Abstract
Multiple myeloma (MM) is a cancer of plasma cells with complex molecular characteristics that evolves from monoclonal gammopathy of undetermined significance, a highly prevalent premalignant condition. MM is the second most frequent hematologic cancer in the United States, and it remains incurable, thereby highlighting the need for new therapeutic approaches, particularly those targeting common molecular pathways involved in disease progression and maintenance, shared across different MM subtypes. Here we report that Wnt/β-catenin is one such pathway. We document the involvement of β-catenin in cell-cycle regulation, proliferation, and invasion contributing to enhanced proliferative and metastatic properties of MM. The pleiotropic effects of β-catenin in MM correlate with its transcriptional function, and we demonstrate regulation of a novel target gene, Aurora kinase A, implicating β-catenin in G2/M regulation. β-catenin and Aurora kinase A are present in most MM but not in normal plasma cells and are expressed in a pattern that parallels progression from monoclonal gammopathy of undetermined significance to MM. Our data provide evidence for a novel functional link between β-catenin and Aurora kinase A, underscoring a critical role of these pathways in MM disease progression.
Introduction
Multiple myeloma (MM) is a neoplasm of plasma cells that infiltrates the bone marrow (BM). Despite recent advances in its treatment, it remains incurable, with a median survival of 6 years.1 MM accounts for more than 10% of all hematologic malignancies and is the second most frequent hematologic cancer in the United States. It is typically preceded by an age-progressive condition termed monoclonal gammopathy of undetermined significance (MGUS), which is present in 1% to 10% of adults older than 25 years of age and progresses to malignant MM at a rate of 0.5% to 3% annually.1,2 This disease is characterized by frequent chromosomal aberrations and mutations in several oncogenes and tumor-suppressor genes.3,4
The Wnt/β-catenin pathway is significant in cancer development because numerous human malignancies such as colorectal, hepatocellular, and breast cancer harbor activating mutations in critical components of this pathway.5,6 Normally, the Wnt signaling pathway is active during embryogenesis, hematopoietic stem cell growth, cell differentiation, and tissue development. β-catenin, a central effector of the Wnt pathway, is involved in both nuclear and cytoplasmic functions.7,8 In the absence of Wnt ligands, β-catenin is targeted by a complex consisting of adenomatous polyposis coli, axin, glycogen synthase kinase-3β, and casein kinase 1α that phosphorylate and mark it for degradation by the ubiquitin-proteasome pathway.9,10 Upon Wnt stimulation, however, the kinase complex is dissociated, and β-catenin is not targeted for destruction. The active form of β-catenin translocates to the nucleus and, in association with lymphoid enhancer factor (LEF)/T-cell factor (TCF) proteins, activates transcription of target genes like c-Myc, cyclin D1, and Axin2, which are involved in cell proliferation, migration, and survival.11-13
Accumulation of nuclear β-catenin also has been documented in MM, although known mutations in the Wnt pathway have not been identified in these cells, which suggests alternate pathways for β-catenin activation.14-16 Wnt/β-catenin signaling also has been implicated in MM–BM stromal-cell interactions, where factors secreted by tumor cells create a positive environment for the growth and development of MM.16,17 Conversely, other reports18-20 have shown that MM tumor cells secrete a potent inhibitor of Wnt signaling, Dickkopf-1 (DKK1), which causes MM-associated bone disease. Understanding the complex molecular genetics of β-catenin activity in disease would provide valuable insight and could help to develop new therapies that target malignancies with aberrant Wnt/β-catenin activity.14
We report that β-catenin plays a crucial role in MM tumor progression and maintenance by influencing cell-cycle progression and proliferation. The reduction of β-catenin levels in MM cells is associated with significant transcriptional changes in Aurora kinase A, a novel Wnt target gene. These transcriptional changes correlated with tumorigenesis, metastasis, and survival in a mouse xenograft model of MM. Overall, our findings highlight the significance of the Wnt/β-catenin activity in this disease and provide molecular insight for developing new therapeutic strategies targeting β-catenin levels or novel Wnt/β-catenin downstream genes.
Methods
Patient samples and cell lines
Patient MM samples were obtained after informed consent was provided in accordance with the Declaration of Helsinki and under the auspices of a Dana-Farber Cancer Institute Institutional Review Board–approved protocol. Cultured MM cell lines were collected from different sources and maintained as previously described.14 Control and Wnt3A CM was collected from supernatants of cultured L cells (CRL-2648) and L-Wnt-3A cells (CRL-2647) obtained from ATCC.
Immunoblotting and immunofluorescence
Immunoblotting and immunofluorescence were performed as previously described.14 Nuclear and cytoplasmic fractions were prepared as previously described.21 Primary antibodies were from the following manufacturers: Zymed: β-catenin (CAT5-H10); Cell Signaling: AurKA (no. 4718), CDC25B (no. 9525), survivin (no. 2802); Santa Cruz: c-MYC (sc-764), green fluorescent protein (GFP; B-2), Actin-HRP (C-11), Cyclin B1 (sc-245), Cyclin A (sc-596); Dako: Ki67 (M7240); and R&D: CDC25A (MAB1648). Secondary antibodies conjugated to horseradish peroxidase (Santa Cruz) for immunoblot and to fluorescein isothiocyanate or Texas Red (5 μg/mL; Southern Biotechnology) for immunofluorescence were used. Immunocytochemical analysis was performed by the use of an epifluorescence microscope (Nikon Eclipse E800; Nikon) with the use of oil-immersion objectives at 60× or 100× magnification and a Photometrics Coolsnap CF color camera (Nikon) and analyzed by the use of MetaMorph Version 4.6.5 software (Molecular Devices).
Tissue microarrays and immunohistochemistry
Tissue microarrays were constructed manually by the use of paraffin-embedded blocks from normal, MGUS, and MM patient BM biopsies (Brigham & Women's Hospital). Immunohistochemistry (IHC) was performed according to standard protocol by the use of specific antibodies for β-catenin (CAT5-H10), AurKA (AHP873; AbD Serotec), CDC25B (no. 9525), and CD138 (281-2; BD Bioscience). Formalin-fixed tissues were paraffin embedded, sectioned at 5 μm, and stained with hematoxylin and eosin. Anti–β-catenin, TdT-mediated biotin 16-dUTP nick-end labeling (TUNEL) stain, and AurKA were visualized with the corresponding biotinylated antibody coupled to streptavidin–peroxidase complex (Vector Labs). All Antibodies, conditions, and reactivities were tested in positive control slides. Histologic micrographs were taken by the use of a Leica DM200 microscope (aperture HC PLANs 10X/22, objective lenses: N PLAN 100×/1.25 oil) and a SPOT Insight QE Model camera with SPOT Advanced acquisition software (Diagnostic Instruments). The microarray data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through GEO Series accession number GSE17385 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17385).
Lentiviral infections of MM cells
Lentiviral pLKO.1 short hairpin (sh)RNA vectors against human β-catenin and AurKA were obtained from the DFCI-Broad RNAi Consortium. β-catenin shRNA no. 2 (CCGGGCTTGGAATGAGACTGCTGATCTCGAGATCAGCAGTCTCATTCCAAGCTTTTT) and AurKA shRNA no. 1 (CCGGACGAGAATTGTGCTACTTATACTCGAG TATAAGTAGCACAATTCTCGTTTTTT) showed consistent knockdown of their respective targets. A puromycin selection marker from the pLKO.1 shRNA vector was replaced by GFP for selection by flow cytometry. Lentiviruses were generated in 293T cells and used to infect target cells. GFP-positive cells were sorted 3 to 6 days after infection and used for in vitro and in vivo assays.
Cell proliferation, cell-cycle, and reporter assays
Cell proliferation was assayed by [3H]-thymidine uptake or MTT assays performed in triplicate and repeated at least twice, as previously described,14 as well as by Trypan blue exclusion with an equal number of control or β-catenin shRNA cells plated and counted in duplicate up to 6 days. The average and SEM of 2 representative experiments are shown. Cell-cycle progression was monitored through propidium iodide staining followed by fluorescence-activated cell-sorting (FACS) analysis. The data represent the average of triplicate analyses repeated twice. Apoptosis was assayed by FACS after staining with annexin V and 7-Amino-actinomycin D (7-AAD). The TCF reporter assays using Dual Luciferase Reporter System (Promega) were performed as previously described.14 In brief, 0.5 × 106 MM1.S cells were transfected with 2.0 μg of TOPFLASH or FOPFLASH or 1.0 μg of AurKA reporter pGL1486 with increasing amounts of β-catenin (0.5-2.0 μg) and 0.01 μg of hRLnull renilla using Fugene6 standard protocol (Roche). Results were normalized to total protein and renilla. Experiments were performed in triplicate and repeated at least twice.
Mouse xenograft experiments
Stable control shRNAGFP and β-catenin shRNAGFP MM1.S cells were generated as described earlier. Eight-week-old NOD.CB17-PrkdcSCID/J mice (The Jackson Laboratory) were intravenously injected with 106 MM1.S-GFP+ cells. GFP tumor nodules were visualized by use of the LT-9500 fluorescent light box (Lighttools Research). All animal experiments were approved by and conformed to the standards of the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute. Experiments were repeated twice independently. Necropsy, visualization, and tissue preparation and analysis were performed as previously described.14
RNA expression profiling
β-catenin expression profiling data on Affymetrix GeneChip for MM primary tumors and normal plasma cells were obtained from the National Center for Biotechnology Information GEO database, accession GSE4452.
RNA was extracted from 5 × 106 MM1.S control or β-catenin shRNA cells with Trizol (Invitrogen), purified by the use of RNeasy kit (QIAGEN), and run on an Affymetrix U133A plus 2.0 array chip. The statistical analyses were performed in R (http://www.r-project.org) with the use of Bioconductor software. Probe set expression values were generated using “plier” (http://bioconductor.org/packages/1.8/bioc/html/plier.html). The potential LEF1/TCF4 targets were obtained from the BROAD GSEA website (http://www.broad.mit.edu/gsea/) and compared with the down-regulated targets obtained from the above analysis. 89 probe sets were down greater than 2-fold with a P value less than .01.
ChIP
Chromatin immunoprecipitation (ChIP) assays were performed by the use of MCF-7 cells according to the protocol previously described.22 The protocol has been well established in the adherent MCF-7 cell line and is used as a prototype for identifying targets under Wnt/β-catenin regulation. In brief, formaldehyde cross-linked chromatin was isolated from 5 × 107 cells followed by IP with rabbit β-catenin antibody (Cell Signaling; no. 9562) or normal rabbit immunoglobulin (IgG) bound to magnetic DynaBeads A and G (Invitrogen) according to the manufacturer's protocol. DNA was eluted from the beads and reverse cross-linked according to the protocol. Real-time polymerase chain reaction was used to analyze the immunoprecipitated DNA with the use of primers against c-Myc TBE1 (positive; fwd-GGTCCACAAGCTCTCCACTT and rev-CGGTTTGCAACAGTCTCG) or putative AurKA TCF4 binding sites identified from the Broad/GSEA website (AurKA A: fwd-CCATAACTTGTGTTAGAGCTGC, rev-GGTAACTGAACATTGTCCATGGGA; AurKA B: fwd-CACTCGCCAGGTAAACAGAAGC, rev-GTCCTTAGTTCGCCTCT GCATC), as well as nonspecific negative control (Neg1, designed to amplify sequences not expected to recruit β-catenin) or water alone. Experiments were repeated twice, and the average fold difference in relative enrichment (ChIP/IgG) over the negative control is represented.
BH3 profiling assays
BH3 profiling assays were performed as described previously.23 In this method, mitochondria are exposed to measured death signals in the form of BH3-domain peptides and the mitochondrial response quantified. Peptides used in this assay were synthesized by Tufts University Core Facility and purified by high-performance liquid chromatography. Release of cytochrome c was determined by a comparison of cytochrome c in the pellet and supernatant quantified by enzyme-linked immunosorbent assay (R&D Systems). The results are representative of at least 3 independent experiments performed in duplicate.
Results
β-catenin is aberrantly expressed in MM, and down-regulation increases sensitivity to chemotherapeutic agents
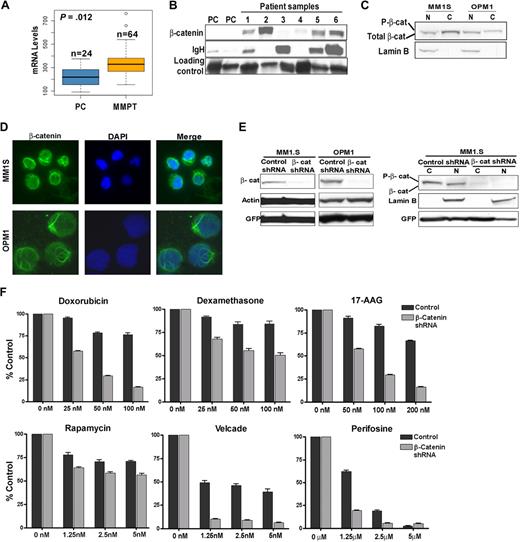
mRNA expression analysis of primary MM patient samples on Affymetrix gene chips revealed a slight increase in β-catenin levels in MM primary tumors compared with normal plasma cells (Figure 1A). However, immunoblots showed that β-catenin is expressed in most patient samples at much greater levels than in normal plasma cells, indicating that β-catenin accumulation through posttranslational mechanisms may be an important contributor to MM pathogenesis (Figure 1B). As documented in other studies,14 β-catenin is also expressed in MM cell lines, which were used in our functional studies by the use loss and gain of function approaches (as discussed in the next paragraph). β-catenin expression in MM1.S and OPM1 cell lines was documented by immunoblotting (Figure 1C) and immunofluorescence (Figure 1D).
β-catenin is aberrantly expressed in MM, and its down-regulation by the use of shRNA knockdown increases sensitivity to chemotherapeutic agents. (A) Affymetrix analysis of β-catenin mRNA expression in normal plasma cells (PC) and multiple myeloma primary tumors (MMPT). (B) Immunoblot of β-catenin protein in PC and MMPT (1-6) cells. Immunoglobulin heavy chain (IgH) and a nonspecific protein were used as loading markers. (C) Nuclear (N) and cytoplasmic (C) protein fractions of MM1.S and OPM1 cells. Lamin B was used as nuclear fraction loading control. (D) Immunofluorescent staining of total β-catenin (green) in MM1.S (60×) and OPM1 (100×) cells. DAPI is shown in blue. (E) Immunoblot of β-catenin knockdown by stable lentiviral shRNA transduction in MM1.S and OPM1 cells. GFP was used as a whole-cell lysate transduction efficiency marker, whereas Actin and Lamin B were used as whole-cell lysate loading and nuclear fraction loading controls markers, respectively. Note reduced β-catenin levels in both cytoplasmic (C) and nuclear fractions (N). (F) MM1.S control or β-catenin shRNA cells were cocultured with BM stromal cells and treated with different drugs followed by MTT assays to measure metabolism.
β-catenin is aberrantly expressed in MM, and its down-regulation by the use of shRNA knockdown increases sensitivity to chemotherapeutic agents. (A) Affymetrix analysis of β-catenin mRNA expression in normal plasma cells (PC) and multiple myeloma primary tumors (MMPT). (B) Immunoblot of β-catenin protein in PC and MMPT (1-6) cells. Immunoglobulin heavy chain (IgH) and a nonspecific protein were used as loading markers. (C) Nuclear (N) and cytoplasmic (C) protein fractions of MM1.S and OPM1 cells. Lamin B was used as nuclear fraction loading control. (D) Immunofluorescent staining of total β-catenin (green) in MM1.S (60×) and OPM1 (100×) cells. DAPI is shown in blue. (E) Immunoblot of β-catenin knockdown by stable lentiviral shRNA transduction in MM1.S and OPM1 cells. GFP was used as a whole-cell lysate transduction efficiency marker, whereas Actin and Lamin B were used as whole-cell lysate loading and nuclear fraction loading controls markers, respectively. Note reduced β-catenin levels in both cytoplasmic (C) and nuclear fractions (N). (F) MM1.S control or β-catenin shRNA cells were cocultured with BM stromal cells and treated with different drugs followed by MTT assays to measure metabolism.
To study the effects of β-catenin in MM disease progression, we undertook a loss of function approach using stable β-catenin shRNA knockdown. Pure populations of stable lentivirally transduced cells were sorted with GFP as a marker. From a panel of 5 β-catenin shRNAs, 2 showed significant β-catenin knockdown, designated shRNA no. 1 and no. 2, (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article, and data not shown). Because shRNA no. 2 was more effective and consistent in knocking down β-catenin, we used this shRNA in further studies. We were able to significantly reduce β-catenin in both OPM1 and MM1.S cells as determined by immunoblotting (Figure 1E). β-catenin levels in both cytoplasmic and nuclear fractions were decreased (Figure 1E right panel). We next investigated the sensitivity of MM cells in response to different chemotherapeutic agents by MTT assay in a MM1.S-BMSC coculture context (Figure 1F). Overall, MM1.S cells with β-catenin knockdown were more sensitive compared with the control cells, particularly those treated with doxorubicin, 17-AAD, dexamethasone, and bortezomib. These results highlight the importance of β-catenin in MM therapeutic susceptibility and encouraged us to further delineate its functional role in MM pathogenesis.
β-catenin promotes proliferation and cell-cycle progression of myeloma cells
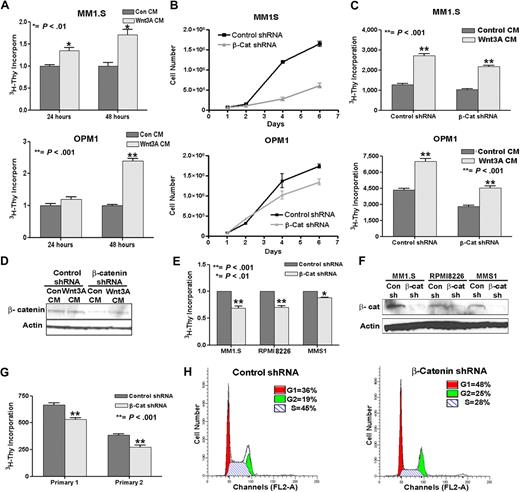
We assessed the functional effects of β-catenin levels in MM cells by demonstrating that treating MM1.S or OPM1 cells with Wnt3A conditioned media (CM)–enhanced cell proliferation (Figure 2A). MM1.S and OPM1 cells stably transduced with either a control (scrambled sequence) or β-catenin shRNA were monitored for cell proliferation. As shown in Figure 2B, β-catenin knockdown in both cell lines led to a significant decrease in proliferation over time compared with control cells. Both β-catenin shRNAs (no. 1 and no. 2) showed similar effects on proliferation in MM1.S cells (supplemental Figure 1B). However, treating β-catenin shRNA cells with Wnt3A CM was able to partially rescue cell proliferation by restoring β-catenin levels, demonstrating the specificity in knocking down β-catenin and the involvement of this pathway in proliferation of MM (Figure 2C-D). In addition to OPM1 and MM1.S cells, other myeloma cell lines RPMI8226 and MMS1 also demonstrated that β-catenin knockdown inhibited proliferation (Figure 2E-F). Primary MM samples stably transduced with control or β-catenin shRNAs were sorted for GFP-positive populations and demonstrated a similar effect on proliferation with β-catenin knockdown (Figure 2G).
β-catenin directly affects proliferation and cell cycle in MM. (A) Wnt3A stimulation of MM1.S (top) and OPM1 (bottom) cells increases proliferation. Cells were treated with control (Con CM) or Wnt3A-conditioned medium (Wnt3A CM) for 24 or 48 hours and assayed for 3H-thymidine incorporation. (B) β-catenin (β-Cat shRNA) knockdown decreased proliferation of OPM1 and MM1.S cells. (C) Partial rescue in proliferation of OPM1 or MM1.S β-catenin shRNA (β-Cat shRNA) cells after 24 hours treatment with Wnt3A CM. In panels B and C, cells had been stably transduced and sorted for GFP-positive cells within 4 days after infection, followed by a growth curve (B) or treatment with control or Wnt3A CM for 24 hours, followed by 3H-Thy proliferation assay (C). The difference in proliferation starts out modestly but is more apparent over a period of 6 days, as is shown in panel B, whereas in panel C the experiment was performed with the use of cells technically at the day 2 time point of panel B. (D) A representative immunoblot shows a slight-but-consistent increase in β-catenin after 24-hour Wnt3A CM treatment in β-catenin shRNA OPM1 cells. (E) β-catenin (β-Cat shRNA) knockdown inhibits proliferation in MM1.S, RPM18226, and MMS1 cells. (F) Immunoblot analysis confirms the reduction of β-catenin in the different MM cells stably transduced with β-catenin shRNA (β-cat). Proliferation assays were performed in triplicate and repeated at least twice. The average and SEM of 2 to 3 experiments are shown. (G) Primary MM patient samples with β-catenin knockdown (β-Cat shRNA) also show decreased proliferation. Proliferation assays were performed in triplicate. The error bars and statistical significance were calculated from the triplicate dataset. (H) Increase in G1 and G2/M cell-cycle phases and decrease in S phase in β-catenin shRNA MM1.S cells compared with control cells. The FACS data represent the average of triplicate analyses repeated twice.
β-catenin directly affects proliferation and cell cycle in MM. (A) Wnt3A stimulation of MM1.S (top) and OPM1 (bottom) cells increases proliferation. Cells were treated with control (Con CM) or Wnt3A-conditioned medium (Wnt3A CM) for 24 or 48 hours and assayed for 3H-thymidine incorporation. (B) β-catenin (β-Cat shRNA) knockdown decreased proliferation of OPM1 and MM1.S cells. (C) Partial rescue in proliferation of OPM1 or MM1.S β-catenin shRNA (β-Cat shRNA) cells after 24 hours treatment with Wnt3A CM. In panels B and C, cells had been stably transduced and sorted for GFP-positive cells within 4 days after infection, followed by a growth curve (B) or treatment with control or Wnt3A CM for 24 hours, followed by 3H-Thy proliferation assay (C). The difference in proliferation starts out modestly but is more apparent over a period of 6 days, as is shown in panel B, whereas in panel C the experiment was performed with the use of cells technically at the day 2 time point of panel B. (D) A representative immunoblot shows a slight-but-consistent increase in β-catenin after 24-hour Wnt3A CM treatment in β-catenin shRNA OPM1 cells. (E) β-catenin (β-Cat shRNA) knockdown inhibits proliferation in MM1.S, RPM18226, and MMS1 cells. (F) Immunoblot analysis confirms the reduction of β-catenin in the different MM cells stably transduced with β-catenin shRNA (β-cat). Proliferation assays were performed in triplicate and repeated at least twice. The average and SEM of 2 to 3 experiments are shown. (G) Primary MM patient samples with β-catenin knockdown (β-Cat shRNA) also show decreased proliferation. Proliferation assays were performed in triplicate. The error bars and statistical significance were calculated from the triplicate dataset. (H) Increase in G1 and G2/M cell-cycle phases and decrease in S phase in β-catenin shRNA MM1.S cells compared with control cells. The FACS data represent the average of triplicate analyses repeated twice.
Interestingly, we also found that reduction of β-catenin led to significant changes in the cell cycle (Figure 2H and data not shown), where a prominent increase in G1 (36% + 0.9% to 48% + 1.9%) and G2 (19% + 0.7% to 24% + 0.6%) and decrease in S (45% + 0.9% to 28% + 1.4%) phases were observed. In addition, flow cytometric analysis by annexin V and 7-AAD staining of cells 8 days after lentiviral infection also showed an increase in cells undergoing apoptosis in cells with β-catenin knockdown (Annexin + 7-AAD = 7.5% + 2.1% to 16.0% + 1.1%; P < .01 and supplemental Figure 2A). Together, our data suggest that knockdown of β-catenin alters cell-cycle progression and influences proliferation of MM cells.
β-catenin target genes are involved in multiple aspects of cell-cycle regulation in MM
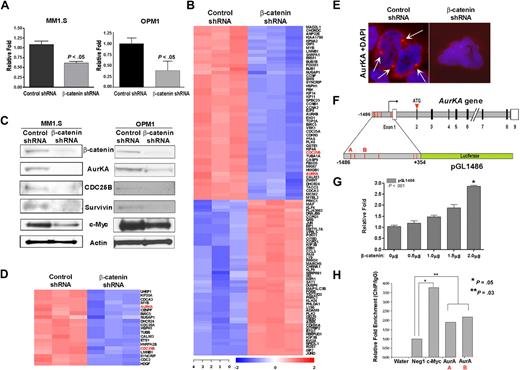
The aforementioned results prompted us to assess the effect of reducing β-catenin on its transcriptional activity in a TCF4-specific reporter assay to further identify β-catenin downstream target genes in MM cells. MM1.S and OPM1 cells stably transduced with control or β-catenin shRNA were transiently transfected with either a TOPFLASH (TCF4 Optimal Promoter-Luciferase) reporter or control (FOPFLASH) and assayed for luciferase activity. As expected, reducing β-catenin diminished the transcriptional activity of the β-catenin-TCF4 complex (Figure 3A).
β-catenin knockdown reveals novel cell-cycle regulators as targets in MM. (A) Decreased Wnt reporter activity in β-catenin shRNA cells. MM1.S and OPM1 cells were transfected with TOPFLASH and assayed for luciferase activity after 48 hours. Control FOPFLASH levels were unaffected. Experiments were performed in triplicate and repeated twice. The results denote the average and SEM of triplicate assays. (B) GEP revealed 2 distinct sets of genes that are most up- or down-regulated by β-catenin knockdown compared with control shRNA MM1.S cells. (C) Microarray target validation by immunoblot show decreased expression of known and novel targets after β-catenin knockdown in MM1.S (left) and OPM1 cells (right). (D) GEP of β-catenin shRNA down-regulated genes containing LEF1/TCF4 binding sites (GCTTTGT/A). Only probe sets expressed in all 3 samples are shown. (E) Immunofluorescence of control or β-catenin shRNA MM1.S cells (100×) for AurKA (red) and DAPI (blue). (F) Diagram of the AurKA gene structure (adapted from GenBank, accession no. AL121914) and AurKA-luciferase reporter pGL1486 (adapted from Tanaka et al24 ). Red lines depict potential TCF4 binding sites, untranslated regions (open box), solid box (open reading frame). (G) Cells transfected with pGL1486 and increasing β-catenin amounts assayed similar to panel A show increased AurKA-luciferase activity. (H) Chromatin IP (ChIP) assays in MCF-7 cells demonstrated direct binding of β-catenin transcriptional complex to AurKA gene promoter sequences. The bar graph represents ChIP/immunoglobulin G (IgG) signal normalized to DNA concentration. A nonspecific primer (Neg1) and water were used as negative controls; known primers against the TBE1 region of the c-Myc promoter (c-Myc) was used as a positive control; AurKA primer pairs A and B were designed around regions of putative TCF4 binding sites of the AurKA promoter, described in panel F (bottom). The average fold difference in relative enrichment (ChIP/IgG) over the negative control is represented.
β-catenin knockdown reveals novel cell-cycle regulators as targets in MM. (A) Decreased Wnt reporter activity in β-catenin shRNA cells. MM1.S and OPM1 cells were transfected with TOPFLASH and assayed for luciferase activity after 48 hours. Control FOPFLASH levels were unaffected. Experiments were performed in triplicate and repeated twice. The results denote the average and SEM of triplicate assays. (B) GEP revealed 2 distinct sets of genes that are most up- or down-regulated by β-catenin knockdown compared with control shRNA MM1.S cells. (C) Microarray target validation by immunoblot show decreased expression of known and novel targets after β-catenin knockdown in MM1.S (left) and OPM1 cells (right). (D) GEP of β-catenin shRNA down-regulated genes containing LEF1/TCF4 binding sites (GCTTTGT/A). Only probe sets expressed in all 3 samples are shown. (E) Immunofluorescence of control or β-catenin shRNA MM1.S cells (100×) for AurKA (red) and DAPI (blue). (F) Diagram of the AurKA gene structure (adapted from GenBank, accession no. AL121914) and AurKA-luciferase reporter pGL1486 (adapted from Tanaka et al24 ). Red lines depict potential TCF4 binding sites, untranslated regions (open box), solid box (open reading frame). (G) Cells transfected with pGL1486 and increasing β-catenin amounts assayed similar to panel A show increased AurKA-luciferase activity. (H) Chromatin IP (ChIP) assays in MCF-7 cells demonstrated direct binding of β-catenin transcriptional complex to AurKA gene promoter sequences. The bar graph represents ChIP/immunoglobulin G (IgG) signal normalized to DNA concentration. A nonspecific primer (Neg1) and water were used as negative controls; known primers against the TBE1 region of the c-Myc promoter (c-Myc) was used as a positive control; AurKA primer pairs A and B were designed around regions of putative TCF4 binding sites of the AurKA promoter, described in panel F (bottom). The average fold difference in relative enrichment (ChIP/IgG) over the negative control is represented.
We then performed gene expression profiling analysis (GEP) of RNA from MM1.S control and β-catenin shRNA cells. Using a Limma moderated t test (Benjamini-Hochberg–adjusted P < .01), we detected 10252 probes clustered into 2 different groups that were differentially expressed in β-catenin shRNA relative to control cells (Figure 3B). Fifty of the most significantly down-regulated and up-regulated genes by β-catenin knockdown in MM1.S are shown in Figure 3B. Interestingly, several of the genes showing altered pattern of expression were involved in different aspects of cell-cycle regulation, such as Aurora kinases A (AurKA) and B, CDC25A and B, and Ki67, which are involved in the G2/M transition. Some of these results were validated by IB in both MM1.S and OPM1 cells (Figure 3C), which showed decreased protein expression of AurKA and CDC25B, as well as known Wnt targets such as c-Myc and survivin. Further analysis revealed that a significant proportion of the down-regulated genes (BH-adjusted P < .01) have LEF/TCF4 (GCTTTGT/A) binding sites in their promoters, suggesting the possibility of being bona fide β-catenin target genes. Both AurKA and CDC25B were among the top 20 down-regulated potential β-catenin target genes (Figure 3D). Decreased AurKA expression in β-catenin knockdown cells was further demonstrated by immunofluorescence (Figure 3E). Analysis of the AurKA promoter sequence revealed several potential LEF1/TCF4 binding sites (Figure 3F red lines). Using an AurKA-luciferase reporter pGL1486,24 we demonstrated a direct correlation between increasing levels of transfected β-catenin and transcriptional activity of the AurKA promoter in MM cells, thereby identifying AurKA as a novel target of β-catenin (Figure 3G). Furthermore, ChIP analysis in MCF-7 cells, with the use of primers designed around the putative TCF4 binding sites of the AurKA promoter, demonstrated the ability of β-catenin to directly recognize and immunoprecipitate regions of the AurKA promoter (AurKA primers A and B), providing evidence that AurKA is a direct target of β-catenin (Figure 3H). These data corroborate the alterations observed in the cell-cycle studies and further implicates β-catenin in regulation of proliferation and cell-cycle progression in MM.
Aurora kinase A as a downstream target of β-catenin is involved in proliferation in MM
On identifying AurKA as a novel target of β-catenin, we explored its function in MM cells and whether β-catenin effects on proliferation were influenced by AurKA levels. As shown in Figure 4A-B, both treatment with Wnt3A CM and stable transfection of β-catenin caused an increase in β-catenin as well as AurKA in MM1.S and OPM1 (supplemental Figure 3A). As noted previously, treatment with Wnt3A CM also correlated with increased proliferation in MM1.S and OPM1 cells (Figure 2A and supplemental Figure 3B). The same proliferative effect also was observed when β-catenin was overexpressed in MM1.S cells (Figure 4C). To evaluate the specificity of AurKA effect on cell proliferation, we generated stable AurKA knockdown in MM1.S and OPM1 cells using lentiviral shRNA (Figure 4D, supplemental Figure 3, and data not shown). Changes in proliferation could be attributed to the effect of AurKA knockdown on cell cycling, which by day 4 after infection demonstrated an increase in G2 (from 19.4% to 29.2%), indicative of aberrant G2/M transition (Figure 4E and supplemental Figure 3C).
Wnt/β-catenin-mediated effects on proliferation are affected by Aurora kinase A levels. (A) Wnt3A CM stimulation of MM1.S cells shows increased β-catenin and AurKA levels. MM1.S cells stably overexpressing β-catenin show increased AurKA levels (B) and enhanced proliferation (C). (D) Immunoblot of stable AurKA shRNA knockdown in MM1.S cells. (E) Cell-cycle profiling of control or AurKA shRNA MM1.S cells show increased G2/M phase. (F) AurKA knockdown in MM1.S decreased proliferation. (G) AurKA shRNA counteracts the positive proliferative effect of β-catenin overexpression or Wnt3A CM stimulation (H). Control (Control GFP) or β-catenin overexpressing cells (β-catenin GFP) were stably transduced with control (■) or AurKA shRNA ([graphic024]) and assayed for proliferation. Proliferation experiments were done in triplicate and repeated twice. The results denote the average and SEM of triplicate assays.
Wnt/β-catenin-mediated effects on proliferation are affected by Aurora kinase A levels. (A) Wnt3A CM stimulation of MM1.S cells shows increased β-catenin and AurKA levels. MM1.S cells stably overexpressing β-catenin show increased AurKA levels (B) and enhanced proliferation (C). (D) Immunoblot of stable AurKA shRNA knockdown in MM1.S cells. (E) Cell-cycle profiling of control or AurKA shRNA MM1.S cells show increased G2/M phase. (F) AurKA knockdown in MM1.S decreased proliferation. (G) AurKA shRNA counteracts the positive proliferative effect of β-catenin overexpression or Wnt3A CM stimulation (H). Control (Control GFP) or β-catenin overexpressing cells (β-catenin GFP) were stably transduced with control (■) or AurKA shRNA ([graphic024]) and assayed for proliferation. Proliferation experiments were done in triplicate and repeated twice. The results denote the average and SEM of triplicate assays.
Similar to β-catenin knockdown, a reduction in AurKA levels also decreased proliferation in MM1.S and OPM1 cells (Figure 4F and supplemental Figure 3D). In addition, the increase in proliferation that was observed after stably overexpressing β-catenin (Figure 4C) was diminished when the cells were subsequently infected with AurKA shRNA, which decreased the levels of AurKA (Figure 4G). A similar, although less dramatic, effect on proliferation also was observed when AurKA shRNA cells were treated with Wnt3A CM compared with control shRNA cells (Figure 4H). As expected, prolonged direct knockdown of AurKA lead to increased MM cell death with respect to the control shRNA cells, with total annexin V staining (sum of annexin V-positive populations in upper left and right quadrants) increasing from 10.1% to 28.5% (supplemental Figure 2B). These data demonstrate that AurKA mediates the effects of β-catenin, at least partially, on cell cycle and proliferation, suggesting an important role for AurKA in the molecular mechanisms of β-catenin–mediated pathogenesis in MM.
β-catenin influences survival and metastasis in an in vivo xenograft model of MM
We next investigated whether β-catenin impacts invasion and survival in a xenograft mouse model of MM. Control shRNA-GFP or β-catenin shRNA-GFP MM1.S cells were transplanted by intravenous injections into sublethally irradiated 8-week-old nonobese diabetic/severely compromised immunodeficient mice. As shown in Figure 5A, Kaplan-Meier survival plots revealed that the survival was significantly increased in mice injected with β-catenin shRNA cells, with a mean survival of 25 days for mice with control shRNA cells (control mice) versus 94.4 days for mice with β-catenin shRNA cells (shRNA mice; P < .002). The control mice developed hind limb paralysis and died by days 23 to 27; however, the shRNA mice survived 3.8 times longer, ranging from 51 to 114 days. Whole-body imaging of GFP-positive tissues revealed that control cells metastasized to their normal homing site in the BM and showed high tumor burden in the head, spine, and long bones (Figure 5B left panel). β-catenin shRNA cells also homed to the BM but showed a dramatic reduction in tumor burden, with fewer and smaller GFP tumor nodules. In addition, the control animals had numerous GFP-positive metastases in other organs such as the liver and kidneys but were almost absent in shRNA animals (Figure 5B-D), indicating marked reduction in the metastatic potential of β-catenin shRNA MM cells.
β-catenin knockdown improves survival in a xenograft mouse model of MM. β-catenin knockdown improves survival in Kaplan-Meier survival curves (A) by decreasing tumor load and metastasis (B). Mice injected with control or β-catenin shRNA-GFP MM1.S cells were analyzed by whole-body imaging. Note decreased tumor GFP nodule number in the spine and liver of β-catenin shRNA mice compared with control mice (white arrows). Histologic and IHC analysis of tumors showed decreased tumor metastasis in the liver (C) and kidney (D), increased numbers of tingible body macrophages (E), and increased TUNEL staining (F), as well as decreased expression of β-catenin (G) and AurKA (H) in engrafted β-catenin shRNA MM1.S cells compared with control MM1.S xenografts. Hematoxylin & eosin stains (panels C, D, and E); IHC stains (panels G and H).
β-catenin knockdown improves survival in a xenograft mouse model of MM. β-catenin knockdown improves survival in Kaplan-Meier survival curves (A) by decreasing tumor load and metastasis (B). Mice injected with control or β-catenin shRNA-GFP MM1.S cells were analyzed by whole-body imaging. Note decreased tumor GFP nodule number in the spine and liver of β-catenin shRNA mice compared with control mice (white arrows). Histologic and IHC analysis of tumors showed decreased tumor metastasis in the liver (C) and kidney (D), increased numbers of tingible body macrophages (E), and increased TUNEL staining (F), as well as decreased expression of β-catenin (G) and AurKA (H) in engrafted β-catenin shRNA MM1.S cells compared with control MM1.S xenografts. Hematoxylin & eosin stains (panels C, D, and E); IHC stains (panels G and H).
Histologic examination of GFP-positive tissues demonstrated an increased tumor burden in the BM of spine in control mice, accounting for the earlier onset of limb paralysis in these animals. Although both tumors showed infiltration into the BM, the shRNA mice showed areas of necrosis and frequent tingible body macrophages within the tumor mass, indicating that tumor cells were more susceptible to be cleared by the host immune system (Figure 5E and data not shown). TUNEL assays showed a noticeable increase in apoptotic activity within the tumor cells of the shRNA mice compared with the control mice (Figure 5F insets). In addition, we performed BH3 profiling analysis to observe whether there were changes in β-catenin shRNA MM1.S cells that caused the mitochondrial apoptotic pathway to be more easily activated.23 Our results revealed that β-catenin shRNA cells undergo mitochondrial permeabilization more readily. Although control cells showed a primarily BCL2 profile, β-catenin knockdown cells exhibited an overlay of BCL2, MCL-1, and BCL-xL profiles (supplemental Figure 4). Therefore, the shift in BH3 profiles might explain the increased cell death sensitivity in the BM tumor masses, perhaps contributing to prolonged survival of these mice.
IHC analysis of BM tissue derived from these mice confirmed decreased expression of β-catenin as well as AurKA, one of the novel targets identified by our in vitro studies (Figure 5G-H).
β-catenin and AurKA expression in the progression of MM
To corroborate the significance of our findings in cell lines and our xenograft murine model of MM, we decided to explore the expression of β-catenin and AurKA in human samples. We performed immunohistochemical analysis using clinically standardized β-catenin and AurKA antibodies on serial sections of a panel of archival BM biopsies of normal individuals (n = 10), patients with MGUS (n = 6), and patients with untreated MM (n = 20). As shown in (Figure 6A), a concurrent increase in the expression of β-catenin and AurKA was observed in plasma tumor cells as the disease progressed from MGUS to MM. A blind immunohistochemical analysis of primary patient samples revealed a significant correlation between β-catenin and AurKA expression in myeloma cells from patients with MGUS and MM, whereas normal plasma cells did not show any significant correlation (supplemental Table 1). Interestingly, the expression of a functional target of AurKA, CDC25B, also was increased from MGUS to MM. A representative example for CDC25B expression is shown (Figure 6A bottom lane). In addition, a reprobe of the immunoblot shown in Figure 1B also revealed a correlation between β-catenin and AurKA expression in tumor samples versus normal plasma cells (Figure 6B).
β-catenin affects expression of target genes in patient sample tissue microarrays. (A) IHC analysis on patient sample tissue microarrays show that AurKA and CDC25B reflect increased β-catenin expression from MGUS to MM compared with normal BM. Costaining of β-catenin (red) and AurKA (brown) in top right panel inset. (B) Immunoblot analysis of AurKA expression normal plasma cells (PC) and MM primary tumors compared with β-catenin expression. Note a correlation between β-catenin and AurKA expression in patient samples 1, 2, 5, and 6.
β-catenin affects expression of target genes in patient sample tissue microarrays. (A) IHC analysis on patient sample tissue microarrays show that AurKA and CDC25B reflect increased β-catenin expression from MGUS to MM compared with normal BM. Costaining of β-catenin (red) and AurKA (brown) in top right panel inset. (B) Immunoblot analysis of AurKA expression normal plasma cells (PC) and MM primary tumors compared with β-catenin expression. Note a correlation between β-catenin and AurKA expression in patient samples 1, 2, 5, and 6.
Overall, these results demonstrated that β-catenin plays important roles in the progression and maintenance of MM in vivo and provide data identifying Aurora kinase A as a significant novel target of β-catenin.
Discussion
MM is a hematologic cancer associated with deregulated Wnt/β-catenin activity. Unlike most epithelial cancers, MM lacks known mutations in critical components of the Wnt pathway, indicating that other mechanisms of Wnt/β-catenin activation may exist.8,25 Here we provide evidence for a role of β-catenin in MM disease progression and maintenance by targeting the AurKA complex, demonstrating a novel role of β-catenin in the regulation of different aspects of MM cell cycle and proliferation. These findings expand on the molecular mechanisms used by β-catenin in contributing to MM pathogenesis as well as other cancers associated with deregulated Wnt activity.
AurKA, a novel Wnt/β-catenin target, is involved cell-cycle regulation in MM
In agreement with previous studies, we have found that Wnt/β-catenin activity is involved in proliferation of malignant plasma cells.14,15,26 However, other studies16,17 indicate a prominent role of aberrant Wnt/β-catenin activity on the surrounding BM microenvironment, underscoring the complex functional network of this pathway in MM pathogenesis. Recent reports18-20 have shown that MM tumor cells secrete a potent inhibitor of Wnt signaling, DKK1, which causes MM-associated bone disease, suggesting the use of DKK1 inhibitors to treat MM-associated bone disease. Here, using a loss of function approach and knocking down the downstream effector of Wnt, we have clearly documented an enhancing effect of β-catenin on MM proliferation. Therefore, our findings are relevant with regard to both understanding the underlying mechanisms involved as well as the treatment of MM and associated bone disease.
Although the role of Wnt/β-catenin in proliferation and cell cycle has been shown before, this effect is mediated at G1 through β-catenin targets c-Myc and Cyclin D1. Here, we demonstrate for the first time that β-catenin also affects other cell cycle phases in MM, which prompted us to investigate novel targets of Wnt/β-catenin that may be involved in the G2/M transition. Our GEP analysis revealed new potential Wnt/β-catenin transcriptional targets involved in various aspects of cell-cycle progression and checkpoint regulation such as CDC25 A/B, cyclins, cyclin-dependent kinases, and AurKA/B, in addition to known targets Survivin, c-Myc, and Cyclin D1.12,13 For instance, the Aurora kinases are involved in the regulation of multiple steps of mitosis, including transit from G2 to cytokinesis, centrosome duplication, formation of bipolar mitotic spindles, chromosome alignment, and spindle checkpoint monitoring. AurKA functions as a mitotic centrosomal kinase required for chromosome maturation and regulation of G2/M phase transition by phosphorylation of many proteins, including CDC25B,27 which we have also found to be regulated by β-catenin in MM. Disruption of AurKA activity in tumor cells leads to accumulation of cells in G2/M and delayed mitotic entry whereas overexpression of AurKA can override G2 checkpoint inhibition.28 Interestingly, the AurKA gene often is amplified in breast29 and colon30 cancers and is associated with aneuploidy, chromosomal aberrations, and tumor progression and behaves as an oncogene in transformation assays.31,32
Indeed, it has recently been shown that inhibition of Aurora kinases leads to significant anti-MM activity33-35 ; however, not much is known about its regulation. Our studies demonstrate that AurKA is a novel target of β-catenin that would directly affect the G2/M transition in cell cycle progression. One of the primary functions of AurKA is the phosphorylation of CDC25B at Ser353 at the centrosome, which in turn activates CyclinB1-Cdk1 to trigger mitosis.36 AurKA is also responsible for G2/M checkpoint recovery through phosphorylation of PLK1.28 Therefore, direct regulation of AurKA by β-catenin would have a significant impact on late stages of cell cycle progression and checkpoint recovery, further supporting the role of β-catenin as a key player in the cell cycle regulation and pathogenesis of MM.
Our expression studies also highlight the effect of β-catenin on the expression of CDC phosphatases, which activate cyclin-dependent kinases at critical stages of the cell cycle.37 Interestingly, CDC25B is phosphorylated by AurKA and is also implicated in the regulation of the transition from G2 to M.27 Transcriptionally, CDC25A and B have been shown to be direct targets of c-Myc,38 itself a known β-catenin target. Similar to AurKA, CDC25A and B also are overexpressed in various cancers. Proteins like AurKA and CDC25B are extremely sensitive to changes in the cell, and disturbance of their normal function can lead to genomic instability and chromosomal alterations, associated with cancer initiation and progression.39 Hence, aberrant expression of β-catenin may cause dysregulated cell-cycle progression and deviant checkpoint regulation, thereby leading to uninhibited proliferation and accumulation of genetic abnormalities, both hallmarks of MM progression.
β-catenin is involved in MM progression and survival
Our studies in a xenograft mouse model of MM demonstrated the significance of β-catenin in MM maintenance. Importantly, they show that a decrease in β-catenin protein caused significant tumor remission and increased survival. Tumors with decreased β-catenin distinctly showed enhanced levels of necrotic and apoptotic cell death within the tumor mass, which may be attributable to inhibition of MM growth and migration, cell-cycle arrest, insufficient interaction with the surrounding environment, and/or changes in apoptotic profiles. This may account for lessening of the tumor burden, delay in hind limb paralysis, and prolonged survival observed in these mice. Furthermore, a decrease in the metastasis of MM cells with β-catenin knockdown was observed, indicating that β-catenin also contributes to the metastatic potential of MM. Our results showed increased survival in the xenograft model of MM and BH3 profiling assays revealed that myeloma cells with decreased β-catenin levels are more “primed” for apoptosis, further underscoring the feasibility of targeting Wnt/β-catenin in the treatment of MM and other hematologic malignancies associated with aberrant Wnt activity.6,14 However, the Wnt/β-catenin pathway is critical for normal cellular functions like hematopoietic stem cell homeostasis,40 which makes therapeutic targeting of the entire pathway or completely abolishing β-catenin function undesirable. One possible option is to interfere with specific interactions of the β-catenin transcriptional complex that would reduce expression of certain target genes without altering others14 (K. Sukhdeo, M.M. Y.Z., D.E.C., K.T., M.G., J.D.-S., H. Ikeda, F. Diaz-Griffero, V. Pena-Cruz, M. Bertagnoli, L. Myeroff, S. Markowitz, K.C.A., D.R.C., manuscript compiled, August 2009). Alternatively, specifically targeting novel downstream targets such as those identified in this work are being investigated in a therapeutic capacity.33-35
In conclusion, we demonstrate that β-catenin and AurKA are critical factors in MM disease progression and maintenance. Our studies provide new molecular insights for further investigation both to elucidate the specific mechanisms involved in MM progression and to develop novel markers for MGUS to MM progression and therapeutic strategies to effectively treat this cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr William Hahn (DFCI, RNAi Consortium [TRC] at the Broad Institute) for lentiviral shRNA vectors, Dr Hans Clevers (Hubrecht Laboratory) for pcDNA β-catenin plasmid, Prof Yoshiaki Ishigatsubo and Dr Atsuhisa Ueda (Yokohoma City University Graduate School of Medicine) for AurKA-luciferase reporter pGL1486, Dr Sonia Vallet for primary patient samples, and Dr Thomas Westerling for assistance with the ChIP experiments. We would also like to thank Dr Victor Pena-Cruz for valuable comments and assistance with the manuscript, and Daisy Moreno, Adalis Maisonet, Mei Zheng, and Nafiun Awal for their excellent technical assistance.
D.R.C. is supported by the Multiple Myeloma Research Foundation (MMRF) Senior Award; K.C.A. is supported by the LeBow Family Fund to Cure Myeloma.
Authorship
Contribution: J.D.-S. and D.R.C. designed and performed research, analyzed data and wrote the manuscript; Y.Z., A.G.L., and K.C.A. analyzed data; G.G., M.G., T.H., K.T., and D.E.C. performed research; M.M. and N.E.C. analyzed data and performed research; and Y.-T.T. and N.R. provided reagents. All authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Daniel R. Carrasco, MD, PhD, Department of Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: ruben_carrasco@dfci.harvard.edu.

![Figure 4. Wnt/β-catenin-mediated effects on proliferation are affected by Aurora kinase A levels. (A) Wnt3A CM stimulation of MM1.S cells shows increased β-catenin and AurKA levels. MM1.S cells stably overexpressing β-catenin show increased AurKA levels (B) and enhanced proliferation (C). (D) Immunoblot of stable AurKA shRNA knockdown in MM1.S cells. (E) Cell-cycle profiling of control or AurKA shRNA MM1.S cells show increased G2/M phase. (F) AurKA knockdown in MM1.S decreased proliferation. (G) AurKA shRNA counteracts the positive proliferative effect of β-catenin overexpression or Wnt3A CM stimulation (H). Control (Control GFP) or β-catenin overexpressing cells (β-catenin GFP) were stably transduced with control (■) or AurKA shRNA ([graphic024]) and assayed for proliferation. Proliferation experiments were done in triplicate and repeated twice. The results denote the average and SEM of triplicate assays.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/13/10.1182_blood-2008-12-194290/4/m_zh89990942480004.jpeg?Expires=1765000204&Signature=lE5gds1zsL~ieNbsDqtKmnDYgkhKO6xsvuOUy6Nn0XWPV~Wf05f2BeC6eCN~AduZYO-WB~sy1ThlldkbcrS7mrXrSpBmq4FdyFuAIYvDvqKx3~M1-dJAR~Su7b3XvC0j3b13Avk4Gmbpg3lkGcC3OdmNGwtf1ljIrYuvRcLAmhT600rfyGlQguIv5pSHgViJSYSI6AyoWREIniMJO6IqZmWYS0hroF233ozsJz6mzxulidntLkwFdh26Flu7MRHZpGjKDC6s0rdK03oGSHhq~tBnI0XcbLvqe9qiJP0C9Oo5-OAftXNWhBjr46q-ctOHe2NDonG8StpSRZYLhu~tjg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal