Abstract
We show the molecular and functional characterization of a novel population of lineage-negative CD34-negative (Lin−CD34−) hematopoietic stem cells from chronic myelogenous leukemia (CML) patients at diagnosis. Molecular karyotyping and quantitative analysis of BCR-ABL transcript demonstrated that approximately one-third of CD34− cells are leukemic. CML Lin−CD34− cells showed kinetic quiescence and limited clonogenic capacity. However, stroma-dependent cultures induced CD34 expression on some cells and cell cycling, and increased clonogenic activity and expression of BCR-ABL transcript. Lin−CD34− cells showed hematopoietic cell engraftment rate in 2 immunodeficient mouse strains similar to Lin-CD34+ cells, whereas endothelial cell engraftment was significantly higher. Gene expression profiling revealed the down-regulation of cell-cycle arrest genes and genes involved in antigen presentation and processing, while the expression of genes related to tumor progression, such as angiogenic factors, was strongly up-regulated compared with normal counterparts. Phenotypic analysis confirmed the significant down-regulation of HLA class I and II molecules in CML Lin−CD34− cells. Imatinib mesylate did not reduce fusion transcript levels, BCR-ABL kinase activity, and clonogenic efficiency of CML Lin−CD34− cells in vitro. Moreover, leukemic CD34− cells survived exposure to BCR-ABL inhibitors in vivo. Thus, we identified a novel CD34− leukemic stem cell subset in CML with peculiar molecular and functional characteristics.
Introduction
Several studies have demonstrated that normal hematopoietic stem cells (HSCs) exist in 2 functional states that can be distinguished based on CD34 expression.1 CD34− cells represent a kinetically and functionally resting stem cell subset that needs to be activated to generate a CD34+ cell population with high proliferative and engraftment potential.2,3 Acquisition of CD34 expression on human HSCs is associated with in vitro hematopoietic activity and cell-cycle recruitment.4 CD34−/CD34+ transition results in metabolic activation and down-regulation of growth-inhibitory pathways, and CD34+ stem cells show a significant up-regulation of self-renewal-, commitment-, and engraftment-related genes.5 No data are presently available on the existence of a stem cell population devoid of CD34 expression in leukemic hematopoiesis.
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder of HSCs that have acquired a BCR-ABL fusion gene. The encoded 210-kDa protein has constitutively elevated tyrosine kinase activity that results in a large variety of biochemical changes in leukemic stem cells (LSCs) mainly inducing growth factor–independent proliferation, inhibition of apoptosis, and alteration of adhesive properties.6 There is an increasing body of evidence indicating that, similar to normal hematopoiesis, a kinetically quiescent stem cell population can be isolated within the CD34+ LSC compartment.7 These G0 CML cells are able to repopulate immunodeficient mice8 and show a different transcriptional profile from dividing CD34+ cells.9 More importantly, the dissection of the stem cell pool in CML has allowed the characterization of subsets of CD34+ LSCs resistant, in vitro and in vivo, to targeted therapies with BCR-ABL inhibitors.10-12 Multiple mechanisms of drug resistance have been described, including kinetic quiescence per se,10 increased expression level of BCR-ABL, and altered expression of ABCB1, ABCG2, and OCT1 cell membrane drug transporter genes in the most primitive CD34+/CD38− LSCs.13 Taken together, these findings stress the importance of gaining further understanding of the molecular and functional properties of the stem cell compartment in CML, in view of the development of more effective therapies.
To this end, in the present study, we assessed the gene expression profile and the functional characteristics of a novel Lin−CD34− stem cell population in CML patients at diagnosis. Our data demonstrate that approximately one-third of Lin−CD34− cells are leukemic and capable of engraftment in immunodeficient mice. Overall, molecular analysis showed 1827 transcripts differentially expressed in CML HSC subpopulations compared with normal counterparts, genes affecting many cellular functions associated with the neoplastic phenotype. Microarray data also highlighted the highest transcriptome differences between CML Lin−CD34− cells and their normal counterparts. Functional studies did also demonstrate the intrinsic resistance of CML Lin−CD34− cells to imatinib mesylate (IM) treatment in vitro and in vivo.
Methods
Cells
Leukemic cells were obtained from 17 chronic phase Ph+ CML patients at diagnosis, before treatment. Additional CML samples were obtained from 7 patients after IM (n = 6) or nilotinib (n = 1) treatment (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Normal samples were leukapheresis products from 15 healthy stem cell donors receiving recombinant human granulocyte colony-stimulating factor (G-CSF; Lenograstim; Sanofi-Aventis). The protocol was approved by the ethical committee of the university hospital, and each patient/donor gave written informed consent in accordance with the Declaration of Helsinki. Hematopoietic stem/progenitor cell purification and phenotypic analyses were performed as previously described.4,5 Aliquots of sorted Lin−CD34−, Lin−CD34+, and Lin+CD34+ were reanalyzed by FACScan (Becton Dickinson) to assess their purities, which were 98.9% (± 1%), 99.2% (± 0.8%), and 98% (± 1%), respectively. A representative example of FACScan analysis is shown in supplemental Figure 1. As reported before,4,5 highly purified Lin−CD34− cells confirmed their hematopoietic origin by the expression of CD45 antigen.
Liquid cultures
Stem cell populations were cultured onto irradiated murine stromal cells (M2-10B4) genetically engineered to produce G-CSF and interleukin-3 (IL-3) at an initial density of 4 × 104 cells/mL in IMDM (Cambrex Bio Science) supplemented with 10% fetal bovine serum (FBS; Invitrogen), l-glutamine, and antibiotics.4 All cultures were maintained at 37°C in humidified 5% CO2 atmosphere in the presence of optimized concentrations of the following rh-cytokines: SCF (50 ng/mL; Amgen), IL-11 (50 U/mL; Endogen), FLT3-ligand (50 ng/mL; Immunex), and megakaryocyte growth and development factor (100 ng/mL; Amgen). Twice a week, the culture medium was replaced by fresh medium, and cytokines and hematopoietic cells were adjusted to the initial cell concentration. Aliquots of cell suspension were cultured after 1 week in methylcellulose and in liquid culture to evaluate the presence of secondary CFU-C. To assess the percentage of CD34+ cells in liquid cultures, the cells were incubated with anti–human CD34–fluorescein isothiocyanate monoclonal antibody (HPCA-2; Becton Dickinson) and then analyzed by FACScan equipment.
For long-term culture initiating cells (LTC-IC) assays, 2 × 104 cells/mL were plated, in quadruplicate onto M2-10B4 cell line for 5 weeks without cytokines and then plated for their secondary CFU-C activity. The number of LTC-IC was calculated as earlier reported.4
In selected experiments, fluorescence-activated cell sorter (FACS)–sorted Lin−CD34− and Lin−CD34+ cells were collected in IMDM (Cambrex Bio Science), added with 10% FBS (Invitrogen), and cultured for 48 hours with or without increasing doses (0.1μM, 1μM, 10μM) of IM (Novartis Pharmaceutical). At the end of incubation, cells were collected, washed, and resuspended in IMDM plus 10% FBS. Viable cells, determined by trypan blue exclusion count, were then plated for CFU-C assays and assessed by quantitative RT-PCR for BCR-ABL transcript.
RNA extraction and microarray analysis
Total cellular RNA was extracted from 2 × 105 cells of each sample using RNeasy Micro kit (QIAGEN) following the protocol supplied by the manufacturer. Disposable RNA chips (Agilent RNA 6000 Nano LabChip kit; Agilent Technologies) were used to determine the concentration and purity/integrity of RNA samples using an Agilent 2100 Bioanalyzer. RNAs originating from 12 healthy donors or from 12 CML patients were pooled to obtain at least 2 μg per sample.
One-cycle target labeling assays, as well as the Affymetrix Human HG-U95Av2 GeneChip array hybridization, staining, and scanning, were performed, using Affymetrix standard protocols (Affymetrix).5 The GeneChip Operating Software absolute analysis algorithm was used to determine the amount of a transcript mRNA (signal), while the GeneChip Operating Software comparison analysis algorithm was used to compare gene expression levels between CML and normal Lin−CD34−, Lin−CD34− and Lin+CD34+ samples.
Present genes were selected as the sequences showing the detection call “P” and signal greater than 100 in at least one sample. Differentially expressed genes were selected as the sequences showing a “change call,” “I” or “D,” and a “signal log ratio” greater than 1 or less than −1 in at least one of these pairwise comparisons: CML versus normal Lin−CD34−, CML versus normal Lin−CD34+, CML versus normal Lin+CD34+, CML Lin−CD34− versus CML Lin−CD34+, CML Lin−CD34− versus CML Lin+CD34+, and CML Lin−CD34+ versus CML Lin+CD34+. The gene list passing this filter was selected as “changing genes.”
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2007 tool (http://david.abcc.ncifcrf.gov/) was used to examine selected lists of genes to identify overrepresentation of functional classes accordingly with gene-ontology classification. All the data have been deposited in the Gene Expression Omnibus MIAME compliant public database (http://www.ncbi.nlm.nih.gov/geo; accession number GSE11675; supplemental Table 2).
CrkL phosphorylation assay
The CrkL Tyr207-phosphorylation assay was performed in untreated and 5μM IM-treated Lin−CD34−, Lin−CD34+, and Lin+CD34+ CML cells after 16 hours in IMDM (Cambrex Bio Science), with 10% FBS (Invitrogen) added. Healthy donor-derived cell populations served as BCR-ABL-negative controls. For each datapoint, 105 cells per sample were labeled for PCrkL as already described.11 Cells were analyzed by a Coulter Epics XL flow cytometer (Coulter Electronics Inc). At least 5000 events were counted for each sample to ensure statistical relevance. The resulting flow cytometry data were normalized according to the mean fluorescent intensity (MIF) so that the untreated control CML Lin+CD34+ cell population for each patient had pCrkl MIF = 1. Then, the results for each sample were calculated as a fraction of this. CML Lin+CD34+ cells' pCrkl MIF levels were similarly set to 1 to compare CML subpopulations with their normal counterparts.
Molecular evaluation of BCR-ABL gene rearrangement
RNA from CML cells was extracted by commercially available kits (RNeasy; QIAGEN), and cDNA synthesis was performed using random hexamer primers and M-MLV reverse transcriptase enzyme. Qualitative real-time polymerase chain reaction (RT-PCR) for BCR-ABL transcript was performed as earlier reported.14,15 Quantitative RT-PCR (RQ-PCR) for BCR-ABL mRNA transcript was performed using a TaqMan gene expression assay (Applied Biosystems) by means of an ABI PRISM 7700 Sequence Detector (Applied Biosystems). Conditions for amplification, hybridization, and fluorescence detection have been already described.16,17 Assays were performed in duplicate. Parallel TaqMan assays were run on each sample for ABL as housekeeping gene, to compensate for different quantities of RNA and cDNA and to exclude negative results derived from samples of poor quality. BCR-ABL and ABL plasmid dilutions (Ipsogen) were used as standards and the final results were calculated as the BCR-ABL/ABL ratio and expressed in percent. RQ-PCR of the BCR-ABL transcript after xenotransplant experiments was performed using human β2-microglobulin (Applied Biosystems) as a housekeeping17 gene because human ABL overlaps with murine ABL using M-BCR Fusion Quant Kit (Ipsogen).
Measurement of human hematopoietic and endothelial engraftment by flow cytometry, RQ-PCR, and RT-PCR
Six- to 8-week-old nonobese diabetic/severe combined immunodeficient β2 Microglobulinnull (NOD/SCID/β2M)18 or NOD/SCID/IL-2Rγnull19,20 mice were sublethally irradiated with 200 cGy and injected intravenously with 104 to 105 purified Lin−CD34+ or Lin−CD34− cells from BCR-ABL-positive CML patients. Sixty to 120 days after transplant, mice were killed, and their blood and marrow collected for the evaluation of human hematopoietic and endothelial engraftment. All procedures involving animals were done in accordance with national and international laws and policies.
Blood and marrow of immunodeficient mice were evaluated by 4-color flow cytometry using a panel of monoclonal antibodies reacting with human CD31, vascular endothelial growth factor-receptor 2 (VEGFR-2; KDR), and CD34 and CD45 antigens. After red cell lysis, cell suspensions were evaluated by a FACSCalibur equipment (Becton Dickinson) using analysis gates designed to exclude dead cells, platelets, and debris. After acquisition of at least 100 000 cells per sample, analyses were considered as informative when adequate numbers of events (ie, > 50, typically 100-200) were collected in the human cell enumeration gates. Percentages of stained cells were determined and compared with appropriate negative controls. Positive staining was defined as being greater than nonspecific background staining. annexin V and 7AAD were used to exclude apoptotic and dead cells.4 The expression of human CD45 in blood and marrow of transplanted mice was confirmed by RT-PCR and Southern blotting.4 The sensitivity and specificity of human CD45 RT-PCR (10−5) was evaluated by serial dilutions of CD45 positive NB4 cells in CD45 negative Hela cells and confirmed by Southern blotting with probes were obtained by 32P-labeled reverse primers.
RQ-PCR was used as previously described to enumerate the copies of the human endothelial-specific transcript VE-Cadherin in the blood and marrow of transplanted mice.21 The sensitivity of the assays was 25 and 10 copies for VE-Cadherin and βactin, respectively. No copies of human VE-Cadherin or βactin were found in nontransplanted mice. To detect BCR-ABL transcript in transplanted mice, bone marrow (BM) samples were processed as reported in “Molecular evaluation of BCR-ABL gene arrangement.”
Progenitor cell assays, FISH analysis, cell cycle analysis, and DNA repair functional assay (Comet assay) methods are described in the supplemental Methods section.
Statistical analysis
The results are reported as the mean plus or minus SD of at least 3 different experiments. Statistical comparisons were performed using the t test, analysis of variance and linear regression when data were normally distributed and the nonparametric analyses of Spearman and Mann-Whitney when data were not normally distributed. P valuse less than .05 were considered as statistically significant.
Results
Isolation, molecular and karyotypic characterization, and engraftment potential of Lin−CD34+ and Lin−CD34− CML cells
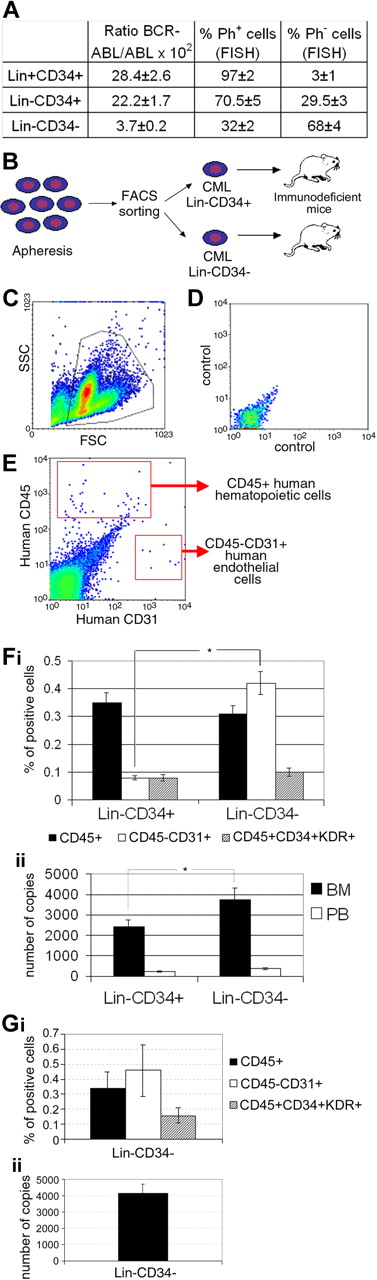
The frequencies of CML Lin−CD34+ and Lin−CD34− cells were 0.29% (± 0.11%) and 0.52% (± 0.12%) of mononuclear cells, respectively, compared with 0.17% (± 0.03%) and 0.16% (± 0.05%) in healthy donors (P = NS). FISH analysis and RQ-PCR for BCR-ABL transcript demonstrated that more primitive hematopoietic cell populations contained higher numbers of residual normal progenitors (Figure 1A). Specifically, Ph+ cells were 97% (± 2%), 70.5% (± 5%), and 32% (± 2%) of total Lin+CD34+, Lin−CD34+, and Lin−CD34− CML cells at diagnosis, respectively. Of note, the finding that only one-third of Lin−CD34− CML cells are leukemic (ie, neoplastic cells are diluted in 70% normal cells) highlights the functional and molecular differences of this cell population versus its normal counterparts.
Molecular and karyotypic characterization and engraftment potential of Lin−CD34+ and Lin−CD34− CML cells. Characterization of CML Lin+CD34+, Lin−CD34+, and Lin−CD34− cell populations purified from the PB of chronic-phase patients at diagnosis. (A) FISH analysis and RQ-PCR for BCR-ABL transcript demonstrating higher numbers of residual normal progenitors in Lin−CD34− cell populations. The results are expressed as mean ± SD of 3 experiments. (B) Study design (number of experiments = 9 with a total of 37 and 22 NOD/SCID/β2M and NOD/SCID/IL-2Rγnull mice, respectively). (C-G) Flow cytometric and molecular evaluation of human cell engraftment in the marrow and blood of NOD/SCID/β2M (F) and NOD/SCID/IL-2Rγnull mice (G) engrafted with 100 000 human cells. Immunodeficient mice were evaluated up to 120 days after transplant. (C) The gate used to exclude platelets, debris, and dead cells in a representative case of xenotransplantation; (D) the negative control; (E) the gates used to identify hematopoietic (human CD45+) and endothelial (human CD45−/CD31+) cells. (Fi) The frequency of human hematopoietic and endothelial cell engraftment in NOD/SCID/β2M mice transplanted with CML Lin−CD34+ and Lin−CD34− cells. (Fii) The number of human VE-Cadherin RNA copies in the blood and marrow of transplanted mice. (Gi-Gii) Human hematopoietic and endothelial cell engraftment in NOD/SCID/IL-2Rγnull mice. RQ-PCR of BCR-ABL mRNA transcript was used to confirm the leukemic origin of engrafted cells in both mice strains. Our results showed a mean of 176 ± 87 BCR-ABL/β2 microglobulin copies in NOD/SCID/β2M mice (mean of 85 ± 32 β2 microglobulin copies) and a mean of 188 ± 62 copies in in NOD/SCID/IL-2Rγnull mice (85 ± 32 β2 microglobulin copies); *P < .05.
Molecular and karyotypic characterization and engraftment potential of Lin−CD34+ and Lin−CD34− CML cells. Characterization of CML Lin+CD34+, Lin−CD34+, and Lin−CD34− cell populations purified from the PB of chronic-phase patients at diagnosis. (A) FISH analysis and RQ-PCR for BCR-ABL transcript demonstrating higher numbers of residual normal progenitors in Lin−CD34− cell populations. The results are expressed as mean ± SD of 3 experiments. (B) Study design (number of experiments = 9 with a total of 37 and 22 NOD/SCID/β2M and NOD/SCID/IL-2Rγnull mice, respectively). (C-G) Flow cytometric and molecular evaluation of human cell engraftment in the marrow and blood of NOD/SCID/β2M (F) and NOD/SCID/IL-2Rγnull mice (G) engrafted with 100 000 human cells. Immunodeficient mice were evaluated up to 120 days after transplant. (C) The gate used to exclude platelets, debris, and dead cells in a representative case of xenotransplantation; (D) the negative control; (E) the gates used to identify hematopoietic (human CD45+) and endothelial (human CD45−/CD31+) cells. (Fi) The frequency of human hematopoietic and endothelial cell engraftment in NOD/SCID/β2M mice transplanted with CML Lin−CD34+ and Lin−CD34− cells. (Fii) The number of human VE-Cadherin RNA copies in the blood and marrow of transplanted mice. (Gi-Gii) Human hematopoietic and endothelial cell engraftment in NOD/SCID/IL-2Rγnull mice. RQ-PCR of BCR-ABL mRNA transcript was used to confirm the leukemic origin of engrafted cells in both mice strains. Our results showed a mean of 176 ± 87 BCR-ABL/β2 microglobulin copies in NOD/SCID/β2M mice (mean of 85 ± 32 β2 microglobulin copies) and a mean of 188 ± 62 copies in in NOD/SCID/IL-2Rγnull mice (85 ± 32 β2 microglobulin copies); *P < .05.
On injection of purified CML Lin−CD34+ or Lin−CD34− cells into sublethally irradiated immunodeficient mice (Figure 1B), we found that both stem cell populations showed hematopoietic potential. Figure 1C through E shows a representative flow cytometric assessment of human hematopoietic (CD45+) and endothelial (CD45−CD31+) cell engraftment in the BM of transplanted mice. Human cell engraftment was confirmed by RT-PCR evaluation of the expression of CD45 (data not shown) and by RQ-PCR to enumerate copies of human endothelial-specific transcript VE-cadherin (Figure 1Fii,Gii). Quantitation of human hematopoietic cell engraftment in NOD/SCID/β2M mice by enumeration of CD45+ cells (Figure 1Fi) indicated that Lin−CD34− CML cells generated a measurable number of human hematopoietic and endothelial mature and progenitor cells in transplanted mice similarly to different subpopulations of CML CD34+ cells (present data and Eisterer et al8 ) and normal Lin−CD34−.4 CD34− cells also showed long-term hematopoietic and endothelial cell engraftment in NOD/SCID/IL-2Rγ null mice (Figure 1Gi-ii). Of note, transplantation of Lin−CD34− CML cells resulted in a significantly better endothelial cell engraftment compared with CML Lin−CD34+ progenitors as demonstrated by the higher number of BM mature CD31+ cells (Figure 1Fi) and the higher number of copies of VE-cadherin transcript (Figure 1Fii). Endothelial cell engraftment was also confirmed by the evaluation of CD45+CD34+KDR+ progenitors in mice transplanted with both CD34+ and CD34− cell populations (Figure 1Fi,Gi).
The RQ-PCR of BCR-ABL mRNA transcript was used to confirm the leukemic origin of engrafted cells in both mice strains. Our results showed a mean of 176 (± 87) BCR-ABL/β2 microglobulin copies in NOD/SCID/β2M mice and a mean of 188 (± 62) copies in NOD/SCID/IL-2Rγ null mice.
Global transcriptome changes in normal and CML stem cell subsets
We then assessed the gene expression profile of all cell populations using Affymetrix HG-U95Av2 GeneChip array, representative of 12 625 transcripts (supplemental Table 2).
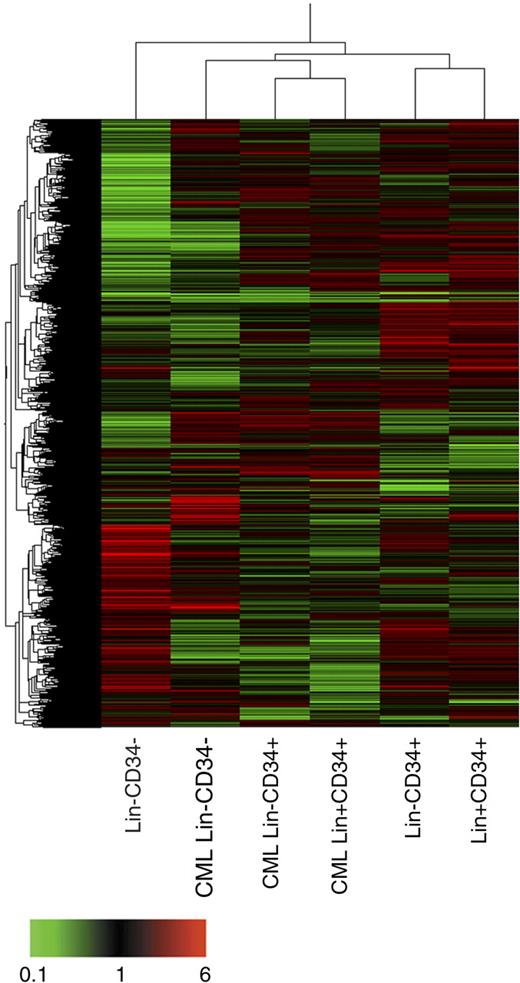
Unsupervised clustering analysis (Figure 2) revealed that CML Lin−CD34− are closer to the leukemic and normal CD34+ subfractions analyzed than to their normal Lin−CD34− counterparts. Next, we addressed the transcriptome differences between the 3 normal and CML stem/progenitor cell phenotypes, ie Lin−CD34−, Lin−CD34+, and total CD34+. As described in “RNA extraction and microarray analysis,” we selected a list of 1827 probe sets as changing genes in the pair-wise comparisons between normal and CML cells (supplemental Table 3). This gene list was uploaded onto DAVID software to identify prevalent categories in the GO-controlled vocabulary family “biological process.” Supplemental Table 4 shows that the prevalent categories increased in CML cells were mitosis, DNA replication, regulation of cell cycle, DNA repair, anti-apoptosis, and angiogenesis. Conversely, the prevalent categories down-regulated in CML samples were antigen presentation; antigen processing, endogenous antigen via MHC class I; antigen processing, exogenous antigen via MHC class II; defense response; and positive regulation of apoptosis. According with these results, we next moved to analyze the processes of cell proliferation, apoptosis, DNA repair, and antigen processing and presentation, together with the expression of genes involved in angiogenesis as a tumor progression-related mechanism. In addition, to study the self-renewal and differentiation capacity of CML cells, we looked at the expression of hematopoietic transcription factors (TFs).
Unsupervised clustering analysis. Clustering has been performed using an unsupervised approach and applying the Pearson correlation equation provided by GeneSpring. A combination of 2 hierarchical clustering analyses (gene tree and condition tree) is shown. The gene tree is shown on left, the condition tree on top. Gene coloring was based on normalized signals as shown at the bottom of the figure.
Unsupervised clustering analysis. Clustering has been performed using an unsupervised approach and applying the Pearson correlation equation provided by GeneSpring. A combination of 2 hierarchical clustering analyses (gene tree and condition tree) is shown. The gene tree is shown on left, the condition tree on top. Gene coloring was based on normalized signals as shown at the bottom of the figure.
Cell-cycle regulators' gene expression and cell-cycle analysis
Analysis of cell-cycle regulator genes (Figure 3A) confirmed previously published datasets22-25 showing the up-regulation of positive cell-cycle regulators22,23 (CCND2, CDK6, and CDK4) and the down-regulation of cell-cycle progression inhibitors23-25 (eg, p21WAF1 and p57KIP226 ) in CML versus normal stem cells. Of note, many growth arrest genes (SERPINF1,27 GAS6,28 RGS2,29 and ZFP3630 ) displayed the strongest down-regulation in CML Lin−CD34− cells compared with their normal counterparts.
Cell-cycle regulators expression and flow cytometric cell-cycle analysis. Expression of cell-cycle regulators (A). Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated transcripts involved in cell-cycle regulation. The normalized signal-based coloring legend is shown at the bottom of the figure. Cell-cycle distribution of normal and CML stem/progenitor cells according to CD34 expression (B). Lin− CD34+ and Lin− CD34− cells were isolated from CML patients at diagnosis and healthy donors and analyzed by flow cytometry for their kinetic status. In panel C, we assessed the clonogenic growth in semisolid medium of CML and normal stem/progenitor cells as well as the LTC-IC activity of 2 CML patients (pt). Our data show a significantly higher frequency of CML Lin−CD34− and Lin−CD34+ cell-derived CFU-C compared with normal samples. Specifically, the clonogenic efficiency of leukemic CD34− cells was 0.4% ± 0.08% versus 0.15% ± 0.05% in normal samples. Interestingly, CML Lin−CD34− cells also showed LTC-IC activity in 2 of 2 patients studied (the results are expressed as the mean ± SD of quadruplicate cultures of secondary CFU-C × 104 cells). As described in “Methods,” FACS-sorted Lin−CD34− cells were also kept in culture for 7 days. After 1 week (t7), cells expressing the CD34 antigen were purified and analyzed for their cell-cycle distribution and BCR-ABL content. As shown in panel D, CML Lin−CD34− cells rapidly up-regulated the CD34 antigen on a cell fraction and progressed through the cell cycle. These findings were associated with the higher values of BCR-ABL gene rearrangement as demonstrated by RQ-PCR. The results of panels B and C are expressed as mean ± SD of 3 (B) and 6 (C) different experiments; *P < .05.
Cell-cycle regulators expression and flow cytometric cell-cycle analysis. Expression of cell-cycle regulators (A). Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated transcripts involved in cell-cycle regulation. The normalized signal-based coloring legend is shown at the bottom of the figure. Cell-cycle distribution of normal and CML stem/progenitor cells according to CD34 expression (B). Lin− CD34+ and Lin− CD34− cells were isolated from CML patients at diagnosis and healthy donors and analyzed by flow cytometry for their kinetic status. In panel C, we assessed the clonogenic growth in semisolid medium of CML and normal stem/progenitor cells as well as the LTC-IC activity of 2 CML patients (pt). Our data show a significantly higher frequency of CML Lin−CD34− and Lin−CD34+ cell-derived CFU-C compared with normal samples. Specifically, the clonogenic efficiency of leukemic CD34− cells was 0.4% ± 0.08% versus 0.15% ± 0.05% in normal samples. Interestingly, CML Lin−CD34− cells also showed LTC-IC activity in 2 of 2 patients studied (the results are expressed as the mean ± SD of quadruplicate cultures of secondary CFU-C × 104 cells). As described in “Methods,” FACS-sorted Lin−CD34− cells were also kept in culture for 7 days. After 1 week (t7), cells expressing the CD34 antigen were purified and analyzed for their cell-cycle distribution and BCR-ABL content. As shown in panel D, CML Lin−CD34− cells rapidly up-regulated the CD34 antigen on a cell fraction and progressed through the cell cycle. These findings were associated with the higher values of BCR-ABL gene rearrangement as demonstrated by RQ-PCR. The results of panels B and C are expressed as mean ± SD of 3 (B) and 6 (C) different experiments; *P < .05.
Different molecular profiles of normal and CML cell populations were reflected in their different kinetic status. As shown in Figure 3B, the percentage of CML Lin−CD34+ cells in G0-phase of the cell cycle (19.4 ± 14.3) was significantly (P < .03) lower than that of other cell populations under study, while the percentage of S-phase cells was significantly higher (7.8 ± 4.5; P < .02). Conversely, leukemic and normal CD34− cells were both kinetically quiescent with no significant differences between the 2 cell populations. However, when we assessed their clonogenic growth in semisolid medium, we observed a significantly (P < .04) higher frequency of CML Lin−CD34− cell-derived CFU-C compared with normal samples (Figure 3Ci) in addition to the well-known higher clonogenic efficiency (P < .05) of CML CD34+ cells (Figure 3Cii). This finding is biologically relevant by considering that only 30% of CML CD34− cells are leukemic and, therefore, are responsible for the difference in the clonogenic efficiency compared with normal samples. In 2 CML cases, we also evaluated more primitive LTC-IC content of CD34+ and CD34− cell populations. As shown in Figure 3Ciii, the number of LTC-IC was 54 (± 15) and 21 (± 14) × 104 CD34+ cells, respectively. The same number of CD34− cells showed 1.3 (± 0.5) and 2.2 (± 1) LTC-IC (Figure 3Civ). Of note, in our experience, normal CD34− cells do not show any LTC-IC activity.4 The leukemic origin of LTC-IC was confirmed by RQ-PCR, performed on secondary colonies collected from semisolid medium, showing a BCR-ABL/ABL ratio of 20.83 and 10.37 for CD34+ cells (number of ABL copies 20 381 and 18 683) and 3.2 and 1.8 for CD34− cells (ABL transcript copies 1800 and 3200).
Furthermore, when cultured on M2-10B4 stromal cells in combination with cytokines, CML Lin−CD34− cells rapidly up-regulated the CD34 antigen on a cell fraction and progressed through the cell cycle. These findings were associated with the higher expression of BCR-ABL gene rearrangement as demonstrated by RQ-PCR (Figure 3D).
Taken together, these results demonstrate that quiescent CML Lin−CD34− stem cells are transcriptionally more similar to proliferating cells than to their normal quiescent counterparts and are ready to enter cell cycle.
Expression of genes linked to tumor progression
As already described for CML mononuclear cells,25 genes involved in single-strand and double-strand DNA break repair were preferentially expressed in CML CD34+ cells (supplemental Figure 2Ai and Aii, respectively). Moreover, our data confirmed the up-regulation of the anti-apoptotic gene BCL223,31 in CML Lin−CD34+ and more mature normal and leukemic Lin+CD34+ and BCLXL32 in CML Lin+CD34+ progenitors, whereas the proapoptotic genes BAX,33 ENO1,31 and MOAP132 5 were down-regulated if compared with normal cells (supplemental Figure 2C).
To confirm the increased DNA repair capacity of CML cells, we tested their ability to repair MMS-induced single-strand DNA breaks and mitoxantrone-induced double-strand DNA breaks34 by means of alkaline and the neutral single-cell gel electrophoresis, respectively (COMET assay; see supplemental Methods). As shown in supplemental Figure 2, the tail length, intensity, and shape were different according to the cell type; 6 hours after drug removal, CML CD34+ cells showed a reduction of approximately 90% in MMS- (Bi and Bii, respectively) and mitoxantrone-induced (Biii and Biv, respectively) DNA damage, whereas normal CD34+ cells showed a reduction of approximately 70% in single- and double-strand breaks (P < .05). These data confirm that CML CD34+ cells have a better ability to repair both double- and single-strand DNA breaks than normal cells.
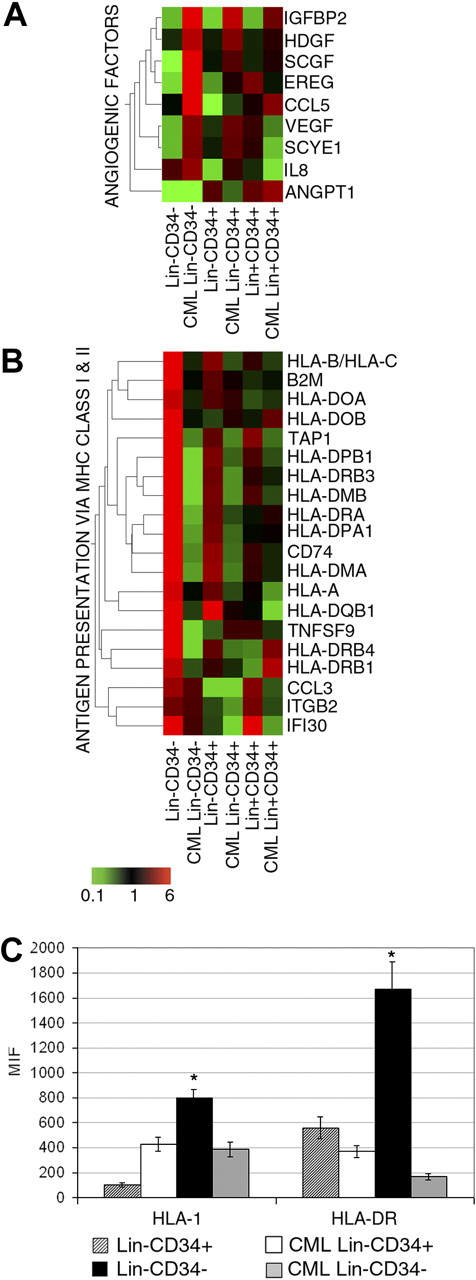
While modulations of genes functionally related to apoptosis22,23,25,35,36 and DNA repair23,25 have already been reported by other groups, our microarray data showed, for the first time, the increased expression of pro-angiogenic factors (VEGFA,37 HDGF,38 RANTES/CCL5,39 and EREG40 ) in CML Lin−CD34− cells versus normal counterparts (Figure 4A).
Expression of genes involved in MHC class I and II and angiogenesis and flow cytometric analysis of MCH class I and II molecules. Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated probe sets belonging to the categories of angiogenesis (A) and antigen presentation and processing via MHC class I and II (B). The signal-based coloring legend is shown at the bottom of the figure. Freshly separated normal and CML Lin−CD34+ and Lin−CD34− cells were analyzed by flow cytometry for the expression of MHC class I and II molecules (C). The results are expressed as mean ± SD of 3 different experiments; *P < .05; MIF, mean intensity fluorescence.
Expression of genes involved in MHC class I and II and angiogenesis and flow cytometric analysis of MCH class I and II molecules. Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated probe sets belonging to the categories of angiogenesis (A) and antigen presentation and processing via MHC class I and II (B). The signal-based coloring legend is shown at the bottom of the figure. Freshly separated normal and CML Lin−CD34+ and Lin−CD34− cells were analyzed by flow cytometry for the expression of MHC class I and II molecules (C). The results are expressed as mean ± SD of 3 different experiments; *P < .05; MIF, mean intensity fluorescence.
Moreover, expression analysis of genes belonging to the categories of endogenous and exogenous antigen processing and presentation via MHC class I and MHC class II, respectively, showed a remarkable decrease in CML cells of several MHC class I (HLA-B, HLA-C, HLA-A, and β2M) and MHC class II molecules, particularly in Lin−CD34− stem cells (Figure 4B). At the same time, we observed in CML cells a consistent down-regulation of TAP1, involved in the antigen presentation via MHC class I,41 and of some costimulatory molecules, such as TNFSF942 and CCL343 (Figure 4B). Whereas the down-regulation of genes belonging to the exogenous antigen presentation category in CML CD34+ cells was already reported,9 our results highlighted the down-regulation of several genes involved in both exogenous (via MHC class II) and, of note, endogenous (via MHC class I) antigen processing and presentation. Consistently, phenotypic analysis demonstrated, at the protein level, the significantly lower expression of MHC class I and II molecules on CML Lin−CD34− cell membrane compared with normal counterparts (Figure 4C).
Self-renewal and myeloid commitment capacity
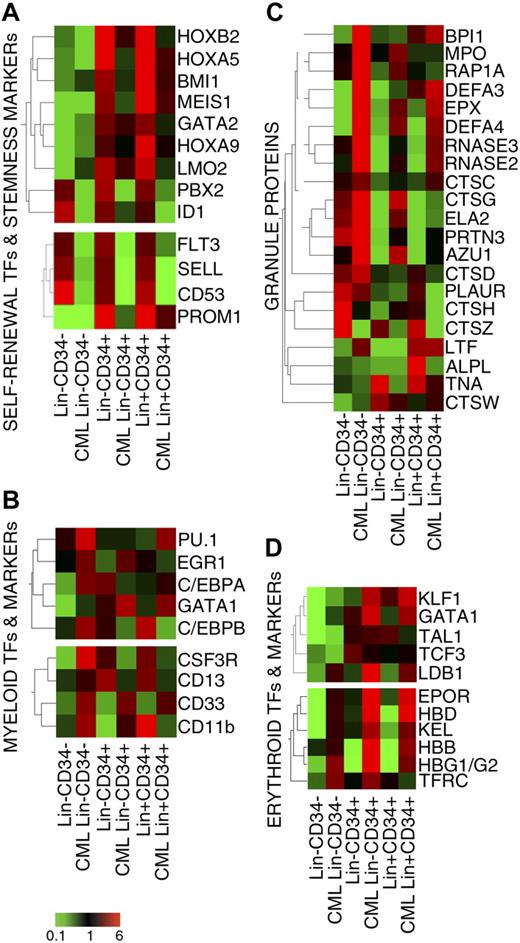
As recently published by Bruns et al,24 microarray results showed the strong down-regulation of most TFs involved in self-renewal (namely GATA-2,44 Bmi1,45 LMO2,46 HoxA5,47 HoxB2,48 HoxA9,49 and its cofactors Meis1 and PBX250 ) in CML versus normal CD34+ cells (Figure 5A). Consistently, transcripts for the HSC marker CD133 (PROM1), as well as L-selectin (SELL) and CD53 molecules, whose expression has been correlated to the maintenance of the stemness features,24 were down-regulated in CML subpopulations (Figure 5A). Genes belonging to the WNT, Hedgehog, and NOTCH pathways were expressed at very low levels in all cell populations (data not shown).
Expression of transcriptional regulators of stemness and myeloid commitment. Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated probe sets belonging to the category of TFs and markers involved in self-renewal and stemness (A), mono-granulocyte lineage commitment (B), neutrophil granule proteins (C), erythroid lineage commitment (D). The signal-based coloring legend is shown at the bottom of the figure.
Expression of transcriptional regulators of stemness and myeloid commitment. Eisen tree map computed using the GeneSpring gene tree and the Pearson correlation equation on the modulated probe sets belonging to the category of TFs and markers involved in self-renewal and stemness (A), mono-granulocyte lineage commitment (B), neutrophil granule proteins (C), erythroid lineage commitment (D). The signal-based coloring legend is shown at the bottom of the figure.
On the contrary, the expression of TFs involved in myeloid differentiation (Figure 5B), such as PU.1/SPI1, EGR-1, C/EBPα, and C/EBPβ,51 was increased in CML versus normal Lin−CD34− cells, suggesting an earlier commitment of LSCs to myelopoiesis. In agreement with this hypothesis, we found a remarkable overexpression of myeloid markers (Figure 5B) and granule proteins (Figure 5C) in the same cell population.
Finally, microarray data supported recent data9,23 showing the overexpression of several TFs involved in erythropoiesis (eg, KLF1 and GATA1) in CML versus normal HSCs (Figure 5D). Similarly, erythropoietin receptor, transferrin receptor, and Hbβ, γ, and δ transcripts were up-regulated in all the CML subpopulations versus their normal counterparts (Figure 5D). Of note, several of the erythroid genes under analysis (namely HBG1/G2, KEL, KLF1, GATA1, LDB1, and TAL1) were already expressed in leukemic Lin−CD34− cells but not on their normal counterparts, thus suggesting an early commitment of CML hematopoiesis to erythroid phenotype.22,24,52
CD34 expression in CML is associated with different sensitivity to IM treatment
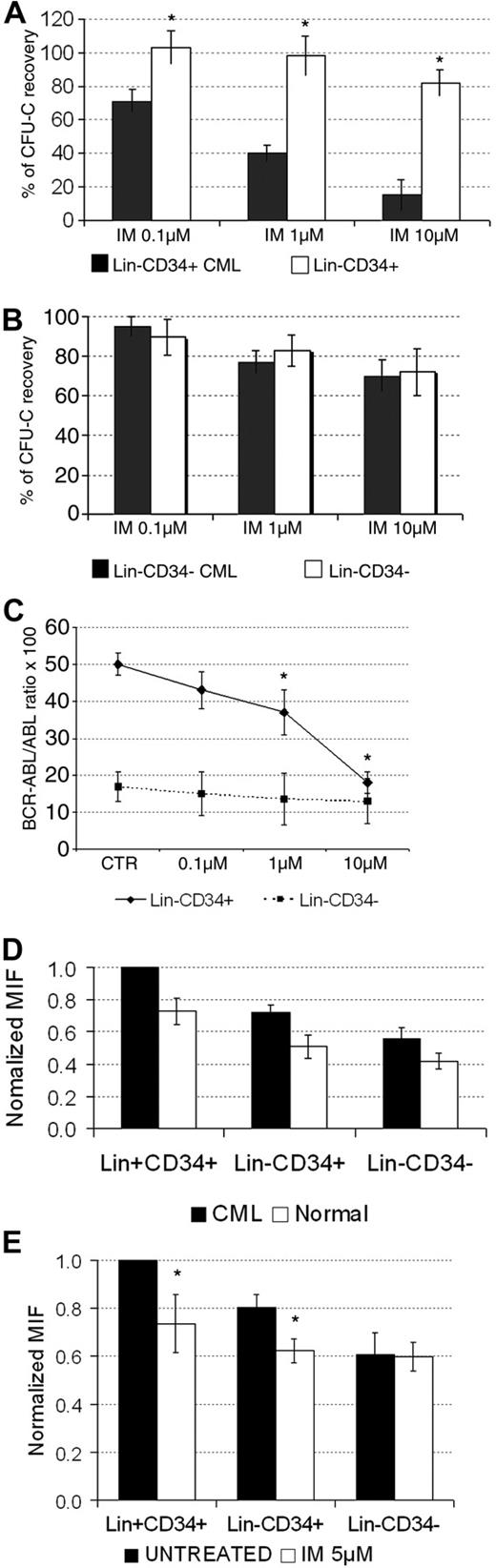
In the last set of experiments, we evaluated whether CML Lin−CD34− cells, showing distinct molecular, functional, and kinetic characteristics compared with Lin−CD34+ cells, have different sensitivity to IM. As shown in Figure 6A, clonogenic CML Lin−CD34+ cells were, as expected, significantly more sensitive to increasing concentrations of IM than normal cells. Conversely, there was no differential inhibitory activity of the drug between normal and leukemic Lin−CD34− cells (Figure 6B). Because CML CD34− cell population includes only 30% of leukemic cells, we performed a quantitative analysis of BCR-ABL transcript (Figure 6C). Our results demonstrated the dose-response effect of IM on CML Lin−CD34+, whereas leukemic Lin−CD34− survived in liquid culture after 48 hours of incubation with the highest dose tested of IM. Thus, Lin−CD34− LSCs appear to be rather insensitive to IM in vitro.
Analysis of the sensitivity of normal and CML Lin+CD34+, Lin−CD34+, and Lin−CD34− cells to IM treatment in vitro. Assessment of the clonogenic activity of normal and CML Lin−CD34+ (A) and Lin−CD34− (B) cells upon treatment with increasing concentrations (0.1μM, 1μM, and 10μM) of IM for 48 hours. The results are expressed as mean values ± SD from 4 independent experiments. The clonogenic efficiency of normal CD34+ and CD34− cells was 2.85% ± 0.7% and 0.27% ± 0.02%, respectively. The clonogenic efficiency of CML CD34+ and CD34− cells was 4.3% ± 1.3% and 0.68% ± 0.24%, respectively. Also in this set of experiments, the clonogenic efficiency of CML progenitor/stem cells was significantly higher (P < .05) than that of normal counterparts; *P < .05 in CML versus normal cells. Quantitative analysis of BCR-ABL transcript in CML Lin−CD34+ and Lin−CD34− upon treatment with increasing concentrations of IM for 48 hours (C). The results are expressed as mean values ± SD from 3 independent experiments. The number of cells analyzed for BCR-ABL transcript was always > 105. The range of ABL copies for CD34+ and CD34− cells was 1737-25 899 and 1000-38 868, respectively. Samples with less than 1000 ABL copies were not considered evaluable and were not analyzed; *P < .05 in untreated versus IM-treated cells. Results of flow cytometric detection of CrkL phosphorylation levels in normal (n = 5) and CML (n = 5) Lin+CD34+, Lin−CD34+ and Lin−CD34− cells (D). CrkL phosphorylation levels were normalized with respect to untreated CML Lin+CD34+ samples, set as MIF = 1. The results are expressed as mean values ± SD; *P < .05 in CML versus normal cells. (E) CML untreated control Lin+CD34+ cells PCrkL MIF levels were similarly set to 1 to compare CML subpopulations (n = 5 experiments) before and after the treatment with IM 5μM for 16 hours. The results are expressed as mean values ± SD; *P < .05 in untreated versus IM-treated cells. MIF indicates mean fluorescence intensity; IM, imatinib mesylate.
Analysis of the sensitivity of normal and CML Lin+CD34+, Lin−CD34+, and Lin−CD34− cells to IM treatment in vitro. Assessment of the clonogenic activity of normal and CML Lin−CD34+ (A) and Lin−CD34− (B) cells upon treatment with increasing concentrations (0.1μM, 1μM, and 10μM) of IM for 48 hours. The results are expressed as mean values ± SD from 4 independent experiments. The clonogenic efficiency of normal CD34+ and CD34− cells was 2.85% ± 0.7% and 0.27% ± 0.02%, respectively. The clonogenic efficiency of CML CD34+ and CD34− cells was 4.3% ± 1.3% and 0.68% ± 0.24%, respectively. Also in this set of experiments, the clonogenic efficiency of CML progenitor/stem cells was significantly higher (P < .05) than that of normal counterparts; *P < .05 in CML versus normal cells. Quantitative analysis of BCR-ABL transcript in CML Lin−CD34+ and Lin−CD34− upon treatment with increasing concentrations of IM for 48 hours (C). The results are expressed as mean values ± SD from 3 independent experiments. The number of cells analyzed for BCR-ABL transcript was always > 105. The range of ABL copies for CD34+ and CD34− cells was 1737-25 899 and 1000-38 868, respectively. Samples with less than 1000 ABL copies were not considered evaluable and were not analyzed; *P < .05 in untreated versus IM-treated cells. Results of flow cytometric detection of CrkL phosphorylation levels in normal (n = 5) and CML (n = 5) Lin+CD34+, Lin−CD34+ and Lin−CD34− cells (D). CrkL phosphorylation levels were normalized with respect to untreated CML Lin+CD34+ samples, set as MIF = 1. The results are expressed as mean values ± SD; *P < .05 in CML versus normal cells. (E) CML untreated control Lin+CD34+ cells PCrkL MIF levels were similarly set to 1 to compare CML subpopulations (n = 5 experiments) before and after the treatment with IM 5μM for 16 hours. The results are expressed as mean values ± SD; *P < .05 in untreated versus IM-treated cells. MIF indicates mean fluorescence intensity; IM, imatinib mesylate.
We then assessed the BCR-ABL kinase status in all stem cell populations, with and without IM, by measuring CrkL phosphorylation (p-CrkL) by flow cytometry. As expected, the baseline intracellular p-CrkL level was higher in all CML cells compared with their normal counterparts (Figure 6D). Perhaps due to normal cell contamination, p-CrkL was reduced in CML Lin−CD34− cells compared with Lin−CD34+ cells. Of note, there was a significant reduction in p-CrkL level in CD34+ cells in response to IM treatment, whereas the phosphorylation status of CrkL in CML Lin−CD34− cells was unchanged (Figure 6E). Finally, we assessed the sensitivity of LSC subsets to tyrosine kinase inhibitors in vivo.53 Therefore, we sorted stem cells, according to CD34 expression, from the BM of CML patients under treatment. As shown in Table 1, we faced 3 different types of response. The patient in hematological, but not molecular, remission (no. 1) showed readily detectable disease in all cell populations. Conversely, patients in persistent complete molecular remission (nos. 5-7) had no disease at any level. Notably, RQ-PCR for BCR-ABL transcript in patients in major molecular remission (nos. 2-4), demonstrated an undetectable level of the disease at the CD34+ cell level, whereas the CD34− cell compartment still harbored leukemic cells.
Molecular evaluation of residual disease in CML patients treated with BCR-ABL inhibitors
| Patient no. . | HR . | CgR . | MR . | BCR-ABL/ABL ratio × 100 . | |||
|---|---|---|---|---|---|---|---|
| BM . | PBL . | Lin−CD34+ cells . | Lin−CD34− cells . | ||||
| 1 | C | P | N | 0.23 | 0.15 | 0.28 | 0.3 |
| 2 | C | C | M | 0.01 | 0.01 | UD | 0.01 |
| 3 | C | C | M | 0.01 | 0.06 | UD | 0.039 |
| 4 | C | C | M | 0.02 | 0.08 | UD | 0.023 |
| 5 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
| 6 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
| 7 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
| Patient no. . | HR . | CgR . | MR . | BCR-ABL/ABL ratio × 100 . | |||
|---|---|---|---|---|---|---|---|
| BM . | PBL . | Lin−CD34+ cells . | Lin−CD34− cells . | ||||
| 1 | C | P | N | 0.23 | 0.15 | 0.28 | 0.3 |
| 2 | C | C | M | 0.01 | 0.01 | UD | 0.01 |
| 3 | C | C | M | 0.01 | 0.06 | UD | 0.039 |
| 4 | C | C | M | 0.02 | 0.08 | UD | 0.023 |
| 5 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
| 6 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
| 7 | C | C | C | UD (nested neg) | UD (nested neg) | UD | UD |
HR indicates hematologic response; CgR, cytogenetic response; MR, molecular response; BM, bone marrow; PBL, peripheral blood; C, complete; P, partial; M, major; N, no response; and UD, undetectable BCR-ABL transcript level.
Complete hematologic response: WBC count of less than 10 × 109/L, platelet count of less than 450 × 109/L, no immature cells (blasts, promyelocytes, or myelocytes) in the PB, and no signs/symptoms related to leukemia (including palpable splenomegaly). Complete cytogenetic response: 0% Ph-positive metaphases, based on results of conventional cytogenetic analysis. Partial cytogenetic response: 1% to 35% Ph-positive metaphases, based on results of conventional cytogenetic analysis. Major molecular response: BCR-ABL/ABL ratio less than 0.1% (international scale). Undetectable BCR-ABL transcript level: BCR-ABL/ABL less than 0.001% (international scale), corresponding to the lowest level of detectability of the method (10−4). Complete Molecular response: samples negative by RQ-PCR were analyzed by nested PCR and a complete molecular response was defined as nested PCR negativity.48 Samples showing less than 1000 copies of ABL transcript were not considered evaluable and are not included in the analysis (range of ABL copies for CD34+ cells was 2998-30 792; range for CD34− cells was 3425-18 693). The number of cells analyzed was (range) 118 000-860 000 and 130 000-1 100 000 for CD34+ and CD34−, respectively.
Discussion
Important advances have been recently made in understanding the molecular mechanisms involved in the neoplastic transformation of Ph-positive CML progenitors. To this end, several gene expression studies have been performed on CD34+ cells to identify the molecular signature of CML stem cells.9,22,24,54 However, no study has been reported so far in which a global molecular and functional analysis of the stem cell compartment of CML is shown.
In this investigation, the isolation of stem cell subsets based on the expression of the CD34 antigen allowed the identification and the molecular and functional characterization of Lin−CD34− Ph-positive, BCR-ABL-positive cells within CML hematopoiesis. Human leukemic Lin−CD34− cells, which represent approximately 30% of total Lin−CD34− cell fraction in CML samples, showed stem cell potential in 2 different xenotransplant animal model superimposable to that of purified subsets of CML cells expressing CD34 antigen.8 Similar to their normal counterparts, CML Lin−CD34− cells were kinetically quiescent and showed low engraftment rate in immunocompromized mice and low expression of self-renewal genes.4,5 However, induction of CD34 antigen in CML HSCs was associated with the recruitment into cell cycle.
Unsupervised clustering analysis demonstrated that leukemic Lin−CD34− cells are transcriptionally different from normal Lin−CD34− cells and more similar to CD34+ subpopulations, suggesting a more mature phenotype of CML Lin−CD34− cells compared with their normal counterparts.
Moreover, microarray data suggest that the contamination of residual normal stem cells does not mask the intrinsic BCR-ABL-dependent transcriptional profile of leukemic Lin−CD34− cells. Despite their modest overall hematopoietic potential, CML Lin−CD34− cells showed enhanced expression of cell cycle–related genes and an increased tendency to enter cell division as demonstrated by their higher clonogenic potential and LTC-IC content compared with normal quiescent counterparts (Lemoli et al4 and present data). Moreover, we found an increased capacity to repair DNA breaks of CML versus normal CD34+ cells. Taken together, these data, coupled with the notion that CD34− cell population includes only 30% of leukemic cells, support the view that, in CML, the expansion of leukemic versus normal hematopoiesis mainly derives from the proliferative advantage of BCR-ABL-positive CD34+ cells and from their resistance to apoptosis with increased cell survival at this stage of maturation.7,55,56
In addition to their increased proliferative potential, CML Lin−CD34− cells differed from normal counterparts for the expression of several genes correlated with tumor progression. Leukemic Lin−CD34− cells up-regulated several proangiogenic factors. Consistently, CML Lin−CD34− cells displayed a higher endothelial cell engraftment compared with leukemic Lin−CD34+ cells and normal HSCs.4 These findings support recent data showing BCR-ABL gene in mature endothelial cells as well as very primitive CD34− stem cells capable of hematopoietic and endothelial lineage differentiation in vitro and in vivo.57,58 Thus, in CML, leukemic transformation may take place in the putative hemangioblast cell.
Molecular and phenotypic analyses also demonstrated the significant down-regulation of MHC class I and II molecules on leukemic Lin−CD34− cells. Therefore, CML stem cells may have the reduced capacity to present foreign and self-antigens (including BCR-ABL–specific peptides) to T cells, and the deficit of antigen presentation may serve as a mechanism of immune evasion.
Furthermore, microarray data showed that several transcriptional regulators of self-renewal, such as Hox genes, Meis1, PBX2,50 GATA-2, Bmi1, and LMO2,44-46 were down-regulated in CML versus normal CD34+ cells. Consistently, the early myeloid commitment of leukemic Lin−CD34− cells was demonstrated by the increased expression of specific TFs and granules proteins. This finding may be clinically relevant as these molecules have been proposed as targets of immunotherapy in CML59 although the poor presenting capacity of Lin−CD34− leukemic cells may limit the efficacy of this approach.
Recent data demonstrated that CD34+ CML stem cells possess several features that significantly decrease their sensitivity to IM and other tyrosine kinase inhibitors.10-12 We extended this observation to leukemic Lin−CD34− cells, which did not show any preferential response to IM treatment in comparison with normal counterparts. This conclusion was supported by clonogenic assays, quantitative analysis of BCR-ABL transcripts, and assessment of p210 activity by evaluation of the phosphorylated CrkL substrate. Perhaps more importantly, molecular evaluation of residual disease in CML patients under treatment with BCR-ABL inhibitors demonstrated the survival of CD34−, but not of CD34+, LSCs in patients in major molecular response. Thus, Lin−CD34− CML cells can be seen as a stem cell subset partially insensitive to targeted therapy that may contribute to disease persistence under treatment. Interestingly, patients in persistent complete molecular remission did not show sign of disease at any level, suggesting that, in some individuals, CML may be fully eradicated by tyrosine kinase inhibitors. Novel therapeutic approaches to kill CD34−/CD34+ stem cells in CML may take advantage from their extensive molecular and functional characterization. In this view, the enhanced expression of angiogenic factors in leukemic Lin−CD34− cells may help designing more effective treatment for minimal residual disease in CML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by the Italian Ministry of Education, University and Scientific Research (MIUR), PRIN 2006 (contract no. 2006057308); the Italian Association for Cancer Research (AIRC); the Italian Association against Leukemia, section of Bologna (BolognAil), Italian National Institute of Health (ISS); and the Sixth EU Framework Program (Integrated Project “Angiotargeting”; contract no. 504743) in the area of “Life sciences, genomics and biotechnology for health” LeukemiaNet. E.B. was a recipient of an AIL fellowship and S.S. was a recipient of an FIRC fellowship.
Authorship
Contribution: R.M.L. designed the research, analyzed the data, and wrote the paper; V.S. and M.F. performed stem cell purification, in vitro clonogenic assays, liquid cultures, flow cytometric analysis, and analyzed the data; E.B. and S.S. performed pCRKL analysis and Comet assay, respectively; E.B., S.S., R.Z., and R.M. performed the microarray studies and analyzed the data; F.B., C.R., I.M.-P., and P.M. performed the xenogenic transplant experiments, molecular analyses on murine samples, and analyzed the data; M.A. and N.T. performed the molecular and cytogenetic analysis, respectively, of bcr-abl gene rearrangement; A.T. was involved in cell-cycle analysis; L.R. performed clonogenic and LTC-IC assays and analyzed the data; F.C. provided CML patient marrow, and blood samples and analyzed the data; G.M., M.B., and S.F. analyzed the data; and R.M. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: M.B. has received research grants and honoraria from and served as a speaker and advisory board member for Novartis, and has received honoraria as a speaker and advisory board member from Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Roberto M. Lemoli, MD, Institute of Hematology and Medical Oncology L & A Seràgnoli, Via Massarenti 9, 40138 Bologna, Italy; e-mail: roberto.lemoli@unibo.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal