Abstract
Molecular mechanisms preserving hematopoietic stem cell (HSC) self-renewal by maintaining a balance between proliferation, differentiation, and other processes are not fully understood. Hyperactivation of the mammalian target of rapamycin (mTOR) pathway, causing sustained proliferative signals, can lead to exhaustion of HSC repopulating ability. We examined the role of the novel ras gene Rheb2, an activator of the mTOR kinase, in colony-forming ability, survival, and repopulation of immature mouse hematopoietic cells. In a cell line model of mouse hematopoietic progenitor cells (HPCs), we found enhanced proliferation and mTOR signaling in cells overexpressing Rheb2. In addition, overexpression of Rheb2 enhanced colony-forming ability and survival of primary mouse bone marrow HPCs. Expansion of phenotypic HSCs in vitro was enhanced by Rheb2 overexpression. Consistent with these findings, Rheb2 overexpression transiently expanded phenotypically defined immature hematopoietic cells after in vivo transplantation; however, these Rheb2-transduced cells were significantly impaired in overall repopulation of primary and secondary congenic transplantation recipients. Our findings suggest that HPCs and HSCs behave differently in response to growth-promoting signals stimulated by Rheb2. These results may have value in elucidating mechanisms controlling the balance between proliferation and repopulating ability, a finding of importance in clinical uses of HPCs/HSCs.
Introduction
Hematopoiesis is a well-coordinated process in which hematopoietic stem cells (HSCs) differentiate to every lineage of mature blood cells as well as renew themselves to prevent exhaustion. Determining how these processes are regulated by elucidating molecular targets and signaling pathways involved will probably be beneficial for transplantation, ex vivo expansion, and other clinical uses of HSCs.
Members of the ras superfamily of small GTPase proteins are known to be involved in various stages of hematopoiesis.1-7 The Ras subfamily, Rheb (ras homolog enriched in brain), contains small (20-kDa) GTPases conserved from yeast to humans. Rheb2 is a member of the mammalian Rheb family, which contains both Rheb1 and Rheb2 proteins. There is evidence that human Rheb proteins are ubiquitously expressed and are elevated in many different types of tumors.8,9 Rheb2 has not been studied extensively in mammalian hematopoiesis, despite subtractive cDNA libraries showing Rheb2 to be preferentially expressed in purified HSC populations,10 suggesting that this protein may have a role in unique HSC functions.
Rheb GTPase proteins activate the mammalian target of rapamycin (mTOR) serine/threonine kinase,11 a kinase essential for growth, proliferation, and survival of cells from various organisms.12,13 Signals from growth factor receptors activate the mTOR pathway by activating the PI3K-Akt-TSC (tuberous sclerosis complex) pathway. mTOR exists in 2 complexes in mammalian cells: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Activation of rapamycin-sensitive mTOR complex 1 by Rheb proteins leads to increased translation, cell growth, and survival.12,13 Activation of rapamycin-insensitive mTOR complex 2, possibly by Rheb proteins, affects cell cytoskeletal dynamics, migration, and other processes.12,13
There is evidence demonstrating that the mTOR pathway is important in hematopoiesis, including controlling normal cell growth and differentiation as well as malignant cell proliferation and survival.14-20 Recently, researchers have demonstrated, using a conditional knockout PTEN mouse model, that hyperactivation of the mTOR pathway impairs the repopulating ability of HSCs via a mechanism in which HSCs are stimulated to proliferate and are subsequently exhausted.21 Indeed, many groups have demonstrated that sustained proliferative stimuli lead to the loss of repopulating ability of HSCs, presumably by altering mechanisms controlling quiescence and/or differentiation.22-25 Because Rheb2 is a known activator of mTOR signaling, we determined whether similar effects occur in cells forced to overexpress Rheb2. We found positive effects of Rheb2 overexpression on hematopoietic progenitor cells (HPC) colony formation, growth, and survival. In addition, phenotypic HSCs were expanded by Rheb2 overexpression in vitro. However, overexpression of Rheb2 significantly impaired the ability of HSCs to repopulate lethally irradiated-recipient mice, despite expanding phenotypically immature cells, consistent with the phenotype found in models of mTOR hyperactivation.
Methods
Retroviral transduction and supernatant production
Full-length mouse Rheb2 cDNA (BC016521) was cloned into the murine stem cell virus retroviral vector Mieg3, which expresses enhanced green fluorescent protein (GFP) and has been previously used to transduce mouse hematopoietic cells.26 A flag-tag was added to the N-terminus of the cDNA sequence. Retroviral supernatants were prepared by transfection of Mieg3 or Mieg3-Rheb2 vectors into Phoenix Ecotropic 293T cells (Nolan Lab) using lipofectamine (Invitrogen). Retroviral supernatants were collected 24 and 48 hours later and stored at −80°C. All supernatants were titered on National Institutes of Health 3T3 cells using polybrene transduction reagent (Chemicon International).
Hematopoietic cell transduction
For all primary mouse hematopoietic cell experiments, mouse bone marrow (mBM) was isolated from femurs of wild-type C57/Bl6 mice using aseptic procedures. Mononuclear cells were isolated using lympholyte M (Cedarlane) according to the manufacturer's protocol. mBM mononuclear cells (MNCs) were prestimulated in α-minimum essential medium containing 20% fetal bovine serum (FBS) and the following growth factors: mouse stem cell factor, human thrombopoietin, and human Flt3 ligand (referred to hereafter as SFT; Biovision) at 100 ng/mL each, for 48 hours. Prestimulated cells in either Mieg3 or Mieg3-Rheb2 supernatants containing SFT were plated on retronectin-coated 6-well plates preloaded with retroviral supernatants as described previously.26 After a 4.5-hour incubation, the supernatants were replaced with fresh α-minimum essential medium plus SFT. Transductions were repeated the next day. Cells were either sorted with fluorescence-activated cell sorting (FACS) or used for transplantation on the following day (total of 4 days after isolation from the mice).
For 32D cell transduction, cells were resuspended in Mieg3 or Mieg3-Rheb2 supernatants containing mouse interleukin-3 (IL-3; 10 ng/mL) and plated on retronectin-coated wells of a 24-well plate preloaded with Mieg3 or Mieg3-Rheb2 supernatants. Transductions were repeated the following day. The cells were FACS-sorted for GFP expression and expanded to create stable, transduced cell lines.
BaF3 cells, for which data are not shown in this manuscript, were transduced in a similar fashion with the exception that 0.25 ng/mL IL-3 was used in all protocols.
Delayed proliferation of mouse hematopoietic cell lines and Western blot for mTOR activity
For delayed IL-3 proliferation assays, 32D cells were washed twice to remove residual IL-3 from the cultures and plated at 105 cells per well. The cells were starved of IL-3 in the presence of 10% FBS at 37°C for 8 hours, after which IL-3 was added to the individual wells at various concentrations. The cells were counted by trypan blue exclusion after 40-hour culture. To determine mTOR activity, 32D cells were starved of IL-3 for 8 hours, after which 10 ng/mL IL-3 plus or minus 50 nM rapamycin (Cell Signaling Technology) were added (phosphate-buffered saline and methanol used as IL-3 and rapamycin controls, respectively) for 20 or 40 minutes, and cells were harvested using M-PER lysis buffer (Pierce) containing phosphatase and protease inhibitors (Calbiochem). Proteins were separated by electrophoresis and blotted with antiphosphoribosomal S6 antibody (Ser235/236; Cell Signaling Technology), anti-β actin (Sigma-Aldrich), or antitotal ribosomal S6 protein (Cell Signaling Technology).
BaF3 cells, for which data are not shown in this manuscript, were analyzed in a similar fashion as 32D cells, except that 0.25-, 0.025-, and 0.0025-ng/mL concentrations of IL-3 were used in the proliferation assays and 0.25 ng/mL IL-3 was used in assays determining mTOR activity.
Real-time PCR detection of Rheb2 cDNA
Bone marrow cells from 10 C57/Bl6 mice were collected. Lineage-negative cells from pooled bone marrow (BM) cells were enriched using lineage Depletion kit (Miltenyi Biotec) as described by the manufacturer. Cells were stained with lineage cocktail antibody conjugated to allophycocyanin (APC), anti-CD34-fluorescein isothiocyanate, Sca-1 phycoerythrin (PE), and ckit-PE-Cy7 antibodies (BD Biosciences). CD34− c-kit+sca-1+ lineage− (KSL) and CD34+KSL cells were sorted using FACS Aria (BD Biosciences). mRNA was isolated and cDNA prepared from lineage-positive, CD34+KSL, and CD34−KSL cells using μMACS cDNA module (Miltenyi Biotec), and DNase treatment was performed during RNA isolation. Expression of Rheb2 in different populations of BM cells was studied using real-time polymerase chain reaction (PCR; Stratagene, Mx3000P) based on SYBR Green monitoring of PCR product accumulation for quantification and normalization of gene expression level. Hypoxanthine phosphoribosyl transferase (HPRT) expression was used as an internal control. Gene primers were designed using Oligo Perfect Designer (Invitrogen).
Mouse myeloid progenitor cell colony assays
mBM MNCs were transduced and GFP+ cells were FACS-sorted. The cells were resuspended in Iscove modified Dulbecco medium (IMDM) after sorting and plated at various cell concentrations in semisolid methylcellulose colony assays. These assays have been previously described.27,28 Briefly, GFP+ cells were plated in 1% methylcellulose containing 30% FBS (Hyclone), 0.1 mM hemin, 5% pokeweed mitogen spleen conditioned medium, 1 U/mL erythropoietin (Amgen), and 50 ng/mL muSCF (Biovision). Colonies were scored after 6- to 8-day incubation in a low-oxygen tension (5% O2) humidified incubator. For expansion cultures, GFP+ cells were plated in complete medium containing SFT (50 ng/mL each); cells were removed from liquid cultures at 0 hours, 48 hours, and 1 week and plated in methylcellulose colony assays. For delayed growth factor colony assays, GFP+ cells were plated in methylcellulose containing FBS, but no other growth factors; the combination of hemin, pokeweed mitogen spleen–conditioned medium, EPO, and muSCF was added to the plates 0, 24, and 48 hours after plating. All mouse studies were approved by the Institutional Animal Care and Use Committee of Walther Oncology Center.
Western blot for mTOR activity in response to SDF-1 stimulation
C57/Bl6 mBM MNCs were harvested and cultured in complete IMDM containing SFT (100 ng/mL) for 48 hours, starved of SFT for 8 hours, and then stimulated with phosphate-buffered saline, SFT (100 ng/mL), or various concentrations of SDF-1 (Biovision) plus or minus 50 nM rapamycin or methanol for 40 minutes. Cellular protein lysates harvested and blotted for phosphorylated ribosomal S6 protein (Serine 235/236; Cell Signaling Technology; or β-actin as a loading control; Sigma-Aldrich).
SDF-1/CXCL12 chemotaxis of 32D cells and CXCR4 expression
Chemotaxis assays were performed as previously described.29 32D cells were washed twice to remove IL-3 and FBS and resuspended in RPMI without FBS or other growth factors but containing 0.5% bovine serum albumin (Sigma-Aldrich). A total of 3 × 105 to 5 × 105 cells were loaded onto the top inserts in a 12-well transwell plate (Sigma-Aldrich) in triplicate for each cell line. RPMI plus 0.5% bovine serum albumin plus or minus SDF-1/CXCL12 (100 ng/mL) was added to the lower chamber of the transwell. Cells were allowed to migrate for 3 hours at 37°C, after which the percentage chemotaxis was calculated using FACS event counts with input cells. Background migration of cells to no SDF-1/CXCL12 was less than 1%.
For CXCR4 expression, 32D cells were stained with anti-CXCR4 antibody conjugated to PE (BD Biosciences PharMingen) and analyzed by FACS.
Expansion of KSL cells in vitro and cell-cycle analysis
mBM MNCs were transduced and cultured. Transduced cells were cultured in duplicate in complete IMDM plus SFT (50 ng/mL) plus rapamycin (50 nM; Cell Signaling Technologies) or vehicle control (methanol) for various times (shown in the figures) after transduction. Cells were stained with Hoechst 33342 DNA binding dye (Invitrogen) at a final concentration of 1 μg/mL, and cells then stained with anti–c-kit, anti–sca-1, and anti-lineage antibodies (BD Biosciences or Calbiochem) for FACS analysis.
In vivo homing and competitive repopulation transplantations
For in vivo short-term homing, 106 transduced, unsorted C57/Bl6 mBM MNCs were injected by tail vein into lethally irradiated recipient BoyJ (CD45.1) mice. BM from bilateral femurs was harvested after 24 hours and analyzed by FACS for GFP expression and anti-CD45.1 and anti-CD45.2 staining (BD Biosciences). For homing of Lin−Sca-1+ (LS) cells, mBM lineage− cells were isolated from C57/Bl6 mice using magnetic separation (Miltenyi Biotec) and transduced as previously described. A total of 5 × 106 cells were injected into lethally irradiated BoyJ recipient mice. BM cells from bilateral femurs, tibia, humeri, and pelvic bones were harvested and analyzed by FACS for GFP expression and anti–sca-1 and anti-lineage (BD Biosciences) staining.
Competitive repopulation assays were performed as previously described.30 Briefly, for short-term repopulation, 4 × 105 transduced, unsorted mBM MNCs from C57/Bl6 mice (CD45.2) were mixed with 4 × 105 BoyJ (CD45.1) BM cells and injected by tail vein into C57/Bl6:BoyJ F1 double-positive recipients (CD45.1:CD45.2), which had been total body irradiated with 9.5 Gy. Peripheral blood (PB) and BM were harvested at 1, 2, and 4 weeks after transplantation and analyzed by flow cytometry for CD45.1-PE and CD45.2-APC (BD Biosciences) antigens. BM was further analyzed by flow cytometry for CD117-PE (c-kit), LyG6-Pe-Cy5.5 (Sca1), and Lineage-APC (Lin) antigens (BD Biosciences and Caltag).
For long-term competitive repopulation, either 4 × 105 (1:1 group) or 1.6 × 105 (0.4:1 group) transduced mBM MNCs from C57/Bl6 mice were mixed with 4 × 105 BM cells from BoyJ mice and injected into irradiated recipient BoyJ mice. PB was harvested at 1, 2.5, 3.5, and 5 months after transplantation to monitor chimerism. After at least 5 months, BM was harvested and stained with CD45.1-PE and CD45.2-APC antibodies for FACS analysis. For secondary transplantation, BM cells from each primary recipient (6 Mieg3 and 5 Rheb2) at 5 months after transplantation were transplanted into 4 BoyJ mouse recipients (2 × 106 cells per recipient) conditioned with 9.5 Gy total body irradiation. PB samples were harvested beginning at 1 month after transplantation and stained with CD45.1 and CD45.2 antibodies for chimerism.
Real-time PCR detection of p21, PTEN, and Gfi-1 expression in 32D cells
RNA was isolated from 32D-Rheb2 and 32D-Mieg3 transduced cells using QIAGEN RNeasy kit according to the manufacturer. cDNA was prepared from 3.5 to 5 μg of RNA using Superscript III reverse transcription kit (Invitrogen) according to the manufacturer. Expression of p21, PTEN, and Gfi-1 genes was measured using real-time PCR (Stratagene, Mx3000P) based on SYBR Green monitoring of PCR product accumulation and normalization of gene expression level. HPRT was used as internal control. All gene expression levels were normalized as fold differences over expression in 32D-Mieg3 cells. Gene primers were designed using Oligo Perfect Designer (Invitrogen).
Western blot for Rheb2 protein
Protein lysates were harvested from National Institutes of Health 3T3, 32D, and BaF3 cells, gel electrophoresis was performed, and blots were developed by chemiluminescence. Rheb2 protein was detected using either an anti-Flag tag antibody (Sigma-Aldrich) or anti-Rheb2 antibody (Abnova).
FACS analysis
FACS data were analyzed using FCS Express software (De Novo Software).
Statistical analysis
The data are presented as mean plus or minus SEM, unless otherwise noted. All differences were analyzed using the Student t test.
Results
Rheb2 overexpression enhances delayed growth factor proliferation and mTOR signaling in 32D cells
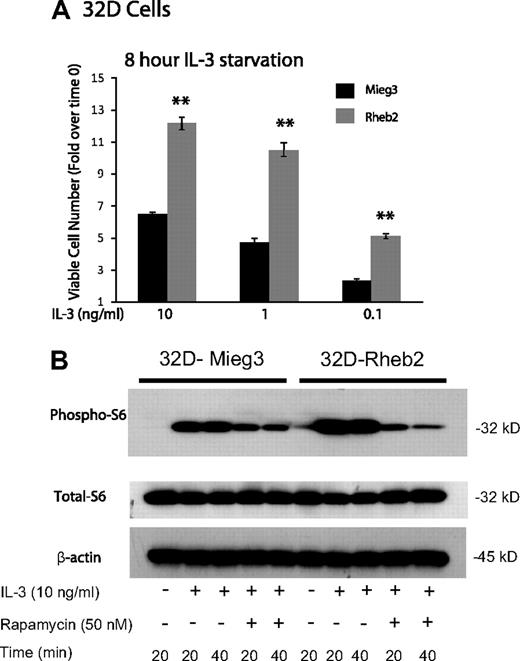
IL-3–dependent mouse myeloid progenitor 32D cells were transduced. Overexpression of Rheb2 was confirmed by Western blotting (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Because IL-3 signaling can activate the mTOR pathway in mouse hematopoietic cell lines,14 we examined the effect of overexpressing Rheb2 on delayed IL-3–induced proliferation and mTOR signaling in 32D cells; 32D cells overexpressing Rheb2 proliferated significantly better than Mieg3 control cells at various IL-3 concentrations after delayed addition (Figure 1A). Signaling through the mTOR pathway was also examined in IL-3–starved 32D cells by measuring phosphorylation of ribosomal S6 protein, a known downstream effector of mTORC1, which can be inhibited by rapamycin.14 In response to IL-3 stimulation, 32D cells overexpressing Rheb2 had increased signaling through the mTOR pathway as indicated by increased phosphorylated ribosomal S6 protein (Figure 1B). Increased phosphorylated S6 protein was observed in Rheb2-transduced cells for at least 2 hours after delayed IL-3 addition (supplemental Figure 2). Furthermore, inhibition of mTORC1 by rapamycin prevented the increased phosphorylation of S6 protein in 32D-Rheb2 cells, demonstrating that mTORC1 is responsible for these findings (Figure 1B). Rheb2-transduced BaF3 cells also exhibited increased IL-3–induced phosphorylation of S6 protein (data not shown), illustrating that these results are not limited to a single hematopoietic cell line or to the myeloid lineage exclusively.
Rheb2 overexpression enhanced IL-3 proliferation and mTOR signaling in a mouse hematopoietic progenitor cell line. (A) Proliferation of transduced 32D cells in response to delayed IL-3 addition (combined data from 2 independent experiments each done in triplicate; mean ± SEM). **P < .01. (B) Western blot of 32D cell lysates after delayed addition of IL-3 ± rapamycin. Total S6 protein and β-actin are shown as loading controls. Western blot is representative of 2 independent experiments.
Rheb2 overexpression enhanced IL-3 proliferation and mTOR signaling in a mouse hematopoietic progenitor cell line. (A) Proliferation of transduced 32D cells in response to delayed IL-3 addition (combined data from 2 independent experiments each done in triplicate; mean ± SEM). **P < .01. (B) Western blot of 32D cell lysates after delayed addition of IL-3 ± rapamycin. Total S6 protein and β-actin are shown as loading controls. Western blot is representative of 2 independent experiments.
Rheb2 overexpression enhances colony-forming ability of primary mBM cells
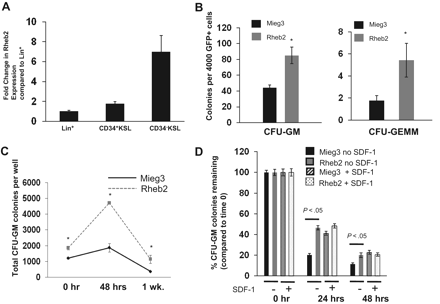
To confirm previous data from subtractive cDNA libraries on Rheb2 expression in hematopoietic cells, we measured Rheb2 expression in primary mBM cell populations using real-time reverse-transcribed PCR. We found that Rheb2 was expressed in lineage+, CD34+KSL, and CD34−KSL populations of BM cells (Figure 2A). Furthermore, Rheb2 was preferentially expressed in the more immature populations of cells (> 6-fold increased expression in CD34−KSL vs Lin+). This confirms previous data on Rheb2 expression in mouse HSCs.10
Rheb2 overexpression enhanced primary mouse hematopoietic progenitor colony formation and survival. (A) Real-time PCR expression of Rheb2 in FACS-sorted subsets of primary mBM cells, 2 or 3 replicates per cell type. (B) Numbers of CFU-GM and CFU-GEMM colonies produced by Mieg3- and Rheb2-transduced mBM cells (3 combined independent experiments, each done in triplicate; mean ± SEM). *P < .05. (C) Numbers of CFU-GM colonies produced in duplicate expanded liquid cultures (mean ± SD). *P < .05. (D) Survival of Mieg3- and Rheb2-transduced mBM cells in delayed growth factor colony assays, representative of 2 experiments each done in triplicate (mean ± SD).
Rheb2 overexpression enhanced primary mouse hematopoietic progenitor colony formation and survival. (A) Real-time PCR expression of Rheb2 in FACS-sorted subsets of primary mBM cells, 2 or 3 replicates per cell type. (B) Numbers of CFU-GM and CFU-GEMM colonies produced by Mieg3- and Rheb2-transduced mBM cells (3 combined independent experiments, each done in triplicate; mean ± SEM). *P < .05. (C) Numbers of CFU-GM colonies produced in duplicate expanded liquid cultures (mean ± SD). *P < .05. (D) Survival of Mieg3- and Rheb2-transduced mBM cells in delayed growth factor colony assays, representative of 2 experiments each done in triplicate (mean ± SD).
GFP+ transduced mBM MNCs were plated in methylcellulose colony assays as detailed in “Mouse myeloid progenitor cell colony assay.”27 Numbers of both CFU-GM (granulocyte-macrophage) and CFU-GEMM (granulocyte-erythroid-macrophage-megakaryocyte) HPCs were significantly increased, almost 2-fold for CFU-GM and greater than 2-fold for CFU-GEMM, by overexpression of Rheb2 (Figure 2B). In addition, Rheb2 overexpression increased absolute CFU-GM progenitor numbers greater than 2-fold at 48 hours compared with Mieg3 in expanded liquid cultures (Figure 2C).
Because the Rheb/mTOR pathway is known to be a positive regulator of survival, we tested the ability of Rheb2 overexpression to enhance survival of transduced HPCs subjected to delayed-growth factor addition. We transduced and plated mBM MNCs at time 0 in methylcellulose colony assays. Growth factors were added to these assays at 0, 24, and 48 hours after plating, and colonies were counted. In addition, we tested whether the presence of the known prosurvival enhancing chemokine SDF-1/CXCL1228 had any additional effect in these assays on Rheb2-transduced cell survival. We found that Rheb2 significantly (> 2-fold at 24 hours) increased the percentage CFU-GM colonies remaining when standardized to the number of colonies at time 0 (Figure 2D). Interestingly, whereas the presence of SDF-1/CXCL12 enhanced the progenitor survival of the Mieg3 cells as expected, the survival of Rheb2 cells was not further enhanced by SDF-1/CXCL12 (Figure 2D).
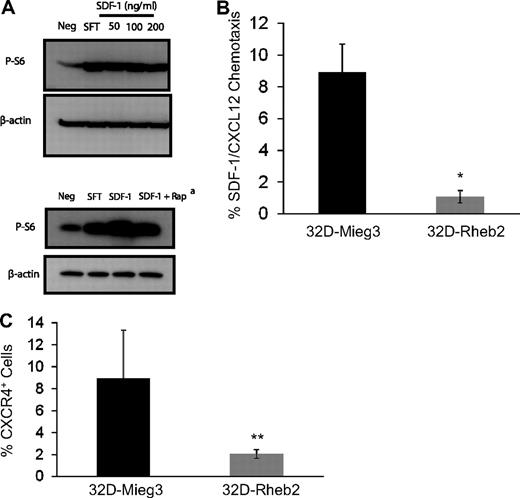
To investigate the relationship between SDF-1/CXCL12 and the mTOR pathway, we stimulated mBM MNCs in vitro with SDF-1/CXCL12 and measured mTOR activation using Western blot (Figure 3A). We found SDF-1/CXCL12 could activate the mTOR pathway in primary mBM MNCs, and this was at least partially reversed by inhibiting mTORC1 by rapamycin (Figure 3A). This suggests that SDF-1/CXCL12 and Rheb2, each being able to activate mTOR, may share some of the same signaling pathways. Using transduced 32D-Rheb2 cells, we determined whether overexpression of Rheb2 affected SDF-1/CXCL12 chemotaxis and expression of its receptor CXCR4. We found significantly impaired SDF-1/CXCL12-mediated chemotaxis in 32D cells overexpressing Rheb2 compared with Mieg3 empty-vector cells (Figure 3B). In addition, we found that the SDF-1/CXCL12 receptor, CXCR4, was down-regulated in 32D cells overexpressing Rheb2 (Figure 3C), suggesting that Rheb2-transduced cells may have blunted physiologic responses to SDF-1/CXCL12 in part because of down-regulation of its receptor.
Overexpression of Rheb2 impairs SDF-1/CXCL12 chemotaxis and CXCR4 expression in 32D cells. (A) Activation of mTOR signaling in response to SDF-1/CXCL12 stimulation in mBM cells, representative of 2 independent experiments. (Bottom panel) Effect of rapamycin at reversing phosphorylation of S6, 100 ng/mL SDF-1/CXCL12 used in this experiment. (B) Transwell chemotaxis of transduced 32D cells to 100 ng/mL SDF-1/CXCL12, combined triplicates from 2 independent experiments. (C) 32D cells, FACS stained for CXCR4 surface expression, combined 2 independent experiments. *P < .02. **P = .05.
Overexpression of Rheb2 impairs SDF-1/CXCL12 chemotaxis and CXCR4 expression in 32D cells. (A) Activation of mTOR signaling in response to SDF-1/CXCL12 stimulation in mBM cells, representative of 2 independent experiments. (Bottom panel) Effect of rapamycin at reversing phosphorylation of S6, 100 ng/mL SDF-1/CXCL12 used in this experiment. (B) Transwell chemotaxis of transduced 32D cells to 100 ng/mL SDF-1/CXCL12, combined triplicates from 2 independent experiments. (C) 32D cells, FACS stained for CXCR4 surface expression, combined 2 independent experiments. *P < .02. **P = .05.
Rheb2 overexpression increases the percentage of KSL mouse cells in liquid culture
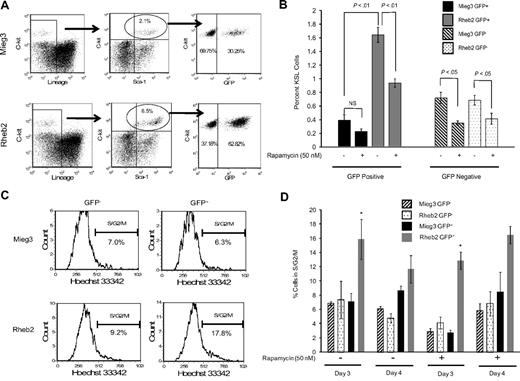
Transduced, unsorted mBM MNCs were expanded in liquid cultures containing either vehicle control or rapamycin (50 nM) and analyzed by FACS to determine the percentage of KSL cells. The FACS plots in Figure 4A show that the percentage of GFP+ cells within the KSL population was higher in cultures of Rheb2-transduced cells compared with Mieg3 cultures. The percentage total GFP+ cells was similar in Mieg3 and Rheb2 cultures, suggesting a specific effect of Rheb2 on phenotypically defined immature cells. In addition, the percentage of GFP+KSL cells in both Mieg3 and Rheb2 cultures analyzed shortly after transduction were similar (0.08% in Mieg3 vs 0.12% in Rheb2), suggesting that the Rheb2 retrovirus did not specifically target the KSL population during transduction. The percentage of Rheb2 GFP+KSL cells in these expanded cultures was greater than 4-fold higher than Mieg3 GFP+KSL cells, whereas the percentage of GFP−KSL cells was similar in both cultures (Figure 4B). The presence of rapamycin significantly reduced the percentage of GFP+KSL cells in the Rheb2 cultures; however, this percentage was still significantly greater than the percentage of Mieg3 GFP+KSL cells (Figure 4B).
Rheb2 overexpression expanded phenotypic HSC (KSL) cells in vitro and increased cell cycling. (A) Representative FACS plots of KSL cells in cultures of Rheb2- and Mieg3-transduced mBM cells. (B) Tabulated data from 2 experiments, each done in duplicate 4 days after transduction (mean ± SEM). (C-D) Representative histograms showing cell cycling of Mieg3- and Rheb2-transduced KSL cells and tabulated data from duplicate cultures; mean ± SD. *P < .05. Rheb2 GFP+ fraction compared with Mieg3 GFP+ fraction in panel D.
Rheb2 overexpression expanded phenotypic HSC (KSL) cells in vitro and increased cell cycling. (A) Representative FACS plots of KSL cells in cultures of Rheb2- and Mieg3-transduced mBM cells. (B) Tabulated data from 2 experiments, each done in duplicate 4 days after transduction (mean ± SEM). (C-D) Representative histograms showing cell cycling of Mieg3- and Rheb2-transduced KSL cells and tabulated data from duplicate cultures; mean ± SD. *P < .05. Rheb2 GFP+ fraction compared with Mieg3 GFP+ fraction in panel D.
The expansion of GFP+KSL cells in in vitro culture was accompanied by an increase in the percentage of cells in S/G2/M cell cycle phases, as measured by Hoechst 33342 DNA staining. Figure 4C shows representative FACS histograms of gated GFP−KSL cells analyzed for G1 and S/G2/M cell cycling status, illustrating that a greater percentage of Rheb2-transduced GFP+KSL cells were found in S/G2/M. Figure 4D shows tabulated data of cells expanded for 3 or 4 days after transduction in the absence or presence of rapamycin. At 3 days of culture, a significantly higher percentage of Rheb2 GFP+KSL cells were found in S/G2/M compared with Mieg3 GFP+KSL cells. This was reproduced at day 4; however, the difference was not significant. This significant increase in cycling was found at 3 days independent of whether rapamycin was in the culture, although the absolute percentages of GFP+KSL cells in S/G2/M decreased because of rapamycin. This again suggests that mTOR plays at least a partial role in Rheb2 effects.
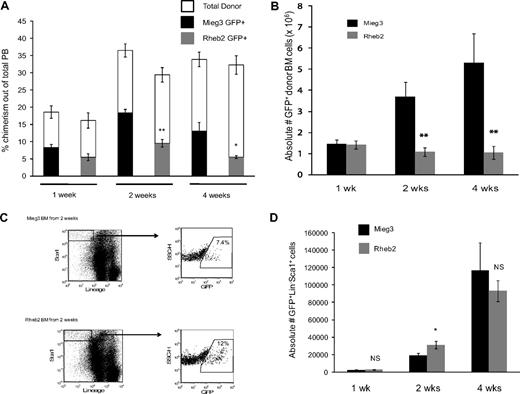
Rheb2 overexpression decreases overall competitive repopulation of transduced cells but expands immature LS cells in vivo in a short-term transplantation model
To examine the effect of overexpressing Rheb2 on in vivo behavior of mouse hematopoietic cells, competitive repopulation assays were performed. This is a widely used assay to measure HSC function.29,30 Figure 5 shows short-term competitive repopulation chimerism in which donor (CD45.2) BM cells were transduced, mixed with competitor (CD45.1) BM cells, and transplanted into recipients (CD45.1/CD45.2). PB and BM samples were harvested at 1, 2, and 4 weeks after transplantation and analyzed by FACS to determine the overall chimerism of donor GFP+ cells. BM cells were further analyzed by FACS for KSL markers. Figure 5A shows the chimerism of GFP+ donor cells in the PB of transplanted animals at 1, 2, and 4 weeks after transplantation. Rheb2 GFP+ cell chimerism was significantly decreased compared with Mieg3 GFP+ cells at 2 and 4 weeks after transplantation, with a greater than 2-fold decrease at 4 weeks (Figure 5A). Donor GFP+ cell BM chimerism showed the same significant trend; Rheb2-transduced cells were significantly impaired in engraftment at 2 and 4 weeks after transplantation, with a 3-fold and 5-fold decrease in donor GFP+ Rheb2 cells compared with Mieg3 cells at 2 and 4 weeks after transplantation, respectively (Figure 5B).
Rheb2 overexpression impaired total donor cell engraftment but transiently expanded phenotypic stem/progenitor cells in vivo in short-term competitive repopulation experiments. (A) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in recipient animals (at least 5 animals in each group at each time point; mean ± SEM). (B) Chimerism of absolute numbers of transduced GFP+ donor cells (CD45.2+) in the BM of recipients (at least 5 animals in each group at each time point; mean ± SEM). (C) Representative FACS plots showing the percentage of GFP+ cells within the immature LS population in recipient animals at 2 weeks after transplantation. (D) The absolute numbers of GFP+LS cells in the BM of recipients (at least 5 animals in each group at each time point; mean ± SEM). *P < .05. **P < .01. NS indicates not significant.
Rheb2 overexpression impaired total donor cell engraftment but transiently expanded phenotypic stem/progenitor cells in vivo in short-term competitive repopulation experiments. (A) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in recipient animals (at least 5 animals in each group at each time point; mean ± SEM). (B) Chimerism of absolute numbers of transduced GFP+ donor cells (CD45.2+) in the BM of recipients (at least 5 animals in each group at each time point; mean ± SEM). (C) Representative FACS plots showing the percentage of GFP+ cells within the immature LS population in recipient animals at 2 weeks after transplantation. (D) The absolute numbers of GFP+LS cells in the BM of recipients (at least 5 animals in each group at each time point; mean ± SEM). *P < .05. **P < .01. NS indicates not significant.
BM cells from these animals were also analyzed for KSL markers. At 1 week after transplantation, we found very dim c-kit staining; and because numbers of events were very low in these animals, especially at 1 and 2 weeks, we have analyzed the data by determining the numbers of LS instead of KSL cells. This population still represents an immature stem/progenitor BM population. At 1 week after transplantation, numbers of GFP+LS cells were similar in animals transplanted with Mieg3- and Rheb2-transduced cells, suggesting that early establishment of engraftment was not affected by Rheb2. At 2 weeks, in contrast to the total GFP+ donor cell chimerism, Rheb2 overexpression expanded absolute numbers of GFP+LS cells in vivo compared with Mieg3 cells (Figure 5C-D). The graph in Figure 5D illustrates that the average absolute number of GFP+LS cells per recipient at 2 weeks after transplantation was significantly higher in Rheb2 versus Mieg3 recipient animals (31 132 ± 4340 vs 19 457 ± 2250 GFP+LS cells in Rheb2 and Mieg3 recipients, respectively; mean ± SEM). This is a dramatic finding given the significantly decreased numbers of total Rheb2 GFP+ cells in the BM (Figure 5B). Importantly, in these experiments, Rheb2 overexpression specifically expanded the GFP+ fraction of the LS population, although not significantly affecting the percentage of GFP− cells within the LS population (data not shown). By 4 weeks after transplantation, the absolute number of Rheb2 GFP+LS cells in transplantation recipients had decreased but was not significantly less than numbers of Mieg3 GFP+LS cells (Figure 5D).
Short-term noncompetitive homing assays were performed to ensure that the in vivo results observed were not simply the result of differences in homing of Rheb2- and Mieg3-transduced cells. The detection of donor cells at very early time points after transplantation, such as 24 hours, is an accepted measure of the ability of the cells to move from the PB into the BM before proliferating. We found similar numbers of Mieg3- and Rheb2-transduced cells in the BM of recipients at 24 hours after transplantation (supplemental Figure 3), suggesting that Rheb2 overexpression did not enhance or impair the ability of the cells to implant in the BM shortly after transplantation. A further homing assay using transduced lineage-negative donor cells demonstrated that there were no significant differences in the absolute numbers of immature GFP+LS cells found in recipients transplanted with Mieg3- or Rheb2-transduced cells at 24 hours after transplantation (274 ± 41 vs 476 ± 108 GFP+LS cells in Mieg3 vs Rheb2 recipients, respectively; mean ± SD).
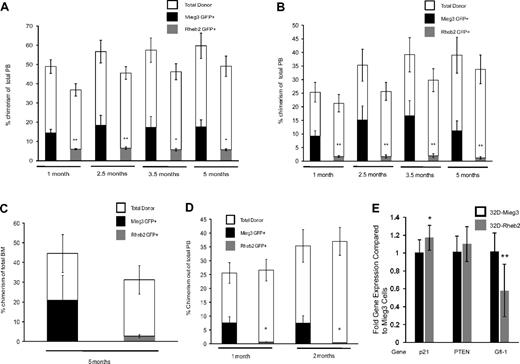
Rheb2 overexpression decreases long-term competitive repopulation in vivo
Additional competitive repopulation assays were performed and analyzed for long-term repopulation. Donor cells (CD45.2) were transduced and mixed with competitor cells (CD45.1) at either a ratio of 1:1 or 0.4:1 donor to competitor cells and transplanted into irradiated recipients (CD45.1). Figure 6A shows PB chimerism of Mieg3- and Rheb2-transduced cells in animals in the 1:1 ratio group up to 5 months after transplantation. At all time points, there was a significant decrease in the PB GFP+ chimerism in animals transplanted with Rheb2 compared with Mieg3-transduced cells (Figure 6A). These decreases were exaggerated in the PB of animals transplanted with a 0.4:1 ratio of donor to competitor cells (Figure 6B). Although decreased, the average level of donor GFP+ cell chimerism remained stable in Rheb2 transplantation recipients throughout the experiment, suggesting that some long-term repopulating cells were present and functional. Figure 6C shows the chimerism of Rheb2- and Mieg3-transduced cells in the BM of primary animals transplanted with a 1:1 ratio of donor to competitor cells (from one of the experiments shown in Figure 6A) harvested 5 months after transplantation, again showing decreased chimerism of Rheb2 compared with Mieg3-transduced cells. BM cells from these primary animals were transplanted into secondary recipient (CD45.1) mice to evaluate the self-renewal capacity of long-term repopulating cells. Figure 6D shows the PB chimerism of secondary recipients followed for 2 months after transplantation. In our experience, this time point gives a good measure of secondary repopulating ability, and chimerism usually does not change significantly when following these animals for longer time periods. Rheb2-transduced cells from primary animals were significantly impaired in engraftment in these secondary recipients (Figure 6D). Overall, these in vivo experiments indicate that Rheb2 overexpression decreases long-term marrow HSC repopulating ability and secondary repopulation capacity.
Overexpression of Rheb2 decreased long-term competitive repopulation in vivo. (A) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in the 1:1 transplantation group (2 independent experiments, at least 11 animals in each group; mean ± SEM). (B) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in the 0.4:1 transplantation group (2 independent experiments, at least 11 animals in each group; mean ± SEM). (C) Primary recipient BM chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in 1:1 group from one of the experiments in panel A (6 Mieg3 and 5 Rheb2 animals; mean ± SEM). (D) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in secondary transplantation recipients (4 secondary recipients per primary animal; mean ± SEM). *P < .05. **P < .02. NS indicates not significant. (E) Real-time PCR analysis of p21, PTEN, and Gfi-1 gene expression in transduced 32D cells. HPRT was used as internal control to standardize expression of all genes in both cell lines. Data are expressed as fold change over levels in 32D-Mieg3 cells, p21 and Gfi-1 are combined results from 3 experiments, each done at least in duplicate, whereas PTEN is combined results from 2 experiments, each done at least in duplicate. *P < .05. **P < .02.
Overexpression of Rheb2 decreased long-term competitive repopulation in vivo. (A) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in the 1:1 transplantation group (2 independent experiments, at least 11 animals in each group; mean ± SEM). (B) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in the 0.4:1 transplantation group (2 independent experiments, at least 11 animals in each group; mean ± SEM). (C) Primary recipient BM chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in 1:1 group from one of the experiments in panel A (6 Mieg3 and 5 Rheb2 animals; mean ± SEM). (D) PB chimerism of transduced GFP+ donor (CD45.2+) cells and total donor (CD45.2+) cells in secondary transplantation recipients (4 secondary recipients per primary animal; mean ± SEM). *P < .05. **P < .02. NS indicates not significant. (E) Real-time PCR analysis of p21, PTEN, and Gfi-1 gene expression in transduced 32D cells. HPRT was used as internal control to standardize expression of all genes in both cell lines. Data are expressed as fold change over levels in 32D-Mieg3 cells, p21 and Gfi-1 are combined results from 3 experiments, each done at least in duplicate, whereas PTEN is combined results from 2 experiments, each done at least in duplicate. *P < .05. **P < .02.
Rheb2 overexpression is associated with decreased Gfi-1 expression in 32D cells
To investigate other possible mechanisms accounting for the decreased HSC repopulation observed in Rheb2-overexpressing cells, we measured the expression of p21, PTEN, and Gfi-1 in transduced 32D cells using real-time PCR. These 3 genes are known to have roles in restricting HSC proliferation.21,23,25 We found that 32D cells overexpressing Rheb2 expressed similar levels of p21 and PTEN (Figure 6E). It should be noted that the difference in p21 expression was significant comparing Mieg3- and Rheb2-transduced 32D cells; however, the fold difference in expression between Rheb2 and Mieg3 was 1.17, a small absolute increase of p21 in Rheb2 cells. The expression of Gfi-1 was decreased significantly in 32D cells overexpressing Rheb2 compared with Mieg3 (fold difference of 0.58, Figure 6E). These data suggest that Gfi-1 expression is down-regulated in 32D cells overexpressing Rheb2 compared with Mieg3, whereas p21 and PTEN are expressed at similar levels.
Discussion
The mTOR pathway is known to play an important role in HSC functions. In the current study, the role of the upstream mTOR activator, Rheb2, is analyzed in mouse hematopoietic progenitor and stem cell populations. Overexpression of Rheb2 significantly enhanced the proliferation of mouse HPCs. In 32D cells, overexpression of Rheb2 significantly increased delayed IL-3 proliferation and signaling through the mTOR pathway. In primary mouse HPCs, overexpression of Rheb2 led to enhanced survival as determined by delayed growth factor addition colony assays. Interestingly, the addition of SDF-1/CXCL12 did not further enhance the survival of Rheb2-transduced cells compared with Mieg3 empty vector-transduced cells. In further experiments, we demonstrated activation of mTOR signaling downstream of SDF-1/CXCL12 in mouse HPCs. In 32D cells, we demonstrated that overexpression of Rheb2 significantly impaired SDF-1/CXCL12 chemotaxis and expression of its receptor CXCR4, suggesting that down-regulation of CXCR4 by Rheb2 in primary cells may explain their SDF-1/CXCL12 insensitivity in delayed growth factor colony assays. In addition, it is possible that the mTOR pathway may be activated by both Rheb2 and SDF-1/CXCL12, and the combination of both molecules does not lead to additive effects on survival. Further studies using primary cells will be needed to fully elucidate Rheb2 effects on this axis. These goals are outside the scope of our current study, but the results presented here provide a basis for further studies.
Overexpression of Rheb2 was sufficient to expand phenotypic HSCs in vitro by a mechanism involving increased cell cycling of the transduced cells (Figure 4). Rapamycin partially reversed this expansion, indicating that mTORC1 is involved in this mechanism. It is possible, especially given the fact that rapamycin only partially inhibited Rheb2 effects, that other non-mTORC1 downstream effectors may be involved in Rheb2 signaling, a process known to occur in response to Rheb1 activity.31 In addition, mTOR signaling is complex, and experiments using rapamycin fail to account for any downstream effects of mTORC2 that is rapamycin insensitive and that could theoretically be altered in Rheb2-overexpressing cells.
Our in vivo results suggest that Rheb2 overexpression initially expands immature hematopoietic cell populations but impairs their overall long-term and secondary repopulation (Figures 5–6). Given that these effects are observed in primary and also secondary recipients, it is probable that Rheb2 overexpression affects an HSC population. Skewed differentiation of these HSCs could explain the effects observed in vivo. However, we determined the expression of various lineage markers (B220, CD3, Ly6G, and CD11) on transduced Rheb2 and Mieg3 PB cells in recipient animals at 4 weeks after transplantation and on transduced BM cells at the conclusion of the competitive repopulation assays, and there were no significant differences, indicating that differentiation is not dramatically skewed by Rheb2 (supplemental Table 1, showing BM data).
We have summarized evidence suggesting that Rheb2 overexpression can expand immature hematopoietic cells. Expansion of HSCs is not always accompanied by preserved self-renewal and repopulating abilities, and there is evidence from other studies to suggest that highly proliferative HSCs have reduced in vivo repopulation.22-25 In addition, a recent study has provided evidence that hyperactivation of the mTOR pathway is detrimental for maintaining normal mouse HSC function, including engraftment.21 This latter study21 showed that hyperactivation of mTOR by PTEN conditional knockout increased the percentage of cycling HSCs and expanded HSCs early after PTEN knockout, which was followed by a subsequent exhaustion of the cells, a phenotype that could be reversed by rapamycin treatment.21 These effects were distinct from the leukemogenic effect of PTEN knockout.21 It is possible that, early after transplantation, Rheb2 overexpression and subsequent hyperactivation of mTOR may exhaust HSCs by a similar mechanism.
To investigate the Rheb2 mechanism further, real-time PCR experiments in transduced 32D cells suggest that p21 and PTEN probably do not have major roles, as their expression is similar in Mieg3 and Rheb2 cells (Figure 6E). However, expression of the transcription factor Gfi1 was significantly decreased in Rheb2-overexpressing 32D cells (Figure 6E). Gfi1 restricts HSC proliferation, and its loss leads to increased cycling, expansion, and exhaustion of HSCs.25 The decreased expression of this gene in Rheb2 cells is consistent with our in vivo phenotype. There are few data on the relationship between mTOR and Gfi1, and it would be interesting to determine in further studies whether downstream signals mediated through the Rheb2-mTOR pathway lead directly to decreased Gfi1 expression. The observation that PTEN was not significantly altered by Rheb2 overexpression could be expected because PTEN is upstream of Rheb2 in the mTOR signaling pathway. A weakness of these experiments is that they were done in cultured cell lines as opposed to primary cells. Obviously, cell lines have altered expression of a variety of proteins involved in cell growth and cycling. However, an advantage of studying gene alterations in 32D cells is that they are relatively homogeneous; thus, differences that may be present in only a small subset of heterogeneous BM hematopoietic cells are easily detected in cell lines. In addition, measuring the expression of multiple genes in transduced, FACS-sorted subpopulations of BM cells is technically challenging. We think our current data provide a rationale for further study into the specific downstream pathways involved in how Rheb2 affects HSC proliferation and exhaustion.
This study reinforces the concept that HPCs and HSCs behave differently, and factors that increase the proliferation of HPCs may have seemingly opposite effects on HSC repopulation and self-renewal.21,23 We propose a role for the novel ras-related GTPase Rheb2 in regulating HPC and HSC biology: an understanding of this protein and its related signaling pathways may be relevant in clinical uses of hematopoietic cells focused on manipulating expansion and self-renewal.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Edward Srour, D. Wade Clapp, and Lawrence Quilliam for advice and support and John Kinzfogl for technical support.
This work was supported by the US Public Health Service (grants RO1 HL56416 and RO1 HL67384) and the National Institutes of Health (Project in PO1 HL53586, H.E.B.; and Training Grant T32 DK7519, T.B.C., H.E.B.).
National Institutes of Health
Authorship
Contribution: T.B.C., W.T., and H.E.B. designed experiments; T.B.C., S.B., and G.H. performed experiments; T.B.C., S.B., and H.E.B. analyzed results; and T.B.C. and H.E.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hal E. Broxmeyer, Walther Oncology Center, Indiana University School of Medicine, 950 West Walnut St, R2-302, Indianapolis, IN 46202-5181; e-mail: hbroxmey@iupui.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal