02 |
| WBC | ||||||
| Less than 100 × 109/L | 1.0 | Reference | 1.0 | Reference | ||
| 100 × 109/L or more | 1.4 | 1.1-1.7 | .003 | 1.4 | 1.1-1.8 | .007 |
| Age | ||||||
| Less than 10 y | 1.0 | Reference | 1.0 | Reference | ||
| 10 y or more | 1.3 | 1.0-1.6 | .04 | 1.4 | 1.1-1.8 | .01 |
| Allogeneic BMT | ||||||
| No | 1.0 | Reference | 1.0 | Reference | ||
| Yes | 0.9 | 0.6-1.3 | .59 | 0.9 | 0.6-1.2 | .41 |
pEFS indicates probability of event-free survival; pOS, probability of overall survival; —, no events; and WBC, white blood cell count.
Multivariate Cox regression analysis limited to the subgroup of patients (n = 328) with a t(9;11)(p22;q23) identified FAB-M5 as an independent favorable prognostic factor for EFS and OS from diagnosis (both HR = 0.4, P < .001). In addition, WBC less than 20 × 109/L was an independent favorable prognostic factor for EFS (HR = 1.6, P = .003). Within the t(9;11) subgroup, allogeneic HSCT as a time-dependent variable did not predict OS (HR = 1.0, P = .99; supplemental Table 5).
Discussion
In this large international retrospective study, we identified novel prognostic subgroups in pediatric 11q23/MLL-rearranged AML. Patients with a t(1;11)(q21;q23) showed favorable outcome independent of other risk factors, whereas those with a t(6;11)(q27;q23), t(10;11)(p12;q23), or t(10;11)(p11.2;q23) showed poor clinical outcome independent of other factors. These results underscore the importance of international collaboration in the investigation of rare diseases or subgroups.
This study is the first to identify t(1;11)(q21;q23) as a subgroup with excellent clinical outcome. The biologic background for this favorable outcome is currently poorly understood, and conflicting results have been reported. Co et al performed AF1q overexpression and knockdown experiments in cell lines and concluded that overexpression resulted in enhanced doxorubicin-induced apoptosis mediated through the BAD pathway, whereas knockdown of AF1q expression resulted in the reverse.27 In another study, in 64 unselected pediatric AML samples, high AF1q expression was independently associated with poor survival.28 Whether MLL-AF1q causes different expression levels of AF1q remains unknown; hence, further studies should address the underlying biology. Given the excellent outcome of this rare subgroup of patients, we suggest that direct screening for this translocation be incorporated in future pediatric AML treatment protocols and that patients with MLL-AF1q should be allocated to the low-risk group (or treatment arm) of these protocols.
In this study, we identified t(6;11)(q27;q23) as an independent predictor of poor prognosis in pediatric AML. In adult AML, the poor outcome of patients with a t(6;11)(q27;q23) has been reported before, and adults with this specific translocation do equally bad (2-year OS, 13%).29 Of interest, children with a t(6;11)(q27;q23) have a higher WBC and older age at presentation than other MLL-rearranged cases. We could not prove that allogeneic HSCT was of benefit in these patients, but numbers were limited. Given the dismal outcome, studies are urgently needed to elucidate the biology of the MLL-AF6 fusion gene, which may lead to the development of new treatment regimens for this subgroup. Meanwhile, screening for this translocation should become part of the standard screening procedures in pediatric AML because it will allow appropriate allocation of these patients to the high-risk treatment arm of pediatric AML protocols and prospectively validate our results.
The cytogenetic abnormalities t(10;11)(p12;q23) and t(10;11)(p11.2;q23) were also identified as independent predictors of an unfavorable prognosis in this study. The t(10;11)(p12;q23) subgroup is the second most common translocation detected in pediatric 11q23/MLL-rearranged AML. An accurate diagnosis of patients with this translocation requires specific screening with FISH because these patients often show heterogeneity in their chromosomal breakpoints and are therefore not identified by conventional karyotyping.30 Similar to patients with t(6;11)(q27;q23), these patients should be allocated to high-risk arm of pediatric AML treatment protocols.
The present study did not confirm the favorable outcome of patients with a t(9;11)(p22;q23), compared with other 11q23/MLL-rearranged subgroups, as previously described in smaller pediatric and adult series.4,9,31 However, patients with t(9;11)(p22;q23) included in NOPHO protocols still had significantly better outcome than other 11q23/MLL-rearranged patients treated on NOPHO protocols. Previously, it was suggested that a high cumulative dosage of cytarabine administered in NOPHO and St Jude Children's Research Hospital protocols contributed to this better outcome, but this relationship could not be confirmed in the present study.4,9 Moreover, no differences in cumulative dosages of anthracyclines or etoposide, which could explain the improved outcome of t(9;11)(p22;q23) on NOPHO protocols, were detected. Therefore, the factor(s) that determines the relatively specific benefit of NOPHO protocols for patients with a t(9;11)(p22;q23) remains unknown. One hypothesis may be that the NOPHO study group has a more ethnically homogeneous population than the other European and American study groups and the outcomes in t(9;11)(p22;q23) may be determined, to some extent, by ethnicity-related genetic factors.32,33
Of interest, within the t(9;11)(p22;q23) subgroup, prognosis was related to the cell type from which the leukemia originated, as patients with acute monoblastic leukemia did significantly better than those with other FAB types. The 5-year pEFS of the group of non–FAB-M5 t(9;11)(p22;q23) was only 31%, which was similar to that of the other 11q23/MLL poor-risk groups discussed earlier. These results are in accordance with the previously reported prognostically favorable outcome of t(9;11)(p22;q23) of the St Jude Children's Research Hospital, where a high frequency (21 of 23 cases) of FAB-M5 was detected and the 2 cases without FAB-M5 did not achieve CR and eventually died.9 In an adult study by Mrozek et al,31 a favorable outcome was detected for t(9;11)(p22;q23) and possibly addressed to intensive postremission therapy. In the latter study, 84% of the adult patients with a t(9;11)(p22;q23) presented with a FAB-M5. This may indicate that the favorable outcome of t(9;11)(p22;q23) in these studies is dominated by the frequency of FAB-M5 patients and stressed the importance of including FAB-type for risk stratification.
The frequency of 11q23/MLL-rearranged AML may have been underestimated in the included study cohorts, as well as in other studies performed in the past because of cryptic MLL rearrangements that cannot be detected by conventional karyotyping. Because the 11q23/MLL-rearranged group is associated with poor outcome, FISH screening for MLL rearrangements at diagnosis has become the standard approach in many AML protocols. The results from this study confirm that this approach is worthwhile, but they also demonstrate the need to screen for specific translocation partners to allow appropriate treatment stratification. This is not possible using FISH analysis alone; therefore, additional methods such as RT-PCR should become part of standardized screening procedures to correctly identify patients with specific low- or high-risk MLL rearrangements.
In conclusion, this unique and very large international retrospective study identified several novel, independent prognostic 11q23/MLL-rearranged subgroups, including the favorable-risk subgroup with a t(1;11)(q21;q23) and the poor-risk subgroups with a t(10;11)(p12;q23), t(10;11)(p11.2, q23), or t(6;11)(q27;q23). We recommend that these subgroups be included in the risk-group stratification process in current and future pediatric AML protocols. The patients with a t(9;11)(p22;q23) represent a heterogeneous subgroup requiring the inclusion of FAB type for accurate risk-group stratification. In addition, the biologic background of the various subgroups of 11q23/MLL-rearranged AML should be further investigated, as these specific subgroups may benefit from specific and targeted treatment components, especially those subgroups with an adverse prognostic outcome, despite current intensive therapy.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Angela McArthur for editorial assistance and Dr Riccardo Masetti and Francesca Predieri for collecting data.
This work was supported by the International Pediatric AML Group of the I-Berlin-Frankfurt-Münster Study Group-SG, which is chaired by Prof. Gertjan J.L. Kaspers (1999-2007) and Dr David Webb (2007-present). This work was further supported in part by the American Lebanese Syrian Associated Charities, the Czech Ministry of Education (MSM0021620813), Children's Oncology Group Chair's Grant U10 CA98543, COG Statistics & Data Center Grant U10 CA98413, and the Swedish Children's Cancer Foundation. The project of B.V.B. is funded by the NWO Netherlands Organization for Scientific Research and the Foundation Childhood Oncology Center Rotterdam.
Authorship
Contribution: B.V.B., R.P., C.M.Z., and M.M.v.d.H.-E. conveyed and planned the study, analyzed the data, and wrote the paper; J.H. and S.C.R. reviewed the karyotypes and wrote the paper; M.Z. performed the statistical analyses and wrote the paper; and T.A.A., A.A., H.B.B., M.C., U.C., M.N.D., E.F., B.G., H.H., C.J.H., N.A.H., G.J.L.K., A.L., N.L., L.L.N., A.M., C.P., D.R., J.E.R., F.O.S., J.S., I.S., S.S., T.T., D.T., D.W., and Z.Z. participated in data collection and in critical review and final approval of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Complete lists of the participants and the involved study groups appear as supplemental Appendices online.
Correspondence: Marry M. van den Heuvel-Eibrink, Erasmus MC–Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Rm Sp 2456, Dr Molewaterplein 60, PO Box 2060, 3000 CB Rotterdam, The Netherlands; e-mail: m.vandenheuvel@erasmusmc.nl.
References
Author notes
*S.C.R. and J.H. contributed equally to this study.
†C.M.Z. and M.M.v.d.H.-E. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal