Abstract
Follicular lymphoma (FL) is genetically characterized by the presence of the t(14;18)(q32;q21) chromosomal translocation in approximately 90% of cases. In contrast to FL carrying the t(14;18), their t(14;18)-negative counterparts are less well studied about their immunohistochemical, genetic, molecular, and clinical features. Within a previously published series of 184 FLs grades 1 to 3A with available gene expression data, we identified 17 FLs lacking the t(14;18). Comparative genomic hybridization and high-resolution single nucleotide polymorphism (SNP) array profiling showed that gains/amplifications of the BCL2 gene locus in 18q were restricted to the t(14;18)-positive FL subgroup. A comparison of gene expression profiles showed an enrichment of germinal center B cell–associated signatures in t(14;18)-positive FL, whereas activated B cell–like, NFκB, proliferation, and bystander cell signatures were enriched in t(14;18)-negative FL. These findings were confirmed by immunohistochemistry in an independent validation series of 84 FLs, in which 32% of t(14;18)-negative FLs showed weak or absent CD10 expression and 91% an increased Ki67 proliferation rate. Although overall survival did not differ between FL with and without t(14;18), our findings suggest distinct molecular features of t(14;18)-negative FL.
Introduction
Follicular lymphoma (FL) represents approximately 30% of all B-cell non-Hodgkin lymphomas and is generally characterized by an indolent clinical behavior with an overall median survival of 8 to 10 years.1,2 On the basis of its characteristic growth pattern with the formation of atypical follicular structures, immunophenotypic features showing frequent expression of CD10 and BCL6 and the presence of ongoing somatic hypermutation (SHM) of the immunoglobulin variable heavy chain genes (IgVH), FL is currently viewed as a germinal center–derived neoplasm.3 According to the number of centroblasts present in the neoplastic infiltrate, FL is subdivided into grades 1 to 3.2 Whereas FL grades 1 to 3A probably constitute a biologic continuum with an increasing number of intermingled centroblasts, FL grade 3B, which is composed of blasts exclusively, shows divergent immunophenotypic and genetic features4 that are more compatible with a follicular variant of diffuse large B-cell lymphoma (DLBCL). An intricate interaction between the neoplastic B cells and bystander cells in the microenvironment of the tumor infiltrate may be of particular relevance in the biology and the clinical course of FL. In particular, gene expression profiling studies have shown bystander cell signatures enriched for genes expressed in T-cell subsets, macrophages, and dendritic cells that are associated with length of survival or the clinical behavior of FL.5,6
The genetic hallmark of FL is the chromosomal translocation t(14;18)(q32;q21) that leads to deregulated expression of the antiapoptotic BCL2 protooncogene in the tumor cells, thus allowing for the acquisition of secondary chromosomal alterations in the germinal center environment where the most nonneoplastic B cells are physiologically destined to undergo apoptosis.7 However, the translocation t(14;18) is not present in all FL cases,8-10 and even with a highly sensitive fluorescence in situ hybridization (FISH) approach, rearrangements of BCL2 can only be detected in up to 90% of FLs.11 In contrast to FL carrying the t(14;18), FLs lacking a BCL2 rearrangement are less well characterized and their pathogenesis remains largely unclear. A subset of t(14;18)-negative FL appears to harbor genetic rearrangements of the BCL6 gene in 3q2710,12,13 or trisomy 3,14 whereas others show BCL2 expression on the immunohistochemical level despite the lack of the t(14;18).10 Moreover, increased expression of IRF4/MUM1, a protein associated with plasma cell differentiation,15 has been described in FL without BCL2 rearrangement.14,16 Cases of FL grade 3B, which may be biologically distinct from typical nodal FL grades 1 to 3A,4,17 were included in previous studies and may have confounded the reported results. This investigation, therefore, focuses on the clinical, genetic, and molecular characterization of t(14;18)-negative FLs within the spectrum of grades 1 to 3A. In a series of 184 well-characterized FL grades 1 to 3A that underwent gene expression profiling by the Leukemia and Lymphoma Molecular Profiling Project in a previous study,6 we wanted to determine the frequency of FLs without BCL2 rearrangement, to characterize their clinical features, and to study differences in gene expression, underlying genetic alterations, and the composition of the microenvironment in contrast to their t(14;18)-positive counterparts. We here show that t(14;18)-negative FLs belong to the biologic spectrum of “classic” FLs, but nevertheless they show distinct molecular features.
Methods
Follicular lymphoma specimens
Frozen as well as formalin-fixed and paraffin-embedded (FFPE) tumor tissues of 184 FL cases from our previous gene expression profiling study6 were available for the current investigation. These included 152 FL grades 1/2 and 32 FL grade 3A. As a validation set, 84 FL cases (80 FL grades 1/2, 4 FL grade 3A) were selected from the files of the Institute of Pathology, University of Würzburg. All FLs showed a predominantly follicular growth pattern and were classified according to the World Health Organization criteria.2 The entire study was approved by the Ethics Committee of the Medical Faculty, University of Würzburg.
Detection of BCL2 rearrangements
DNA extracted from frozen tissue of all 184 FL specimens was used in a polymerase chain reaction (PCR) to detect BCL2 rearrangements at both the major breakpoint region and the minor cluster region according to a standard protocol.18
In PCR-negative cases with available FFPE tissue, FISH was performed with break-apart probes for the BCL2 and BCL6 gene loci (Abbot). In the context of this study, cases with a BCL2 breakpoint are referred to as t(14;18)-positive FL, because translocation partners other than IgH are exceedingly rarely affected in FL.2,3 To evaluate FISH assays, the signal constellation in 200 randomly selected cells was analyzed with a Zeiss Axioskop2 microscope with a cut-off of 8% for both the BCL2 and BCL6 break-apart probes as determined by FISH experiments in reactive tissues.
Comparative genomic hybridization analysis
Conventional comparative genomic hybridization (CGH) was performed according to a standard protocol19 in 184 FLs with DNA extracted from the same frozen tissue specimens that were used for previous gene expression profiling experiments.6 For graphical representation we used the imaging tools provided by the SKY/M-FISH and CGH database from which complete CGH data are available (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/sky/).
Immunohistochemistry
Immunostaining was performed on FFPE tissues according to standard protocols. Staining for BCL2 (Clone 124; 1:400; DAKO) was applied to all 184 FL cases as well as the 84 FL cases from the validation series. A broad panel of antibodies, including CD10 (NCL-CD10 270; 1:100; Novocastra), Ki67 (MIB-1; 1:800; DAKO), IRF8 (polyclonal; 1:200; Santa Cruz Biotechnology), IRF4/MUM1 (MUM-1p; 1:800; DAKO), GRZMB (GrB-7; 1:80; Monosan), FOXP3 (1:50; kindly provided by Dr Giovanna Roncador, Madrid, Spain), CD57 (1:800; BD Biosciences), and BCL6 (Clone pG/B6p; 1:20; DAKO) was stained in the validation series of 84 FL cases with the BCL6 staining being also applied to a subset of the 184 initial FL specimens.
Statistical evaluation
To compare the gene expression profiles of FL cases with and without the presence of the t(14;18) within the initial set of 184 cases,6 a 2-sided t test and gene set enrichment analysis (GSEA; http://www.broad.mit.edu/gsea) was performed as described,20,21 using 81 previously published lymphoma-associated gene expression signatures22 (http://lymphochip.nih.gov/signaturedb) as well as 836 regulatory motif gene sets (c3) from the Molecular Signatures Database.21 If the nominal P value and the tail-area false discovery rate (tail-FDR) were .05 and 0.25 or less, respectively, the corresponding gene set was assessed as significantly enriched. A GSEA integrated leading edge analysis was performed with significantly enriched gene sets to extract genes that account for the enrichment score.
To correlate gene expression data and CGH results, chromosomal regions were considered that showed alterations in at least 5 FL cases, and cases were coded as normal or altered. Probe sets from the HG-U133A and U133B gene expression arrays (Affymetrix) and positional gene sets (c1) from the Molecular Signatures Database, mapping to these regions, were selected and tested for an association by a t test and GSEA approach. To account for multiple comparisons, local FDR (for the t test) and tail-FDR (for GSEA) were calculated for significant P values, and those with a local FDR less or equal to .01 or a tail-FDR less or equal to 0.25 were considered truly statistically significant. The tail-FDR provides an estimate of the probability that a gene set or signature with a given normalized enrichment score represents a false-positive finding.23 According to the results of the t test, chromosomal regions that showed an excess of probes significantly associated with gene expression were determined with the use of a Poisson model. Annotations to gene families were accomplished with the use of the Broad Institute annotation platform (http://www.broad.mit.edu/gsea/msigdb/annotate.jsp). For survival analysis, the Statistical Package for the Social Science software (Version 15.0; SPSS Inc) was used. Specifically, to evaluate survival differences between t(14;18)-positive and t(14;18)-negative tumors, Kaplan-Meier analysis was performed, and survival curves were compared by the log-rank test. Frequencies of various clinical parameters were compared with the Fisher exact test. P values less than .05 were considered significant.
High-density single nucleotide polymorphism array analysis
Single nucleotide polymorphism (SNP) array studies were performed in 11 t(14;18)-negative FLs with the 250k NSP array (Affymetrix) according to the manufacturer's instructions. Data files were generated with the Gene Chip Operating Software and the Gene Chip Genotyping Analysis Software (Affymetrix) with the use of the Dynamic Modeling algorithm (threshold, 0.33). Unpaired analysis with an independent reference set of 16 laboratory-internal controls and 15 controls provided by Affymetrix/Hapmap project (www.affymetrix.com/www.hapmap.org) was performed. DNA copy number was analyzed with the use of the copy number analysis tool (CNATv4.0; Affymetrix) and the copy number analyzer for gene chip (CNAGv3.0) applying the AsCNAR algorithm (http://www.genome.umin.jp/). Only alterations consisting of more than 20 consecutive SNPs were counted.
Clonality (Genescan) analysis and analysis of ongoing SHM
Genescan analysis and analysis of ongoing SHM was performed on selected FL cases (for details see supplemental information, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
Study cohorts
The previously published gene expression dataset of 184 FLs6 provided the basis for the current study. By CGH, 180 of 184 FL cases could be successfully hybridized, of which 127 FLs showed detectable alterations.
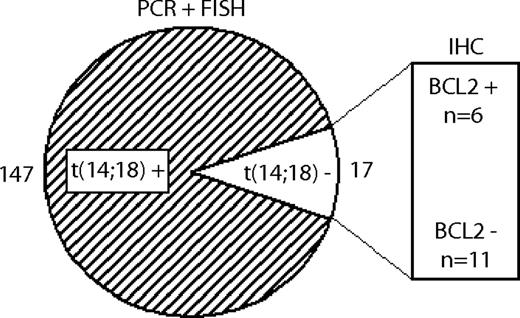
PCR analysis to detect BCL2 rearrangements was performed in all 184 FL cases and showed clonal bands in 90 cases. Of the remaining 94 FLs, 74 had FFPE tissue samples available that were subjected to FISH analysis, resulting in 57 samples with t(14;18) and 17 samples without detectable t(14;18). Thus, information on the t(14;18) status was available in 164 FLs [147 FLs with and 17 FLs without t(14;18)]. All 17 t(14;18)-negative cases and 17 randomly selected t(14;18)-positive cases were investigated by Genescan analysis, and the SHM status was evaluated in a subset of cases (supplemental information). SNP array analysis could be performed in 11 FLs without t(14;18). The study set with both CGH data and information on the t(14;18) status comprised 102 FL cases with and 10 cases without t(14;18).
The validation set for immunohistochemistry (IHC) experiments consisted of 84 additional, preselected FLs [42 FLs with and 42 FLs without t(14;18)]).
Chromosomal alterations in FLs detected by CGH
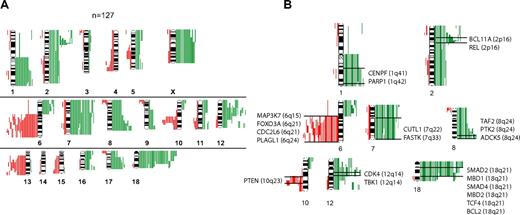
Chromosomal gains or losses or both in 127 FLs are summarized in Figure 1A. Chromosomal gains were frequently found in 1q, 2p, 7, 8q, 12q, 18q, and X, whereas chromosomal losses occurred most frequently in 6q, 10q, and 13q. These results are well in line with published reports.8,9 When correlating the presence of minimally altered chromosomal regions (MCRs) with overall survival of patients with FL, the presence of 18q21 amplifications correlated with inferior outcome. Only 4 tumors, however, carried this alteration, but all 4 patients died in fewer than 5 years (P = .002).
Comparative genomic hybridization in follicular lymphoma. (A) Chromosomal gains and losses in 127 FL cases showing altered karyotypes by comparative genomic hybridization (CGH). Gains are displayed in green bars and losses are displayed in red bars. (B) Selected genes that show up-regulation in follicular lymphoma (FL) with chromosomal gains in respective regions (1q, 2p, 7q, 8q, 12q and 18q) or down-regulation in FL with chromosomal losses in respective regions (6q, 10q).
Comparative genomic hybridization in follicular lymphoma. (A) Chromosomal gains and losses in 127 FL cases showing altered karyotypes by comparative genomic hybridization (CGH). Gains are displayed in green bars and losses are displayed in red bars. (B) Selected genes that show up-regulation in follicular lymphoma (FL) with chromosomal gains in respective regions (1q, 2p, 7q, 8q, 12q and 18q) or down-regulation in FL with chromosomal losses in respective regions (6q, 10q).
Correlation of genetic alterations with gene expression
The analysis of associations between distinct chromosomal alterations and the expression levels of genes localized in each of these regions (supplemental Table 1) showed a total of 2465 probe sets that were significantly altered in gene expression, according to the t test and a local FDR of less than 0.01 (supplemental Table 2). Of those, more than 900 probe sets were significantly expressed at P values less than .001 when a Bonferroni correction was applied. With few exceptions, chromosomal gains were associated with increased expression, and chromosomal losses were associated with decreased expression of genes localized in these regions. The probe sets showed by the t test approach could be assigned to 195 altered chromosomal bands, 41 of which showed a significant excess of association according to a Poisson model (supplemental Table 3). Genes that showed significantly altered gene expression levels were assigned to the gene families “oncogenes,” “tumor suppressor genes,” “transcription factors,” “translocated genes,” “cytokines,” and “kinases.” Some of these annotated genes in the chromosomal regions 1q, 2p, 6q, 7q, 8q, 10q, 12q, and 18q are displayed in Figure 1B (for complete data, see supplemental Table 4A-B; supplemental Figure 1A-M). Notably, we obtained similar results with GSEA, thus validating the findings by a different mathematical approach (data not shown). Next, we analyzed whether the presence of any of the MCRs was associated with the expression level of the 2 prognostically relevant bystander signatures in FL, the immune response 1 (IR1) and immune response 2 (IR2) signatures.6 Interestingly, each MCR was associated with a decreased expression signature of IR1 with almost half of these associations having a P value less than .05, whereas no statistical correlations could be detected between the gene expression level of IR2 and any of the MCRs.
Definition of FL subgroups according to the presence of the t(14;18) and BCL2 protein expression
As detailed in the first paragraph of “Results,” of 164 FLs with available information 147 cases carried the t(14;18), whereas it was lacking in 17 cases. Among the 17 t(14;18)-negative FLs, 11 cases were negative for BCL2 expression on the immunohistochemical level, whereas 6 cases were positive (Figure 2). The incidence of the t(14;18) did not differ between the FL grades 1/2 and 3A (90% vs 86%, respectively).
Definition of FL subgroups with and without translocation t(14;18). 147 FLs showed evidence of the t(14;18) by PCR or FISH techniques, whereas 17 FLs were t(14;18)-negative. Within the t(14;18)-negative subgroup, 11 FLs were negative for BCL2 on the protein level, and 6 were positive, as determined by IHC.
Definition of FL subgroups with and without translocation t(14;18). 147 FLs showed evidence of the t(14;18) by PCR or FISH techniques, whereas 17 FLs were t(14;18)-negative. Within the t(14;18)-negative subgroup, 11 FLs were negative for BCL2 on the protein level, and 6 were positive, as determined by IHC.
Clonal IgVH rearrangements and ongoing SHMs were detected in all cases tested (supplemental information).
FLs with and without t(14;18) differ in the presence of 18q11- q21 gains or amplifications
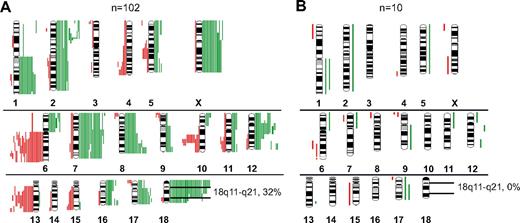
By CGH analysis, various genetic alterations were present at different frequencies between t(14;18)-positive and t(14;18)-negative FLs. Gains in chromosome 7 (19.5%), 8q (16%), X (13.5%) and losses in 13q (14%) and 10q (12.5%) were only encountered in t(14;18)-positive FLs (Figure 3A-B). Notably, gains or amplifications of 18q11-q21 occurred in 32% of t(14;18)-positive FLs but were not detectable in t(14;18)-negative FLs (P = .032). No alteration was solely restricted to t(14;18)-negative FLs (Figure 3B). By CGH, a higher percentage of FL cases with t(14;18) showed genetic alterations compared with FLs without t(14;18) (70% vs 47%; P = .1). To test whether rearrangements of the BCL6 gene occurred more frequently in t(14;18)-negative FLs, as reported previously,4,10,12,13 we applied a BCL6 break-apart probe to 15 t(14;18)-negative and 39 t(14;18)-positive FLs. The frequency of BCL6 rearrangements, however, did not differ significantly between the 2 groups (18% vs 27%; P = .475; data not shown).
Chromosomal gains and losses in FLs with and without translocation t(14;18) detected by CGH. (A) Gains (green bars) and losses (red bars) in t(14;18)-positive FL. (B) Gains and losses in t(14;18)-negative FL.
Chromosomal gains and losses in FLs with and without translocation t(14;18) detected by CGH. (A) Gains (green bars) and losses (red bars) in t(14;18)-positive FL. (B) Gains and losses in t(14;18)-negative FL.
High-density SNP array analysis of t(14;18)-negative FLs
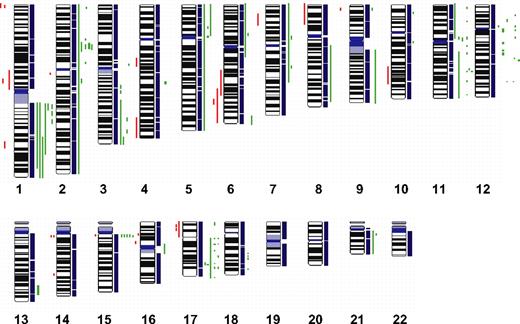
To define chromosomal gains and losses of t(14;18)-negative FLs at higher resolution, we studied 11 cases by SNP arrays. The results that are summarized in Figure 4 confirmed the alterations previously identified by CGH with only a few minor exceptions but also revealed additional alterations. Strikingly, 4 t(14;18)-negative FLs (36%) carried small gains or amplifications in the chromosomal region 2p16, including the BCL11A and REL loci that were not evident in the conventional CGH analysis. Likewise, additional gains or amplifications in 3q, 8q, 12q, and 17q were detected in single cases. Gains or amplifications in 18q11-q21 that were present in 32% of t(14;18)-positive FLs, but not in t(14;18)-negative FLs by conventional CGH, could be uncovered in one single t(14;18)-negative FL by SNP array analysis (Figure 4); interestingly, this gain did not include the BCL2 locus. However, no alterations that were specific for the t(14;18)-negative FL subgroup could be detected with this high-resolution approach.
High-density SNP array profiling in t(14;18)-negative FL. Copy number gains (green bars) and losses (red bars) in 11 t(14;18)-negative FLs determined by high-resolution 250K SNP array analysis.
High-density SNP array profiling in t(14;18)-negative FL. Copy number gains (green bars) and losses (red bars) in 11 t(14;18)-negative FLs determined by high-resolution 250K SNP array analysis.
FLs with and without t(14;18) differ in gene expression profiles
The comparison of gene expression profiles between FLs with and without t(14;18) showed 1562 differentially expressed probe sets between the 2 groups with the use of a 2-sided t test (P < .001). As expected, BCL2 was the most differentially expressed gene (P < .001), with a higher expression in t(14;18)-positive cases. Subsequent GSEA analysis displayed major differences between the 2 groups. In particular, germinal center B-cell (GCB)–associated signatures were enriched in the t(14;18)-positive subgroup, whereas activated B-cell (ABC)–like signatures were enriched among t(14;18)-negative FLs, as were NFκB-, post-GCB–, T-cell–, cell cycle–, proliferation-, and interferon-associated signatures (Table 134-44 ). Importantly, the IR1 signature that had been previously associated with improved survival times in FL was significantly enriched among t(14;18)-negative FLs, whereas the IR2 signature was not significantly enriched in one of the subgroups.
Gene set enrichment analysis (GSEA) with 81 lymphoma-associated signatures in 147 t(14;18)-positive and 17 t(14;18)-negative FL cases
| Signatures . | Enriched in t(14;18)+ . | Enriched in t(14;18)− . | P . | FDR q value . |
|---|---|---|---|---|
| GCB | .01 | < 0.1 | ||
| Rosenwald et al, 2002, ref. 32 | Yes | No | ||
| Dave et al, 2006, ref. 34 | Yes | No | ||
| ABC | .02 | < 0.1 | ||
| Wright et al, 2003, ref. 35 | No | Yes | ||
| NFKB | ≤ .02 | ≤ 0.1 | ||
| Lam et al, 2005, ref. 36 | No | Yes | ||
| Post-GCB | .02 | < 0.1 | ||
| Shaffer et al, 2002, ref. 37 | No | Yes | ||
| Weller et al, 2004, ref. 38 | No | Yes | ||
| Wright et al, 2003, ref. 35 | No | Yes | ||
| IR1 | .03 | < 0.1 | ||
| Dave et al, 2004, ref. 6 | No | Yes | ||
| T cell | < .01 | 0.14 | ||
| McHugh et al, 2002, ref. 39 | No | Yes | ||
| Kovanen et al, 2003, ref. 40 | No | Yes | ||
| Cell cycle | < .01 | 0.2 | ||
| Shaffer et al, 2001, ref. 41 | No | Yes | ||
| Proliferation | ≤ .02 | ≤ 0.1 | ||
| Su et al, 2004, ref. 42 | No | Yes | ||
| Rosenwald et al, 2003, ref. 43 | No | Yes | ||
| Interferon | < .01 | < 0.1 | ||
| Baechler et al, 2003, ref. 44 | No | Yes |
| Signatures . | Enriched in t(14;18)+ . | Enriched in t(14;18)− . | P . | FDR q value . |
|---|---|---|---|---|
| GCB | .01 | < 0.1 | ||
| Rosenwald et al, 2002, ref. 32 | Yes | No | ||
| Dave et al, 2006, ref. 34 | Yes | No | ||
| ABC | .02 | < 0.1 | ||
| Wright et al, 2003, ref. 35 | No | Yes | ||
| NFKB | ≤ .02 | ≤ 0.1 | ||
| Lam et al, 2005, ref. 36 | No | Yes | ||
| Post-GCB | .02 | < 0.1 | ||
| Shaffer et al, 2002, ref. 37 | No | Yes | ||
| Weller et al, 2004, ref. 38 | No | Yes | ||
| Wright et al, 2003, ref. 35 | No | Yes | ||
| IR1 | .03 | < 0.1 | ||
| Dave et al, 2004, ref. 6 | No | Yes | ||
| T cell | < .01 | 0.14 | ||
| McHugh et al, 2002, ref. 39 | No | Yes | ||
| Kovanen et al, 2003, ref. 40 | No | Yes | ||
| Cell cycle | < .01 | 0.2 | ||
| Shaffer et al, 2001, ref. 41 | No | Yes | ||
| Proliferation | ≤ .02 | ≤ 0.1 | ||
| Su et al, 2004, ref. 42 | No | Yes | ||
| Rosenwald et al, 2003, ref. 43 | No | Yes | ||
| Interferon | < .01 | < 0.1 | ||
| Baechler et al, 2003, ref. 44 | No | Yes |
FDR indicates false discovery rate; GCB, germinal center B cell; and ABC, activated B cell.
These findings suggest biologic differences between the 2 subgroups that may be related to the stage of differentiation of the neoplastic B cells, oncogenic pathways that are operative and the composition of the microenvironment in these tumors. In agreement with these findings, PAX5 regulatory motifs were enriched in t(14;18)-positive FLs and MYC/MAX and NFκB regulatory motifs were enriched in the t(14;18)-negative subgroup (supplemental Table 6).
Finally, we also compared gene expression profiles between 147 FLs carrying the t(14;18) and 6 FLs that were t(14;18) negative, but showed BCL2 expression by immunohistochemistry. Interestingly, GCB-associated signatures remained enriched among t(14;18)-positive FLs, whereas ABC-, NFκB-, and proliferation-associated signatures were enriched with FLs lacking the t(14;18). This result suggests that the expression of BCL2 in t(14;18)-negative FLs may not be sufficient to alter the gene expression phenotype toward the profile of t(14;18)-positive FLs (data not shown).
Differences in clinical parameters between FLs with and without t(14;18)
The major clinical variables of age, Eastern Cooperative Oncology Group performance status, sex, tumor grade, stage, extranodal sites, B symptoms, and lactate dehydrogenase levels within FL cohorts with and without t(14;18) are provided in supplemental Table 7. Patients with t(14;18)-negative FL had more frequently lower stage (62% vs 27% in t(14;18)-positive FL; P = .008), whereas no differences were observed between the 2 groups in the other clinical variables and overall survival (supplemental Table 7; supplemental Figure 2).
Immunohistochemical validation of gene expression data in an independent cohort of FLs with and without t(14;18)
In an attempt to validate findings derived from the comparison of gene expression profiles between t(14;18)-positive and t(14;18)-negative FLs we performed IHC in an independent series of 84 FLs [42 FL with and 42 FL without t(14;18)]. These cases were stained for the germinal center–associated markers CD10, BCL6, and IRF8, as well as for IRF4/MUM1, a marker associated with the postgerminal center stage of B-cell differentiation as well as a target of the NFκB pathway. Because GSEA analysis had indicated differences in the proliferative activity of the tumor cells between both subgroups, Ki67 staining was performed. GRZMB, a marker of cytotoxic cells, was one of the most differentially expressed genes in the comparison of gene expression profiles and was therefore included in the panel of antibodies. Finally, FOXP3, a marker of T-regulatory cells, and CD57 that is expressed in follicular T-cells were selected, based on the well-established biologic and prognostic relevance of the nonmalignant microenvironment in FL tumor samples.
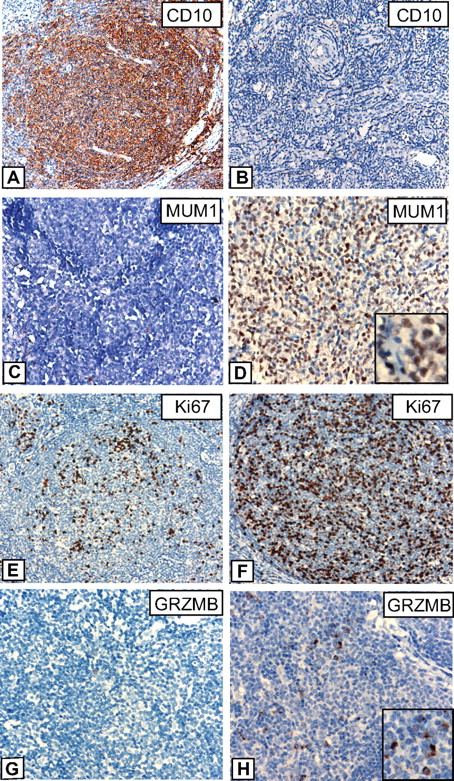
IHC results are summarized in Table 2, and examples of various stainings are provided in Figure 5. Striking differences were observed for CD10; although all FLs with the t(14;18) were strongly positive for this marker, CD10 expression was lacking or only weakly present in 32% of FLs without t(14;18), which is in line with the results of gene expression profiling that showed an enrichment of GCB-associated genes in FLs with t(14;18). Interestingly, low or absent CD10 expression was restricted to t(14;18)-negative FLs without BCL2 expression, whereas t(14;18)-negative FLs that, nevertheless expressed BCL2 on the protein level, were all strongly CD10-positive (data not shown). By contrast, IRF4/MUM1 expression, Ki67 labeling, and expression of GRZMB were higher in t(14;18)-negative FLs in agreement with observed differences in RNA expression levels as measured by microarray analysis. The data discussed in this study have been deposited in Gene Expression Omnibus from the National Center for Biotechnology Information (NCBI) and are accessible through GEO Series accession no. GSE16131.
Immunohistochemical (IHC) results in a validation set of 84 FLs, including 42 t(14;18)-positive and 42 t(14;18)-negative FL
| Antibodies (IHC) . | t(14;18)+ . | t(14;18)− . | P . |
|---|---|---|---|
| CD10 | < .01 | ||
| Negative | 0/42 (0) | 12/38 (31.6) | |
| Positive | 42/42 (100) | 26/38 (68.4) | |
| BCL6 | — | ||
| Positive | 40/40 (100) | 38/38 (100) | |
| IRF4/MUM1 | .039 | ||
| No greater than 10% | 42/42 (100) | 31/35 (88.6) | |
| Greater than 10% | 0/0 (0) | 4/35 (11.4) | |
| IRF8 | NS | ||
| Negative | 3/39 (7.7) | 1/38 (2.6) | |
| Positive | 36/39 (92.3) | 37/38 (97.4) | |
| Ki67 | < .01 | ||
| No greater than 25% | 17/41 (41.5) | 3/33 (9.1) | |
| Greater than 25% | 24/41 (58.5) | 30/33 (90.9) | |
| GRZMB | < .01 | ||
| No greater than 3% | 29/36 (80.6) | 16/35 (45.7) | |
| Greater than 3% | 7/36 (19.4) | 19/35 (54.3) | |
| FOXP3 | NS | ||
| No greater than 5% | 19/40 (47.5) | 11/36 (30.55) | |
| Greater than 5% | 21/40 (52.5) | 25/36 (69.4) | |
| CD57 | NS | ||
| No greater than 15% | 33/40 (82.5) | 35/37 (94.6) | |
| Greater than 15% | 7/40 (17.5) | 2/37 (5.4) |
| Antibodies (IHC) . | t(14;18)+ . | t(14;18)− . | P . |
|---|---|---|---|
| CD10 | < .01 | ||
| Negative | 0/42 (0) | 12/38 (31.6) | |
| Positive | 42/42 (100) | 26/38 (68.4) | |
| BCL6 | — | ||
| Positive | 40/40 (100) | 38/38 (100) | |
| IRF4/MUM1 | .039 | ||
| No greater than 10% | 42/42 (100) | 31/35 (88.6) | |
| Greater than 10% | 0/0 (0) | 4/35 (11.4) | |
| IRF8 | NS | ||
| Negative | 3/39 (7.7) | 1/38 (2.6) | |
| Positive | 36/39 (92.3) | 37/38 (97.4) | |
| Ki67 | < .01 | ||
| No greater than 25% | 17/41 (41.5) | 3/33 (9.1) | |
| Greater than 25% | 24/41 (58.5) | 30/33 (90.9) | |
| GRZMB | < .01 | ||
| No greater than 3% | 29/36 (80.6) | 16/35 (45.7) | |
| Greater than 3% | 7/36 (19.4) | 19/35 (54.3) | |
| FOXP3 | NS | ||
| No greater than 5% | 19/40 (47.5) | 11/36 (30.55) | |
| Greater than 5% | 21/40 (52.5) | 25/36 (69.4) | |
| CD57 | NS | ||
| No greater than 15% | 33/40 (82.5) | 35/37 (94.6) | |
| Greater than 15% | 7/40 (17.5) | 2/37 (5.4) |
Values are n (%).
NS indicates not significant.
Immunohistochemistry in FLs with and without t(14;18). Representative stainings for CD10, IRF4/MUM1, Ki67, and Granzyme B (GRZMB) in FL grade 1/2 cases with t(14;18) (A, C, E, G) and without t(14;18) (B, D, F, H). (A-B) Images were captured at magnification ×200, and (C-H) at magnification ×400 with the use of an Olympus, Color View, BX50 microscope, the Color View digital camera, and the analysis work soft imaging system (all Olympus).
Immunohistochemistry in FLs with and without t(14;18). Representative stainings for CD10, IRF4/MUM1, Ki67, and Granzyme B (GRZMB) in FL grade 1/2 cases with t(14;18) (A, C, E, G) and without t(14;18) (B, D, F, H). (A-B) Images were captured at magnification ×200, and (C-H) at magnification ×400 with the use of an Olympus, Color View, BX50 microscope, the Color View digital camera, and the analysis work soft imaging system (all Olympus).
Discussion
FL is generally characterized by the presence of the t(14;18) chromosomal translocation, resulting in overexpression of the BCL2 protein. Approximately 90% of FLs carry this hallmark alteration, whereas, in the remainder, BCL2 rearrangements are not detectable, even with highly sensitive techniques.11 In our study, 17 of 164 FLs grades 1 to 3A lacked the t(14;18), confirming the previously reported frequency of this particular subgroup among all FLs. The characterization of t(14;18)-negative FLs by gene expression profiling, CGH, and SNP array analysis and by IHC showed molecular features that are distinct from t(14;18)-positive FLs, but nevertheless show that t(14;18)-negative FLs belong to the spectrum of classic FL.
Our CGH analysis in the whole series of 184 FLs identified recurrent gains in the chromosomal regions 1q, 2p, 12q, and 18q, as well as losses in 6q, 10q, and 13q, at similar frequencies as previously reported,9,24 indicating that the present series includes a representative spectrum of genetic alterations typically observed in FL. We specifically excluded FL grade 3B cases from our investigation, based on the accumulating evidence that these tumors may be molecularly distinct from FL grades 1 to 3A.4,25
Comparing the genetic alterations in the t(14;18)-positive and t(14;18)-negative FL subsets, 18q gains or amplifications were surprisingly restricted to the t(14;18)-positive FL subset. This result is in contrast to a previous study in which gains in 18q were frequently observed in t(14;18)-negative FL.10 It is unlikely that this discrepancy is due to methodologic issues, because the CGH results in our study were confirmed by FISH analysis in which the break-apart probe approach to detect rearrangements of the BCL2 locus would usually result in additional fluorescence signals in case of the presence of more than 2 copies of the BCL2 locus. Moreover, high-resolution SNP array analysis in 11 t(14;18)-negative FL cases also confirmed the CGH results with the exception of 1 single case, in which a small gain in 18q was discovered that, however, did not encompass the BCL2 locus. In summary, 18q gains or amplifications, including the BCL2 locus, were not detected in our series of t(14;18)-negative FLs. Given that the BCL2 protein is nevertheless expressed in a subset of t(14;18)-negative FL (6 of 17 cases), we conclude that mechanisms other than an increase of the BCL2 gene dosage account for the BCL2 protein expression in t(14;18)-negative FL, for example, an overexpression of transcriptional regulatory elements or the modification of the corresponding binding sites by histone acetylation.26,27 We were also unable to confirm the frequent occurrence of trisomy 3 and a gain or amplification at 3q2714,28 in t(14;18)-negative FLs in our series. This discrepancy, however, might be explained by the exclusion of FL grade 3B cases in our series, or by a potential difference in the genetic constitution of FL cases in Asia and Western countries. Likewise, t(14;18)-negative FL in our series did not show an increased load of genetic alterations compared with t(14;18)-positive FL (data not shown), in contrast to a recent study by Nanjangud et al.29 Again, that study included FL grade 3B cases that frequently lack the t(14;18) and show a high karyotypic complexity.4,10,25
The comparison of gene expression profiles between t(14;18)-positive and t(14;18)-negative FLs in our series showed signatures that point to subtle differences in the developmental stages of the neoplastic B cells, the usage of divergent oncogenic pathways, and the composition of the microenvironment. Specifically, GCB-cell signatures were enriched in t(14;18)-positive FL, whereas ABC-like and post-GCB signatures were found to be overexpressed in t(14;18)-negative FL. Although these results initially suggested that t(14;18)-positive FL may correspond to the germinal center stage of B-cell differentiation and the t(14;18)-negative FL to the postgerminal center stage, in analogy to the subdivision of DLBCL into GCB and ABC subtypes, the findings of the IgVH gene mutational analysis argues against this hypothesis. Notably, all 5 tested t(14;18)-negative FLs showed evidence of ongoing SHM, a feature of GCB cells. We therefore suggest that a subset of t(14;18)-negative FL may show the phenotype of a late GCB cell that has not yet exited the germinal center stage of differentiation. Support for this idea comes from the IHC analysis in a validation series of 84 FLs. Although all FLs carrying the t(14;18) showed strong CD10 expression in the tumor cells, one-third of t(14;18)-negative FLs had weak or no CD10 expression at all (P < .01). Of interest, CD10 was weak or absent only in FLs that lacked both the t(14;18) and BCL2 expression at the protein level, whereas CD10 expression was strong in all BCL2-expressing FLs without the t(14;18). In line with the results for CD10 expression, IRF4/MUM1, a marker of late or post-GCB differentiation, was expressed in 4 t(14;18)-negative FLs, providing another piece of evidence of a late GCB stage of differentiation of some t(14;18)-negative FLs.
GSEA analysis in the expression data of the t(14;18)-positive and t(14;18)-negative FL subsets also identified proliferation-associated signatures as differentially expressed. Specifically, these signatures were more highly expressed among t(14;18)-negative FLs, a finding that could be confirmed by Ki67 immunohistochemistry in our independent validation series. Ninety-one percent of t(14;18)-negative FLs showed Ki67 labeling in greater than 25% of the tumor cells, whereas only 59% of t(14;18)-positive FLs reached this level of proliferative activity. This difference is statistically highly significant (P < .01), and it is important to point out that the distribution of the FL grades 1/2 and 3A was equal between the groups. More precisely, our validation set contained only 4 cases of FL grade 3A, 2 of which carried the t(14;18). Considering the known correlation between increased Ki67 labeling and a higher grade of FL cases, a bias in the composition of the 2 FL subgroups in our validation cannot account for the observed differences in the proliferation rate. We, therefore, conclude that an increased proliferative activity is an inherent biologic feature of t(14;18)-negative FL. Because FL grade 3B cases had been excluded from our study and because only a few cases showed expression of IRF4/MUM1, we did not observe a significant association between increased Ki67 staining and a higher level of IRF4/MUM1 expression, as reported previously.30
Gene expression signatures also point to an increased level of NFκB activity within the subset of t(14;18)-negative FL cases, as evidenced by increased expression levels of well-known NFκB target genes. In addition, the composition of the microenvironment, an important biologic and prognostic feature in FL,6 appears to differ between the subgroups, as evidenced by an enrichment of the IR1 and T-cell signatures in t(14;18)-negative FL. Moreover, GRZMB, a cytotoxic molecule, was more highly expressed in the t(14;18)-negative FL subgroup, and this finding could be validated immunohistochemically, showing an increased number of cytotoxic cells expressing GRZMB in t(14;18)-negative FL.
Conceptually, the question arises whether lymphomas lacking the translocation t(14;18) as well as BCL2 and occasionally also CD10 expression by immunohistochemistry, represent true FL rather than, for example, misclassified marginal zone B-cell lymphomas (MZLs) with marked follicular colonization or, simply, reactive follicular hyperplasias. The following features strongly suggest that t(14;18)-negative FLs belong to the spectrum of true FL. First, from a morphologic point of view, the t(14;18)-negative FL in our series showed classic features of FL with a predominance of centrocytes intermingled with few, transformed blasts. A prominent proliferation in the marginal zones, monocytoid B cells, and/or evidence of follicular colonization was not obvious in our cases. Second, by immunohistochemistry, t(14;18)-negative FL strongly expressed the germinal center-associated markers BCL6 (in all cases) and IRF8 (in all but one case) as well as CD10 in 68% of the cases, which would be unusual features in MZL.31 Third, genetically, CGH and SNP array analyses uncovered gains and amplifications of the REL locus in 2p16 in 5 of 17 t(14;18)-negative FL cases. This alteration is frequently present in t(14;18)-positive FL and other germinal center–derived B-cell lymphomas, for example, the GCB subtype of DLBCL.32 Moreover, characteristic genetic alterations present in MZL, such as trisomies 3, 7, and 18, were not encountered in our cases. Fourth, Genescan analysis showed clonally rearranged immunoglobulin genes in all 17 t(14;18)-negative FL cases, strongly arguing against a reactive condition. Finally, gene expression–based algorithms for the classification of lymphomas assigned all t(14;18)-negative FL cases in our series to the category of FL and not to the categories of reactive hyperplasia or marginal zone B-cell lymphoma (Leukemia and Lymphoma Molecular Profiling Project, A.R., L.M.S., unpublished data, December 2008).
In the present cohort of cases, no difference in overall survival was observed between the t(14;18)-positive and t(14;18)-negative FL subgroups, which also held true for other clinical parameters (Eastern Cooperative Oncology Group performance status, sex, involvement of extranodal sites, B symptoms, or lactate dehydrogenase). One potential exception might be the more frequently observed low disease stage among t(14;18)-negative FL cases, which, however, needs to be validated in larger and more homogeneously treated cohorts.
It is important to note that FL subsets that frequently lack the t(14;18) and show distinct genetic and clinical features were not included in the present study. Specifically, pediatric follicular lymphomas, cutaneous follicle center lymphomas, and the recently described FL subgroup with a predominantly diffuse growth pattern and frequent deletions in the chromosomal region 1p3633 were excluded from our investigation, for which only FL with a predominantly follicular growth pattern were selected.
In summary, our study provides evidence that t(14;18)-negative FLs belong to the biologic spectrum of FL, but show distinct genetic features as well as gene expression and immunohistochemical profiles that differ from their t(14;18)-positive counterparts. Future studies will have to identify the t(14;18)-negative FL subset in large, prospective clinical trials and address the question whether the distinct molecular features of this subset have an effect on clinical parameters, the clinical course, or the response to current treatment protocols.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all members of the Leukemia and Lymphoma Molecular Profiling Project who are not coauthors of this study, Dr Heike Horn for helpful discussions, and Theodora Nedeva and Doris Hetzer for technical assistance.
This work was supported by the Interdisciplinary Center for Clinical Research (IZKF), University of Würzburg, Germany (A.R., E.L., and E.H.); the Robert-Bosch-Stiftung (Stuttgart, Germany; G.O.); the Spanish Ministry of Science (Madrid, Spain; SAF 05/5855; E.C.); the Instituto de Salud Carlos III, Red Temática de Investigación del Cáncer (2006RET2039; E.C.); and the National Cancer Institute (NCI) Strategic Partnering to Evaluate Cancer Signature (grant UO1-CA 114778).
National Institutes of Health
Authorship
Contribution: E.L. performed research, analyzed and interpreted the data, performed statistical analysis, and wrote the manuscript; I.S. performed research, analyzed and interpreted the data, and performed statistical analysis; S.B., A.Z., and E.M.H. performed research and analyzed the data; G.W. and V.M. performed statistical analysis; R.D.G., W.-C.C., R.M.B., L.M.R., D.D.W., J.D., E.S.J., A.L., J.F., L.M.S., and H.-K.M.-H. collected and analyzed the data; E.C. and G.O. designed research and supervised the study; and A.R. designed research, supervised the study, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Rosenwald, University of Würzburg, Institute of Pathology, Josef-Schneider-Str 2, 97080 Würzburg, Germany; e-mail: rosenwald@mail.uni-wuerzburg.de.
References
Author notes
*E.L. and I.S. contributed equally to this work.
†E.C., G.O., and A.R. are co–senior authors of the study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal