Abstract
We studied patients with myeloid neoplasm associated with ringed sideroblasts and/or thrombocytosis. The combination of ringed sideroblasts 15% or greater and platelet count of 450 × 109/L or greater was found in 19 subjects fulfilling the diagnostic criteria for refractory anemia with ringed sideroblasts (RARS) associated with marked thrombocytosis (RARS-T), and in 3 patients with primary myelofibrosis. JAK2 and MPL mutations were detected in circulating granulocytes and bone marrow CD34+ cells, but not in T lymphocytes, from 11 of 19 patients with RARS-T. Three patients with RARS, who initially had low to normal platelet counts, progressed to RARS-T, and 2 of them acquired JAK2 (V617F) at this time. In female patients with RARS-T, granulocytes carrying JAK2 (V617F) represented only a fraction of clonal granulocytes as determined by X-chromosome inactivation patterns. RARS and RARS-T patient groups both consistently showed up-regulation of ALAS2 and down-regulation of ABCB7 in CD34+ cells, but several other genes were differentially expressed, including PSIP1 (LEDGF), CXCR4, and CDC2L5. These observations suggest that RARS-T is indeed a myeloid neoplasm with both myelodysplastic and myeloproliferative features at the molecular and clinical levels and that it may develop from RARS through the acquisition of somatic mutations of JAK2, MPL, or other as-yet-unknown genes.
Introduction
Within myeloid neoplasms, myelodysplastic syndromes (MDSs) are characterized by ineffective hematopoiesis and peripheral cytopenia,1 whereas myeloproliferative neoplasms (MPNs) are typically associated with overproduction of mature blood cells.2 However, the existence of conditions with overlapping features is well established. In fact, the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues comprises the category of myelodysplastic/MPNs.3 This category includes chronic myelomonocytic leukemia, atypical chronic myeloid leukemia (BCR-ABL1 negative), juvenile myelomonocytic leukemia, and myelodysplastic/MPNs, unclassifiable (MDS/MPN, U). These latter have been defined as clonal conditions that at the time of initial presentation have some features supporting a diagnosis of MDS and other features more consistent with MPN. One of these conditions is the provisional entity defined as refractory anemia with ringed sideroblasts (RARS) associated with marked thrombocytosis (RARS-T).4
Ringed sideroblasts are erythroblasts with iron-loaded mitochondria visualized by Prussian blue staining as a perinuclear ring of blue granules.5 The iron deposited in these perinuclear mitochondria is stored in mitochondrial ferritin, which is encoded by the FTMT gene.6 Ringed sideroblasts are the distinctive feature of sideroblastic anemias, a heterogeneous group of inherited and acquired disorders.7,8 The presence of ringed sideroblasts in the bone marrow (BM; 15% or more) is a marker of the MDS defined as RARS.9 Ringed sideroblasts, however, can be occasionally found also in myeloproliferative disorders such as essential thrombocythemia or primary myelofibrosis.10,11
Mutations in JAK2, MPL, or both have been detected in a substantial portion of patients with RARS-T,12-22 suggesting a potential relation between these mutant genes and the MDS characterized by ringed sideroblasts.23,24 At the same time, the very presence of these mutations has raised the question of whether RARS-T is a form of essential thrombocythemia rather than a separate disorder.25
To gain a deeper insight into the pathophysiology of RARS-T, we studied a cohort of patients with myeloid neoplasms and investigated the relation between thrombocytosis, ringed sideroblasts, JAK2 or MPL mutations, and CD34+ cell gene expression profiles.
Methods
Patients and clinical procedures
These investigations were approved by the local Ethics Committee (Fondazione IRCCS Policlinico San Matteo). The procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000, and samples were obtained after subjects provided written informed consent in accordance with the Declaration of Helsinki.
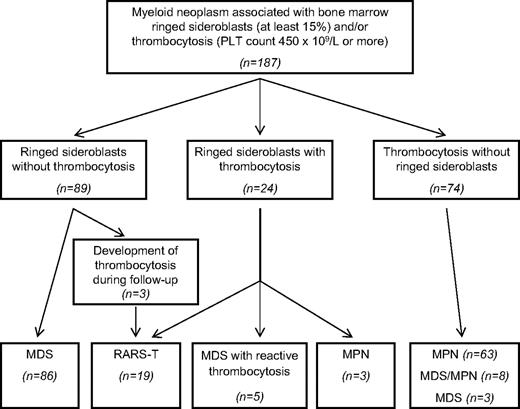
We studied patients diagnosed with BCR/ABL1-negative myeloid neoplasm at the Department of Hematology Oncology, University of Pavia Medical School, Fondazione IRCCS Policlinico San Matteo, between 2001 and 2006. All these patients had BM aspiration, biopsy, or both as a routine diagnostic procedure. Subjects having 15% or greater ringed sideroblasts, thrombocytosis (platelet count ≥ 450 × 109/L), or both were included in this study. For MPN, only patients with essential thrombocythemia or primary myelofibrosis were considered, whereas those with polycythemia vera were excluded from the study. Overall, 187 patients were enrolled (Figure 1), and all diagnoses were revised according to the 2008 WHO classification of tumors of hematopoietic and lymphoid tissues.3
Patient selection criteria and final diagnosis after a clinicopathologic review process. We selected patients with the following characteristics: (1) a diagnosis of BCR/ABL-negative myeloid neoplasm with the exclusion of polycythemia vera; (2) ringed sideroblasts of 15% or greater, platelet (PLT) count of 450 × 109/L or greater, or both. Overall, 187 patients were identified fulfilling these selection criteria: (1) 89 patients had ringed sideroblasts of 15% or greater and a PLT count less than 450 × 109/L at the time of diagnosis; (2) 24 patients had ringed sideroblasts of 15% or greater and a PLT count of 450 × 109/L or greater (range, 453-1420 × 109/L); (3) 74 patients had a PLT count of 450 × 109/L or greater (range, 458-1308 × 109/L) with less than 15% ringed sideroblasts in the BM. After the clinicopathologic review process of the original cohort of 187 patients, 94 were found to be affected with MDS, 66 with MPN, and 27 with MDS/MPN (19 cases of RARS-T; 5 cases of MDS/MPN, U without ringed sideroblasts; and 3 cases of chronic myelomonocytic leukemia). In 5 patients with MDS, thrombocytosis was associated with an inflammatory condition and regressed later on, so that a diagnosis of reactive thrombocytosis was made.
Patient selection criteria and final diagnosis after a clinicopathologic review process. We selected patients with the following characteristics: (1) a diagnosis of BCR/ABL-negative myeloid neoplasm with the exclusion of polycythemia vera; (2) ringed sideroblasts of 15% or greater, platelet (PLT) count of 450 × 109/L or greater, or both. Overall, 187 patients were identified fulfilling these selection criteria: (1) 89 patients had ringed sideroblasts of 15% or greater and a PLT count less than 450 × 109/L at the time of diagnosis; (2) 24 patients had ringed sideroblasts of 15% or greater and a PLT count of 450 × 109/L or greater (range, 453-1420 × 109/L); (3) 74 patients had a PLT count of 450 × 109/L or greater (range, 458-1308 × 109/L) with less than 15% ringed sideroblasts in the BM. After the clinicopathologic review process of the original cohort of 187 patients, 94 were found to be affected with MDS, 66 with MPN, and 27 with MDS/MPN (19 cases of RARS-T; 5 cases of MDS/MPN, U without ringed sideroblasts; and 3 cases of chronic myelomonocytic leukemia). In 5 patients with MDS, thrombocytosis was associated with an inflammatory condition and regressed later on, so that a diagnosis of reactive thrombocytosis was made.
RARS-T was defined according to the following WHO criteria4 : (1) refractory anemia associated with erythroid dysplasia and ringed sideroblasts of 15% or greater, (2) less than 5% blasts in the BM, (3) platelet count of 450 × 109/L or greater, (4) presence of large atypical megakaryocytes similar to those observed in BCR/ABL1-negative MPN, and (5) absence of del(5q), t(3;3)(q21;q26) or inv(3)(q21q26). Representative BM smears from a patient with RARS-T are shown in Figure 2.
BM smears from a patient with RARS-T. (Left) BM ringed sideroblasts after Perls staining (100×/1.1 NA oil objective). (Right) BM smears showing atypical megakaryocytes (May-Grünwald/Giemsa staining, 10×/0.25 NA). Bone marrow smears were analyzed through an Axioscope 2 Plus microscope (Carl Zeiss). Images were acquired through an Axiocam MRc 5 camera (Carl Zeiss) and processed through Axiovision release 4.6.3 imaging solution software (Carl Zeiss). Reproduced from Hellström-Lindberg and Cazzola.24 (Appendix: color figures at http://asheducationbook.hematologylibrary.org/cgi/content/full/2008/1/491.)
BM smears from a patient with RARS-T. (Left) BM ringed sideroblasts after Perls staining (100×/1.1 NA oil objective). (Right) BM smears showing atypical megakaryocytes (May-Grünwald/Giemsa staining, 10×/0.25 NA). Bone marrow smears were analyzed through an Axioscope 2 Plus microscope (Carl Zeiss). Images were acquired through an Axiocam MRc 5 camera (Carl Zeiss) and processed through Axiovision release 4.6.3 imaging solution software (Carl Zeiss). Reproduced from Hellström-Lindberg and Cazzola.24 (Appendix: color figures at http://asheducationbook.hematologylibrary.org/cgi/content/full/2008/1/491.)
Cytogenetic analysis was performed at diagnosis using standard G-banding with trypsin-Giemsa staining, and karyotypes were classified using the International System for Cytogenetic Nomenclature Criteria.
Sample collection and cell separation
Mononuclear cells (MNCs) were separated from peripheral blood (PB) or BM samples by standard density gradient centrifugation (Lympholyte-H; CEDARLANE Laboratories Ltd). T lymphocytes were further isolated from PB MNCs by immunomagnetic adsorption on MiniMACS separation columns using an anti-CD3 antibody (Miltenyi Biotec GmbH). Granulocytes were purified from the PB red cell pellet after lyses of erythrocytes with hypotonic solution. Granulocytes and T lymphocyte purity was regularly greater than 95% on cytologic assessment. CD34+ cells were separated from BM MNCs using immunomagnetic beads coated with the monoclonal antibody CD34 (Miltenyi Biotec GmbH). The purity of CD34+ separated cells was assessed by flow cytometry using anti-CD34 monoclonal antibodies (Becton Dickinson) and was greater than 95% in all cases. Genomic DNA was extracted from granulocytes, T lymphocytes, and CD34+ cells by following standard protocols for human tissue (PUREGENE Blood Core Kit B; Gentra Systems Inc). Total RNA was prepared from all cell fractions using TRIzol Reagent according to the manufacturer's protocol (Invitrogen).
JAK2 and MPL mutation analyses
A quantitative real-time polymerase chain reaction (PCR)–based allelic discrimination assay was used for the detection of the JAK2 (V617F) mutation in circulating granulocytes, as previously described in detail.26 The sensitivity of this assay is approximately 0.2% of mutant alleles.
MPL mutation scanning was performed using high-resolution melting (HRM) analysis. A 209 base pair (bp) amplicon was generated using primers located in MPL intron 9 (5′-GCCGAAGTCTGACCCTTTTT-3′) and intron 10 (5′-ACAGAGCGAACCAAGAATGCCTGTTTACA-3′). Each 20-μL PCR contained 100 ng DNA, 0.5 U HotStart Taq polymerase together with 1× buffer containing 1.5mM MgCl2 (QIAGEN), 800μM dNTPs, 0.3 μM of each primer, and 1× EvaGreen (Idaho Technologies) as the intercalating dye. The cycling and HRM analysis were conducted on a RotorGene 6000TM real-time analyzer (Corbett Life Sciences), applying the following thermal protocol: an initial hold at 95°C for 15 minutes, 50 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 10 minutes, then a melt from 85°C to 95°C rising at 0.1°C per second. Normalization bars were between 88°C and 88.5°C for the leading range and 92.5°C and 93° for the tailing range. MPL mutations were confirmed by direct genomic sequencing.
Direct genomic sequencing was used for the identification of JAK2 exon 12 mutations, as previously described in detail.27
Analysis of X-chromosome inactivation patterns
Clonal analysis of hematopoietic cells was performed through the study of methylation pattern and allelic expression of HUMARA, PGK, and IDS genes on PB granulocytes and CD34+ cells, whereas T lymphocytes were used as a control tissue.
The HUMARA assay was performed as previously described in detail with minor modifications.28 For the PGK assay, DNA was digested with HpaII or RsaI and subjected to 35 cycles at 58°C using 1.5mM MgCl2, 0.5μM of each primer, and 0.4 U of DNA polymerase (Finnzymes). Primer sequences were 5′-AGCTGGACGTTAAAGGGAAGCGGGTCGTTA-3′ for the sense primer and 5′-TACTCCTGAAGTTAAATCAACATCCTCTTG-3′ for the antisense primer.29 PCR products were digested with 10 U of BstXI (New England BioLabs) at 55°C and analyzed on agarose gel. In a heterozygote, BstXI digestion after PCR yields bands of 530 bp, representing the allele lacking the polymorphic site, as well as of 433 and 97 bp, representing the allele containing the site. To carry out the IDS assay, granulocyte and T lymphocyte genomic DNA and mRNA sequences containing nucleotide 438 (C/T polymorphism) were amplified,30 and the resulting fragments were digested with HpaII and analyzed on agarose gel. In heterozygous patients 2 bands were observed: 151 and 126 bp or 158 and 133 bp using DNA or RNA, respectively.
Band intensities were quantitated by densitometric scanning. In the HUMARA and PGK assays, for each sample a cleavage ratio (CR) between alleles was calculated by dividing the ratio between band intensities of HpaII predigested samples (upper band/lower band) by the same ratio in undigested samples. If the ratio was less than 1, the reciprocal value was considered. In the IDS assay, the CR was obtained by dividing the ratio between band intensities of allele transcripts by the same ratio of DNA samples. A CR of 3.0 or greater was adopted as cutoff to distinguish between balanced and unbalanced X-chromosome inactivation, and XCIPs were defined as follows: (1) polyclonal XCIP in subjects with CR less than 3 in both granulocytes and T lymphocytes; (2) clonal XCIP in subjects with CR of 3 or greater in granulocytes and CR less than 3 in T lymphocytes; (3) ambiguous or skewed XCIP in subjects with CR of 3 or greater in both granulocytes and T lymphocytes.
Gene expression profiling
The quality of the RNA samples was evaluated using Agilent Bioanalyzer 2100 (Agilent Technologies). For each sample, 50 ng of total RNA was amplified and labeled with the 2-Cycle cDNA Synthesis and the 2-Cycle Target Labeling and Control Reagent packages (Affymetrix).31 Biotin-labeled fragmented cRNA (10 μg) was hybridized to GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix), covering more than 47 000 transcripts, representing 39 000 human genes. Hybridization was performed at 45°C for 16 hours in Hybridization Oven 640 (Affymetrix). Chips were washed and stained in a Fluidics Station 450 (Affymetrix) and scanned with a GeneChip Scanner 3000 (Affymetrix).
Cell intensity calculation and scaling were performed using GeneChip Operating System. Data analysis was performed using GeneSpring 7.3.1 (Agilent Technologies). The GeneChip Operating System software was used to perform quality control after scaling the signal intensities of all arrays to a target of 100. The values obtained for scale factors, background levels, percentage of present calls, 3′/5′ GAPDH ratio, and intensities of spike hybridization controls were within the acceptable range for all samples. Affymetrix CEL files were preprocessed by sing Robust MultiChip Analysis.32 Hierarchical clustering was performed with GeneSpring software using Pearson correlation. DAVID was used to identify enriched biologic themes, in particular gene ontology terms.33
Hematopoietic progenitor colony assay
The growth of colonies (CFU-E, BFU-E, CFU-GM) from peripheral MNCs was evaluated using Methocult GF (StemCell Technologies).
Flow cytometric enumeration of circulating CD34+ cells
Circulating CD34+ cells were enumerated by flow cytometry with the use of a single-platform assay as previously described,34 following the cell-gating guidelines recommended by the International Society for Hematotherapy and Graft Engineering and the subsequent modifications of the European Working Group of Clinical Cell Analysis.
Statistical analysis
Statistical analyses were performed using both Statistica 7.0 (StatSoft) and Stata 9 (Stata Corporation). Numerical variables were routinely summarized by their median and ranges. Both parametric (ANOVA, multivariate analysis) and nonparametric analysis (Kruskal-Wallis test, Mann-Whitney U test, Spearman rank correlation) were used. Actuarial probability of overall survival, event-free survival, and leukemia-free survival were estimated using the Kaplan-Meier product limit method. All analyses were carried out with Statistica 7.0 (StatSoft) and Stata 9 (Stata Corporation) software.
Results
Patient selection and identification of cases of RARS-T
The selection procedure that was applied to the cohort of 187 patients with myeloid neoplasm is summarized in Figure 1.
Sixteen of 24 patients with BM ringed sideroblasts (≥ 15%) and thrombocytosis (platelet count ≥ 450 × 109/L) fulfilled the criteria for the diagnosis of RARS-T. Three patients with RARS had low-to-normal platelet counts at diagnosis, but they developed increasing counts with appearance of large atypical megakaryocytes during follow-up and were then found to fulfill the criteria for diagnosis of RARS-T. Therefore, 19 cases of RARS-T were identified in the study cohort.
Clinical characteristics of the 19 patients with RARS-T are summarized in Table 1 and compared with those of 24 patients with RARS belonging to the same patient cohort. Apart from platelet count and abnormal megakaryocytes, patients with RARS-T had higher hemoglobin levels and white blood cell counts and lower serum ferritin.
Clinical characteristics of patients with RARS-T compared with those of patients with RARS
| Variable . | Diagnosis . | P . | |
|---|---|---|---|
| RARS . | RARS-T . | ||
| No. of patients | 24 | 19 | |
| Median age, y (range) | 67 (33-84) | 59 (27-90) | NS |
| Median Hb level, g/dL (range) | 9.0 (6.4-12.1) | 10.1 (6.0-12.6) | .04 |
| Median WBC count, ×109/L (range) | 5.0 (2.9-7.8) | 6.8 (2.9-13.0) | < .001 |
| Median PLT count, ×109/L (range) | 271 (113-471) | 688 (475-1420) | < .001 |
| Median serum ferritin, ng/mL (range) | 669 (173-1800) | 443 (65-2010) | .04 |
| Median serum erythropoietin, mU/mL (range) | 89 (24-1000) | 37 (12-1000) | NS |
| Median percentage of ring sideroblasts (range) | 63 (23-94) | 51 (15-91) | NS |
| Abnormal megakaryocytes, proportion (%) | 1/24 (4) | 15/19 (79) | < .001 |
| Bone marrow fibrosis, proportion (%) | 0/24 (0) | 1/19 (5) | NS |
| Cytogenetic abnormalities, proportion (%) | 10/23 (43) | 2/19 (11) | .07 |
| JAK2/MPL mutations, proportion (%) | 0/24 (0) | 11/19 (58) | < .001 |
| Variable . | Diagnosis . | P . | |
|---|---|---|---|
| RARS . | RARS-T . | ||
| No. of patients | 24 | 19 | |
| Median age, y (range) | 67 (33-84) | 59 (27-90) | NS |
| Median Hb level, g/dL (range) | 9.0 (6.4-12.1) | 10.1 (6.0-12.6) | .04 |
| Median WBC count, ×109/L (range) | 5.0 (2.9-7.8) | 6.8 (2.9-13.0) | < .001 |
| Median PLT count, ×109/L (range) | 271 (113-471) | 688 (475-1420) | < .001 |
| Median serum ferritin, ng/mL (range) | 669 (173-1800) | 443 (65-2010) | .04 |
| Median serum erythropoietin, mU/mL (range) | 89 (24-1000) | 37 (12-1000) | NS |
| Median percentage of ring sideroblasts (range) | 63 (23-94) | 51 (15-91) | NS |
| Abnormal megakaryocytes, proportion (%) | 1/24 (4) | 15/19 (79) | < .001 |
| Bone marrow fibrosis, proportion (%) | 0/24 (0) | 1/19 (5) | NS |
| Cytogenetic abnormalities, proportion (%) | 10/23 (43) | 2/19 (11) | .07 |
| JAK2/MPL mutations, proportion (%) | 0/24 (0) | 11/19 (58) | < .001 |
RARS-T indicates refractory anemia with ringed sideroblasts associated with marked thrombocytosis; RARS, refractory anemia with ringed sideroblasts; NS, not significant; Hb, hemoglobin; WBC, white blood cell; and PLT, platelet.
We compared the clinical course of patients with RARS, RARS-T, and refractory cytopenia with multilineage dysplasia and ringed sideroblasts included in this study. No significant difference in overall survival and leukemia-free survival was observed between these groups. A comparison was then made considering progression to a more-advanced MDS, or evolution into acute leukemia, or death as events. There was no significant difference between RARS and RARS-T groups, whereas the event-free survival was significantly shorter for patients with refractory cytopenia with multilineage dysplasia and ringed sideroblasts compared with the other groups (P < .05).
Three patients with MPN (all affected with primary myelofibrosis) had ringed sideroblasts (≥ 15%) and thrombocytosis (platelet count ≥ 450 × 109/L; Figure 1). No peculiar clinical feature was identified in these patients compared with the remaining patients with primary myelofibrosis.
JAK2 and MPL mutation analysis in circulating granulocytes and BM CD34+ cells
One hundred fifty-five patients of the original cohort were tested for JAK2 (V617F). Forty-eight (73%) of 66 patients with a final diagnosis of MPN (essential thrombocythemia or primary myelofibrosis) carried the mutation in circulating granulocytes. In particular, 2 of 3 patients with MPN with ringed sideroblasts carried the mutation (Figure 1).
JAK2 (V617F) was detected in 10 (53%) of 19 patients with RARS-T and in 2 of 5 patients with MDS/MPN, U. The median percentage of mutant alleles in patients with RARS-T was equal to 10.4% (range, 1.1%-27.7%). Patients with RARS-T were tested also for JAK2 exon 12 and MPL mutations through direct sequencing and HRM, respectively. Two patients carrying JAK2 (V617F; 5.8% and 10.2% of mutant alleles, respectively) also carried the MPL (W515L) mutation (approximately 20% and 25% of mutant alleles, respectively). A patient who was negative for JAK2 (V617F) was found to carry JAK2 delE543D544, that is, a JAK2 exon 12 mutation,27 with approximately 10% of mutant alleles.
We analyzed the mutation status of isolated BFU-Es in 1 of the 2 patients carrying both the JAK2 (V6517F) and MPL (W515L) mutations. Three of 5 BFU-Es grown from PB MNCs were found to carry MPL (W515L) exclusively, whereas the remaining 2 had JAK2 (V617F) exclusively. All colonies were heterozygous for these mutations.
Two patients with an initial diagnosis of RARS (platelet counts of ≥ 55 and 397 × 109/L, respectively), who were negative for JAK2 and MPL mutations at clinical onset, showed a progressive increase in platelet count starting from 18 and 32 months after diagnosis, respectively. A new search for JAK2 (V617F) at this time was positive with 0.4% and 1.5% mutant alleles, respectively. In the first patient, a third sequential assessment performed 8 months after the potentially ambiguous second test (0.4%), showed a clear-cut positivity (21.8% mutant alleles), associated with a further increase in the platelet count that allowed a conclusive diagnosis of RARS-T.
Six patients with RARS-T carrying JAK2 (V617F) in circulating granulocytes were tested for the mutation in purified BM CD34+ cells. The mutation was detected in all these cases, and the proportion of mutant alleles was substantially similar to that observed in circulating granulocytes. By contrast, JAK2 or MPL mutations were never detected in T lymphocytes.
Overall, 11 (58%) of 19 patients with RARS-T carried JAK2 or MPL mutations in circulating granulocytes, whereas these somatic mutations were not detected in any of the patients with RARS (Table 1; P < .001).
X-chromosome inactivation patterns
Of 11 informative female patients with RARS, 6 had clonal XCIP based on studies of circulating granulocytes and T lymphocytes, and these clonal XCIPs were confirmed in purified CD34+ cells from 4 of these women. Four patients with RARS had skewed XCIP, and the remaining patient had polyclonal XCIP.
Of 8 informative female patients with RARS-T, 7 had clonal XCIP based on studies of circulating granulocytes and T lymphocytes, and these patterns were confirmed in purified CD34+ cells from 5 of these women. The remaining patient with RARS-T had skewed XCIP.
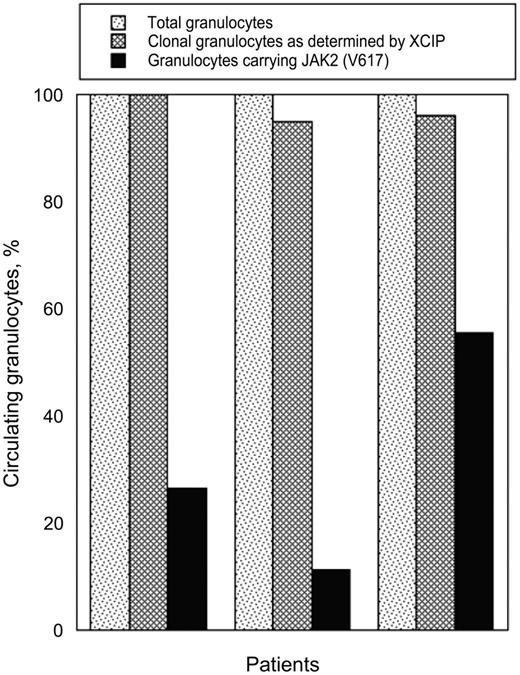
Three of the 7 patients with RARS-T with clonal XCIP carried JAK2 (V617F). On the basis of cleavage ratios between alleles, the proportion of clonal granulocytes ranged from 95% to 100% (Figure 3). By contrast, the proportion of JAK2 (V617F) mutant alleles ranged from 5.6% to 27.7% Assuming that positive cells were heterozygous for the mutation (ie, the scenario compatible with the largest clone for any given percentage of mutant alleles), the proportion of granulocytes carrying JAK2 (V617F) ranged from 11.2% to 55.4% (median value, 27%). Therefore, clonal granulocytes represented almost the totality of circulating granulocytes, whereas granulocytes carrying the JAK2 (V617F) mutation represented only a fraction of clonal granulocytes in these women.
Clonal analysis of hematopoiesis in 3 female patients with RARS-T. The proportion of clonal granulocytes was derived from the cleavage ratio between alleles. Calculation of the proportion of granulocytes carrying JAK2 (V617F) assumes that the mutation was present at the heterozygous state, that is, the scenario compatible with the largest clone for any given percentage of mutant alleles. Should the mutation have been present at the homozygous state, the proportion of granulocytes carrying JAK2 (V617F) would be half of that shown.
Clonal analysis of hematopoiesis in 3 female patients with RARS-T. The proportion of clonal granulocytes was derived from the cleavage ratio between alleles. Calculation of the proportion of granulocytes carrying JAK2 (V617F) assumes that the mutation was present at the heterozygous state, that is, the scenario compatible with the largest clone for any given percentage of mutant alleles. Should the mutation have been present at the homozygous state, the proportion of granulocytes carrying JAK2 (V617F) would be half of that shown.
Gene expression profiles
When we compared gene expression profiles of CD34+ cells from 12 patients with RARS and 6 patients with RARS-T, 255 genes were found to be differentially expressed (Figure 4). Within the 165 up-regulated genes, the most-represented genes included genes related to transcription regulation, cell proliferation, and cytoskeleton organization. Within the 90 down-regulated genes, those involved in cell cycle arrest and cell adhesion showed the greatest representation.
Gene expression profiles of CD34+ cells. Hierarchical clustering of the differentially expressed genes between the CD34+ cells from 6 patients with RARS-T (red symbols under profiles) and the CD34+ cells from 12 patients with RARS (blue symbols under profiles). Each row represents a single Affymetrix probe set, and each column represents a separate patient sample.
Gene expression profiles of CD34+ cells. Hierarchical clustering of the differentially expressed genes between the CD34+ cells from 6 patients with RARS-T (red symbols under profiles) and the CD34+ cells from 12 patients with RARS (blue symbols under profiles). Each row represents a single Affymetrix probe set, and each column represents a separate patient sample.
Forty-six genes were found to be differentially expressed in all 6 cases of RARS-T, and the 10 most significant differentially expressed genes are shown in Table 2. In addition, CXCR4, a gene encoding a CXC chemokine receptor specific for stromal cell–derived factor-1, CTSG, encoding cathepsin, and LTF, encoding lactotransferrin, were markedly down-regulated in patients with RARS-T. CDC2L5, encoding a member of the cyclin-dependent serine/threonine protein kinase family, was up-regulated in patients with RARS-T.
Gene expression profiles of CD34+ cells
| Affymetrix ID . | Gene . | Map . | P . | Average ratio . | |
|---|---|---|---|---|---|
| RARS-T . | RARS . | ||||
| 232363_at | PSIP1 (LEDGF) | 9p22.2 | < .001 | 1.20 | 0.71 |
| 229712_at | SNAPC3 | 9p22.2 | < .002 | 1.39 | 0.83 |
| 219221_at | FLJ35036 | 3q23 | < .002 | 2.18 | 1.14 |
| 210926_at | ACTB | 7p15-p12 | < .002 | 0.79 | 1.30 |
| 224920_x_at | MYADM | 19q13.42 | < .002 | 0.69 | 1.05 |
| 1557905_s_at | CD44 | 11p13 | < .002 | 0.58 | 1.08 |
| 243910_x_at | TIP120A | 12q14 | < .002 | 1.37 | 0.70 |
| 219007_at | NUP43 | 6q24.3 | < .002 | 1.58 | 0.98 |
| 210916_s_at | CD44 | 11p13 | < .002 | 0.61 | 1.06 |
| 204461_x_at | RAD1 | 5p13.2 | < .002 | 1.32 | 0.87 |
| Affymetrix ID . | Gene . | Map . | P . | Average ratio . | |
|---|---|---|---|---|---|
| RARS-T . | RARS . | ||||
| 232363_at | PSIP1 (LEDGF) | 9p22.2 | < .001 | 1.20 | 0.71 |
| 229712_at | SNAPC3 | 9p22.2 | < .002 | 1.39 | 0.83 |
| 219221_at | FLJ35036 | 3q23 | < .002 | 2.18 | 1.14 |
| 210926_at | ACTB | 7p15-p12 | < .002 | 0.79 | 1.30 |
| 224920_x_at | MYADM | 19q13.42 | < .002 | 0.69 | 1.05 |
| 1557905_s_at | CD44 | 11p13 | < .002 | 0.58 | 1.08 |
| 243910_x_at | TIP120A | 12q14 | < .002 | 1.37 | 0.70 |
| 219007_at | NUP43 | 6q24.3 | < .002 | 1.58 | 0.98 |
| 210916_s_at | CD44 | 11p13 | < .002 | 0.61 | 1.06 |
| 204461_x_at | RAD1 | 5p13.2 | < .002 | 1.32 | 0.87 |
The 10 most significant differentially expressed genes between patients with RARS-T and with RARS are listed.
RARS-T indicates refractory anemia with ringed sideroblasts associated with marked thrombocytosis; and RARS, refractory anemia with ringed sideroblasts.
However, no difference in the expression of ALAS231 and ABCB735 was observed between patients with RARS and with RARS-T. ALAS2 was up-regulated in both groups (median expression, 17.23 and 11.08, respectively), whereas ABCB7 was down-regulated (median expression, 0.62 and 0.57, respectively).
Interestingly, there was no significant difference in gene expression profiles between the 4 patients with RARS-T who carried JAK2 (V617F) and the remaining 2 patients who did not.
Flow cytometry enumeration of circulating CD34+ cells
We enumerated circulating CD34+ cells in patients with MDS, RARS-T, and MPN. Median values were 0.9 × 106/L (range, 0.2-7.4 × 106/L) in MDS, 3.1 × 106/L (range, 0.4-10.3 × 106/L) in RARS-T, and 4.1 × 106/L (range, 0.4-12.8 × 106/L) in MPN. Overall, a significant difference in the circulating CD34+ cell count was found between the 3 groups (P = .001). Two-group comparisons showed a significant difference between MDS and MPN (P < .001), whereas that between MDS and RARS-T was borderline (P = .059).
Discussion
In the WHO description of RARS-T, Vardiman et al4p86 underlined that it is not clear whether this condition is “a distinct entity, one end of the spectrum of RARS, a progression of RARS due to an additional acquired genetic abnormality, or less likely, the occurrence of 2 rare diseases in the same patient.” An additional possibility that has been recently proposed is that RARS-T is a form of essential thrombocythemia, based on the observation that the 2 disorders have similar hematologic phenotype, JAK2 and MPL mutation status, and prognosis.25
Within MDSs, RARS is unique in that it is associated with mitochondrial iron overload,7 mitochondrial ferritin overproduction,6 and abnormal expression of genes of erythroid iron metabolism in CD34+ cells.31,35 With respect to this latter feature, we consistently observed up-regulation of several genes in the heme biosynthesis pathway,31 including ALAS2, the gene mutated in X-linked sideroblastic anemia.36 By contrast, ABCB7, the gene encoding a membrane-associated protein that exports iron from the mitochondrion to the cytoplasm and whose mutants are responsible for X-linked sideroblastic anemia with cerebellar ataxia,37 was found to be down-regulated.35 Although RARS is a benign condition,38,39 mainly characterized by abnormalities in the erythroid lineage, previous observations40 and the clonal analysis of hematopoiesis performed in this study clearly indicate that it is a clonal myeloid disorder. So far, however, no causative somatic mutation has been identified in patients with RARS.
Findings of the present study clearly indicate that RARS-T also is a clonal disorder of hematopoiesis; almost all female patients studied showed clonal XCIP in granulocytes or CD34+ cells and polyclonal XCIP in T lymphocytes. In addition, in subjects carrying JAK2 (V617F), mutant alleles were present in only a fraction of clonal granulocytes (Figure 3). This suggests that the somatic mutation had occurred in a preexisting clone of myelodysplastic hematopoietic cells and that its occurrence gave rise to a myelodysplastic/myeloproliferative subclone. Previous studies in patients with MPN also showed that in a proportion of these subjects JAK2 (V617F) occurs on the background of clonal hematopoiesis caused by a somatic mutation in an as-yet-unknown gene.41,42
Additional observations reinforce the opinion that RARS-T may develop from the occurrence of a JAK2 or MPL mutation (or of another as-yet-unknown gene) in a patient with RARS. First, 2 patients in our study cohort initially had a typical RARS and did not carry any mutation of JAK2. They later presented increasing platelet counts, and JAK2 (V617F) was detected at this time. A third patient progressed from RARS to RARS-T without acquiring JAK2 or MPL mutations, perhaps through the acquisition of another molecular lesion involving STAT5 signaling. Second, colony studies in a patient who had both JAK2 (V617F) and MPL (W515L) showed that these mutations belonged to different subclones, which had probably originated independently from a preexisting clone. That multiple molecular events can characterize the clinical course of RARS-T is also supported by the recent identification of uniparental disomy of chromosome 1p and biallelic MPL (W515L) mutation in a few patients with this condition.22
The patients with RARS and RARS-T had gene expression features in common. Notably the 2 patient groups both had up-regulation of ALAS231 and down-regulation of ABCB7,35 distinctive “sideroblastic” features at the molecular level. However, despite these common characteristics, the gene expression profiling data in our study clearly separates the 2 groups. The most probable explanation is that the RARS-T patient group had a different gene expression profile because of additional secondary mutations such as the ones described in this study. However, 8 of the 19 patients in the RARS-T group (including 2 patients studied for gene expression profiles) had no identifiable mutations of JAK2 and MPL. One possibility is that these patients had small mutant clones that were not detected by the methods we used, in particular those for identifying mutations of MPL or JAK2 exon 12, although such clones would probably have been too small to affect the gene expression profiling result. These considerations suggest that other mutant genes, in addition to JAK2 and MPL, may be involved in the pathogenesis of myeloproliferative features in patients with RARS-T. In this regard, it should be noted that approximately one-third of patients with essential thrombocythemia or primary myelofibrosis has no identifiable mutations of JAK2 and MPL.43
The 2 most differentially expressed genes between RARS and RARS-T were PSIP1 (LEDGF) and SNAPC3. These 2 genes were both expressed at higher levels in RARS-T than in RARS. Interestingly the 2 genes are located adjacent to each other at chromosome 9p22, within a common region of uniparental disomy in polycythemia vera that also encompasses JAK2. PSIP1 (LEDGF) and SNAPC3 have overlapping functions, both playing a role in the transcription of RNA pol II. Moreover, PSIP1 (LEDGF) has been previously implicated in leukemia and cancer and recently was found to critically associate with MLL,44 which is a frequent target for recurrent translocations in acute leukemia.
We also found that CDC2L5, a gene known to promote megakaryocyte development in BM cultures,45 was up-regulated in patients with RARS-T. On the contrary, CXCR4 had a lower expression in RARS-T compared with RARS. This gene was previously found to be down-regulated in primary myelofibrosis,46 thereby probably contributing to the abnormal stem cell trafficking that is typically observed in this condition. Indeed, we found marginally elevated CD34+ cell counts in patients with RARS-T compared with patients with MDS.
Apart from patients with RARS-T, the combination of ringed sideroblasts of 15% or greater and platelet count of 450 × 109/L or greater was observed in 3 patients with a clinical picture of primary myelofibrosis, 2 of whom carried JAK2 (V617F). A possible explanation is that patients with MPNs may acquire additional somatic mutations, including the as-yet-unknown mutation responsible for mitochondrial iron overload. However, this latter appears to be a very rare event, based on the findings of this study. In fact, only 3 (5%) of 66 patients with an initial diagnosis of essential thrombocythemia or primary myelofibrosis were found to have 15% or more ringed sideroblasts in the BM.
RARS-T has several features of essential thrombocythemia, including the low mutant allele burden that is typically found in patients with this latter condition.26 This is consistent with the observation that low levels of JAK2 (V617F), as those associated with small clones of hematopoietic cells that are heterozygous for the mutation, are mainly responsible for thrombocytosis and an essential thrombocythemia-like phenotype.47 However, the clinical course of RARS-T appears to be less favorable than that of essential thrombocythemia.48 On the contrary, findings of this study and previous observations48,49 suggest that it is not markedly different from that of RARS. Thus, from a clinical point of view it appears to be important to distinguish RARS-T from essential thrombocythemia.
In conclusion, our observations indicate that RARS-T is a myeloid neoplasm with both myelodysplastic (RARS-like) and myeloproliferative (essential thrombocythemia-like) features at the molecular and clinical levels and that it may develop from a preexisting RARS through the acquisition of somatic mutations of JAK2, MPL, or other as-yet-unknown genes. Thus, the current designation of MDS/MPN appears to accurately reflect the underlying biology of RARS-T.4
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from AIRC (Associazione Italiana per la Ricerca sul Cancro), Fondazione Cariplo, Fondazione IRCCS Policlinico San Matteo, MIUR/PRIN, and Alleanza Contro il Cancro (all in Italy; M.C.), by grants from the Leukemia Research Fund (United Kingdom; J.B.), and by the Swedish Cancer Society (E.H.-L.).
Authorship
Contribution: L.M. and M.C. designed research, analyzed data, and wrote the paper; M.G.D.P., E.B., E.T., E.R., F.P., and R.I. collected clinical and morphologic data; D.P., A.P., A.G., and A.B. performed the experiments; and L.C., J.B., J.S.W., and E.H.-L. analyzed experimental data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario Cazzola, Department of Hematology Oncology, University of Pavia Medical School and Fondazione IRCCS Policlinico San Matteo, 27100 Pavia, Italy; e-mail: mario.cazzola@unipv.it.