Abstract
Abstract 389
Normal hematopoiesis and acute myeloid leukemia (AML) are organized as hierarchies with stem cells, which possess extensive self-renewal and proliferative capacity, at the apex. Although there is definitive evidence from experimental models for the existence of leukemic stem cells (LSC) in some human leukemias, the relevance of LSC to human disease progression is still lacking. While chemotherapeutic treatment of AML patients typically results in disease remission, the majority of patients will eventually relapse and succumb to the disease, indicating that residual LSC are not eliminated by current treatment. We hypothesize that stem cell derived gene expression profiles may be more clinically relevant than those derived from examination of bulk leukemia samples. Here we show the clinical significance of novel stem cell related expression profiles derived from 25 functionally validated human leukemia stem cell populations and 6 normal hematopoietic stem cell populations.
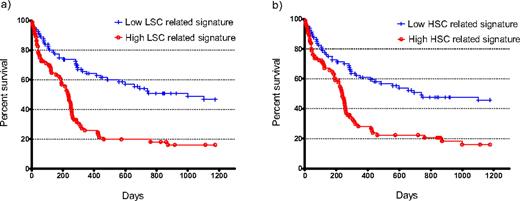
Little is currently known about the molecular regulatory networks that govern human LSC or hematopoietic stem cells (HSC). Therefore, we have carried out global mRNA gene expression profiling of FACS sorted subpopulations of cells enriched for human stem cells, progenitor cells and mature cells from 16 AML primary patient samples and 3 cord blood samples to investigate these pathways. Similar to normal hematopoietic stem cells, leukemia stem, progenitor and mature cells can be sorted using CD34 and CD38 markers. Due to the heterogeneous nature of AML, it is vital that quantitative functional assays are used to characterize the LSC and progenitor activity in each sorted fraction. In vitro cell suspension cultures and methylcellulose colony formation assays were performed to characterize progenitor and blast populations. Importantly, we applied a novel and improved in vivo SCID leukemia initiating cell assay to substantiate the presence of LSC activity in each sorted fraction of 16 AML patient samples. With this enhanced assay, LSC were detected in the expected CD34+/CD38- population. However, in the majority of AML samples, LSC were detected in at least one additional fraction, demonstrating the importance of functional validation when interpreting global gene expression profiles of sorted stem cell populations. LSC and HSC specific signatures were identified following a statistical analysis that compared fractions with stem cell activity against those without (25 LSC vs 29 non-LSC; 6 HSC vs 6 non-HSC). When applied to an independent gene expression data set from 160 cytogenetically normal AML samples, a 25 probe LSC signature was the strongest predictor of overall survival (p<0.0001, HR=2.6, 95%CI 1.8-4.0, median survival 236 vs 999 days; Figure 1a). Furthermore, the 225 probe HSC specific signature derived from normal cells also provided a strong predictor of survival (p<0.0001, HR=2.3, 95%CI 1.5-3.4, median survival 238 vs 741 days; Figure 1b). We queried the gene expression-based chemical genomic database Connectivity Map with the LSC-related gene list and found a negative correlation between the genes in the LSC profile and the expression of genes that are transcriptionally induced following treatment with common chemotherapeutic compounds such as doxorubicin, suggesting resistance to chemotherapy as one possible mechanism for the correlation of the stem cell signatures with survival. Together these data support the hypothesis that the biological determinants that underlie stemness in both normal and leukemic cells are predictors of poor outcome, and are potential targets for novel therapy.
Expression of stem cell-related signatures predicts overall survival. The median expression of a) the 25 probe LSC signature or b) the 225 probe HSC signature was determined in 160 cytogenetically normal AML samples and used to stratify the patients into two groups of 80. All the patients had been treated with intensive double-induction therapy and consolidation chemotherapy. Expression was scaled to 0 for each probe. A Kaplan-Meier plot was generated and statistical significance determined by log-rank test.
Expression of stem cell-related signatures predicts overall survival. The median expression of a) the 25 probe LSC signature or b) the 225 probe HSC signature was determined in 160 cytogenetically normal AML samples and used to stratify the patients into two groups of 80. All the patients had been treated with intensive double-induction therapy and consolidation chemotherapy. Expression was scaled to 0 for each probe. A Kaplan-Meier plot was generated and statistical significance determined by log-rank test.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal