Abstract
To investigate molecular events involved in the regulation of lymphoid lineage commitment, we crossed λ5 reporter transgenic mice to Rag1-GFP knockin mice. This allowed us to subfractionate common lymphoid progenitors and pre-pro-B (fraction A) cells into λ5−Rag1low, λ5−Rag1high, and λ5+Rag1high cells. Clonal in vitro differentiation analysis demonstrated that Rag1low cells gave rise to B/T and NK cells. Rag1high cells displayed reduced NK-cell potential with preserved capacity to generate B- and T-lineage cells, whereas the λ5+ cells were B-lineage restricted. Ebf1 and Pax5 expression was largely confined to the Rag1high populations. These cells also expressed a higher level of the surface protein LY6D, providing an additional tool for the analysis of early lymphoid development. These data suggest that the classic common lymphoid progenitor compartment composes a mixture of cells with relatively restricted lineage potentials, thus opening new possibilities to investigate early hematopoiesis.
Introduction
The understanding of how multipotent hematopoietic stem cells (HSCs) differentiate into B-lymphoid cells has been derived largely through the identification of surface markers, which has allowed for the prospective isolation of progenitor cells at defined stages of development. To this end, different protocols have been independently developed,1-3 and they all allow for the identification of B-lineage cells representing defined developmental stages. One strategy has been based on the fact that the earliest B-cell progenitors express a lineage-restricted splice form of the pan-hematopoietic marker CD45, denoted B220, together with expression of CD43 (fraction A [FrA]).1 This purification protocol was further refined by the inclusion of the surface antigens CD93 (AA4.1) and CD4,4 allowing for additional subfractionation of the earliest B-lineage cells. This work served as a foundation for the understanding of stage-specific gene expression patterns and immunoglobulin recombination, revealing a sequential order of events associated with functional characteristics of the progenitor cells.1,4,5 However, the use of an increasing number of defined surface markers has suggested that B220 and CD43, originally used as markers for committed B-lineage cells, are not sufficient to categorically identify cells as belonging to the B-lymphoid lineage. Therefore, it has been proposed that the actual commitment event does not occur until the CD19+ stage of development.6 The surface expression of CD19 is linked to the functional expression of the transcription factor PAX5,7,8 which is critically involved in lineage restriction of early lymphoid cells.9,10 However, recent reports have indicated that B-lineage commitment may occur in the absence of PAX511 and before the expression of CD19,12 suggesting that B-lineage commitment may occur in earlier CD19− progenitor cells. Even though most immature progenitor cells essentially lack expression of lymphoid surface markers and transcription of lymphoid genes,13-15 lymphoid primed multipotent progenitors (LMPPs)16 display a significant expression of lymphoid associated genes, such as Rag1, Dntt, and sterile immunoglobulin transcripts.15,17,18 At this stage of development, cells do not appear to express genes specific for B- or T-lineage cells, suggesting that the transcription of lymphoid genes in LMPPs is associated with lineage priming rather than commitment to any specific lineage.15 However, in common lymphoid progenitors (CLPs),19 low levels of B lineage–associated genes, such as Ebf1, Pax5, Cd79a, and Pou2af1, can be detected.12,20-22 Expression of these genes is critically dependent on EBF1 because these transcripts are absent in CLPs from mice lacking a functional Ebf1 gene.22,23 Even though the expression of Ebf1 can be detected in the majority of CLPs, only a small fraction of CLPs express other B lineage–specific genes.12 These genes are, however, expressed in a coordinated manner, and use of a reporter transgenic mouse carrying the human CD25 (hCD25) gene under the control of the Igll1 (λ5) promoter (λ5Tg mice)24 allowed for the isolation of a CD19− population with reduced T-lineage potential. This suggested that the CLP compartment contains lineage-restricted subpopulations of cells and that lineage restriction occurs before expression of classic B-lineage surface markers.
To increase our understanding of lineage restriction within the lymphoid progenitor compartment, we crossed λ5Tg24 mice to Rag1-GFP reporter mice,25 which allows for purification of early lymphoid restricted progenitors.26 This allowed us to subfractionate the early lymphoid compartments into 3 defined stages with differential lineage potentials, leading us to suggest that the previously defined CLP/FrA compartments are heterogeneously composed of lineage-restricted progenitor cells. Thus, we think that B-lineage commitment is a stepwise process where “true” CLPs initially lose NK-cell potential to become a B/T cell–restricted progenitor, and then mature into a B cell–restricted progenitor.
Methods
FACS staining and purification of bone marrow cells
Animal procedures were performed with approval from the local ethics committee at Lund University (Lund, Sweden). Bone marrow was harvested from 10- to 15-week-old hCD25 λ5 promoter transgenic (λ5)Tghet)24 and hCD25(λ5)Tghet Rag1-GFPhet mice.25
For analysis and cell sorting of bone marrow cells, Fc-blocked (CD16/CD32, 2.4G2) cells were further stained with antibodies against FLT3 (A2F10), CD11b (M1/70), GR1 (RB6-8C5), TER119 (Ter119), CD3ϵ (145-2c11), CD11C (N418), LY6C (AL-21), NK1.1 (PK136), SCA1 (D7), CD19 (1D3/6D5), Ly-6D (49-H4), KIT (2B8), IL-7R (A7R34), CD45R/B220 (RA3-6B2), hCD25 (BC96), and propidium iodine (PI). For progenitor isolation, bone marrow cells were subjected to magnetic-activated cell sorting column enrichment of KIT+ cells using anti-CD117 immunomagnetic beads (Miltenyi Biotec) before antibody staining. Analysis and cell sorting were done on a FACSAria (BD Biosciences).
In vitro evaluation of NK-, B-, and T-cell potential by OP9/OP9DL1 coculture
For evaluation of NK-, B-, and T-cell potential, single cells were deposited (using a FACSAria) directly into 96-well plates containing preplated (2000 cells/well) OP9/OP9DL1 cells. NK cultures (on OP9 stroma layers) were supplemented with 20 ng/mL interleukin 15 (IL-15), 40 ng/mL IL-2, 10 ng/mL KIT ligand (KL), 10 ng/mL fms-like tyrosine kinase 3 ligand (FLT3L), and 10 ng/mL IL-7. B cultures (on OP9 stroma layers) were supplemented with 10 ng/mL KL, 10 ng/mL FLT3L, and 10 ng/mL IL-7. T cultures (on OP9DL1 stroma layers) were supplemented with 10 ng/mL KL, 10 ng/mL FLT3L, and 10 ng/mL IL-7. All cytokines were acquired from PeproTech. Cultures were substituted with fresh cytokines every 14 days. OptiMEM supplemented with 10% fetal calf serum, 50 μg/mL gentamicin, and 50μM β-mercaptoethanol was used for maintaining the OP9/OP9DL1 stroma cell lines as well as for cocultures.
Cocultures were evaluated by fluorescence-activated cell sorter (FACS) staining with CD19 (1D3), B220 (RA3-6B2), and PI for B OP9 cocultures, CD19 (1D3), NK1.1 (PK136), and PI for NK OP9 cocultures, and CD19 (1D3), CD90.2/Thy1.2 (53-2.1), and PI for T OP9DL1 cocultures. All cocultures were evaluated at day 14 or 15, except for LMPP OP9DL1 cocultures, which were evaluated at day 20 or 21. Samples were analyzed on a FACSCalibur (BD Biosciences).
In vitro evaluation of developmental kinetics by OP9-coculture
A total of 200 cells from investigated populations were seeded onto preplated OP9 stroma cells in OptiMEM supplemented with 10 ng/mL KL, 10 ng/mL FLT3L, and 10 ng/mL IL-7 (as described for B cultures in “In vitro evaluation of NK-, B-, and T-cell potential by OP9/OP9DL1 coculture”). Individual wells were stained with CD11b (M1/70), GR1 (RB6-8C5), CD19 (6D5), hCD25 (BC96), and 7AAD before FACS analysis after 30, 54, 87, 150, and 250 hours of coculture.
In vitro evaluation of myeloid potential
We have modified a previously described method to evaluate myeloid potential.15,16 In brief, 150 cells were sorted in 3 mL of medium (OptiMEM supplemented with 10% fetal calf serum, 50 μg/mL gentamicin, 50μM β-mercaptoethanol, 25 ng/mL KL, 25 ng/mL FLT3L, 5 ng/mL IL-3, 5 ng/mL human colony-stimulating factor 1 (M-CSF), 5 ng/mL human colony-stimulating factor 2 (GM-CSF), and 10 ng/mL colony-stimulating factor 3), and 20 μL plated into each well of two 60-well plates (Nunc Minitrays). Wells were scored after 5 days (using an inverted light microscope) for clonal growth and clone size. Clone size was scored based on the coverage of the tissue culture plate well and, for smaller clones, the number of cells. Clones covering more than 50% of the well were placed in the 100% to 50% category, whereas clones covering between 10% and 50% of the well were placed in the 50% to 10% category. Smaller clones composed of more than 10 cells but covering less than 10% of the well were placed in the 10%-10 cells category and the smallest clones containing fewer than 10 cells were placed in the 10-2 cells category. At least 240 cells per population were plated in 2 independent experiments and at least 16 clones picked randomly from each population for clonal evaluation by reverse-transcribed polymerase chain reaction (RT-PCR). RT-PCRs were essentially done as described for single-cell RT-PCRs (for primers used, see “Gene expression analysis of single cells by multiplex RT-PCR”).
Quantitative RT-PCR
Quantitative RT-PCR analysis of sorted cells was performed as previously described.22 Assays-on-Demand probes used were: Pax5; Mm00435501_m1, Hprt; Mm00446968_m1 and Ebf1; Mm00432948_m1, Pou2af1;Mm00448326_m1, Rag1; Mm01270936_m1. Sequences for the Assay-by-Design primers/probe used for λ5 (Igll1) detection were: forward 5′GGAACAACAGGCCTAGCTATGG, reverse 5′CTCCCCGTGGGATGATCTG, probe 5′CCGGCAGCTCCTGTTC. All experiments were performed in triplicate, and differences in cDNA input were compensated by normalizing against HPRT expression levels.
Affymetrix gene expression and data analysis
RNA was extracted from purified adult bone marrow (BM) subsets as described for quantitative RT-PCR. RNA was labeled and amplified according to the Affymetrix GeneChip Expression Analysis Technical Manual and hybridized against the MOE430 2.0 Affymetrix gene expression arrays chip. Chips were scanned using a GeneChip Scanner 3000. Probe level expression values were calculated using RMAexpress,27 and further analysis was done using dChip28 (www.dchip.org).
Gene expression analysis of single cells by multiplex RT-PCR
Multiplex single-cell RT-PCR analysis was performed as previously described.15,29 Multiplex PCR primers used were: Hprt (1) GGGGGCTATAAGTTCTTTGC; (2) GTTCTTTGCTGACCTGCTGG; (3) TGGGGCTGTACTGCTTAACC; (4) TCCAACACTTCGAGAGGTCC. Rag1 (1) CCAAGCTGCAGACATTCTAGCACTC; (2) CAGACATTCTAGCACTCTGG; (3) GCTTGACTTCCCATCAGCATGGA; (4) CAACATCTGCCTTCACGTCGATCC. Pax5 (1) CTACAGGCTCCGTGACGCAG; (2) ATGGCCACTCACTTCCGGGC; (3) GTCATCCAGGCCTCCAGCCA; (4) TCTCGGCCTGTGACAATAGG. IgII1 (λ5) (1) AGTTCTCCTCCTGCTGCTGC; (2) GGGTCTAGTGGATGGTGTCC; (3) CAAAACTGGGGCTTAGATGG; (4) CCCACCACCAAAGACATACC. Ebf1 (1) CCCTCTTATCTGGAACATGC; (2) CTACTCCCTGTATCAAAGCC; (3) TGTACGACAGTGTGACTTCC; (4) TAAGGATCACTTCCTTTGGC. CD3ϵ (1) CCTCAGAAGCATGATAAGC; (2) GTTGACATCTGTATCACTCTGG; (3) ACTGCTCTCTGATTCAGGC; (4) AGCAAGGTTCTAGGACACG. Mpo (1) CGCTTCTCCTTCTTCACTGG; (2) ACTGGCCTCAACTGCGAGAC; (3) GTGTATTGACAGCCAGCAGC; (4) CTGCCATTGTCTTGGAATCG; Csf3r (1) ACAGGAGTGTGAACTTCGCT; (2) TACCAGCCACAGCTCAAAGG; (3) ACGTGTCCAGTCTGATGGTG; (4)TTGCTTCTTCTGACACCACG. Ptcra (1) CTCTACCATCAGGCATCG; (2) GGCAGTGCCCTAGACGCC; (3) CTCCTGGCTGTCGAAGATTCC; (4) GAAGCAGTTTGAAGAGGAGC. Pou2af1 (1) ACGCCCAGTCACATTAAAGAAG; (2) AGCCAGTGAAGGAGCTACTGAG; (3) ATGTTCCCTCCTCTGTCACTGT; (4) CTGTCACTGTGGAAGCAGAAAC.
Primers 1 and 4 constitute the outer primer pair, whereas primers 2 and 3 constitute the inner nested primer pair. All microarray data can be found in the Gene Expression Omnibus public database (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE19142.
Results
Expression of Rag1-GFP and λ 5 promoter–controlled hCD25 allows for a sub-fractionation of the CLP and FrA populations in mouse BM
Even though surface marker expression has been used with great success to identify subpopulations in the BM, the addition of reporter transgenes allows for a more refined analysis. To investigate early B-lineage specification events, we crossed a reporter transgenic mouse expressing hCD25 under the control of the mouse λ5 (Igll1) promoter24 with a mouse carrying a GFP reporter gene under the control of the endogenous Rag1 locus.25,26 The expression of the hCD25 transgene has previously been reported to be associated with B-lineage restriction,12 whereas the expression of the Rag1-GFP reporter has been associated with a lymphoid bias of early progenitor cells.26 To simultaneously investigate the expression of the reporters in early hematopoietic progenitor cells as well as in CLPs19,30 and FrA1 cells, we devised a staining strategy for the visualizing FrA cells based on their expression of FLT3 (Figure 1B; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). This allowed us to follow the reporter gene expression from the multipotent stem cell populations, via the LMPP, into the CLP and later into B220+ populations. This revealed that the Rag1-GFP reporter was expressed at a low level in a subpopulation of the LIN−/lowSCA1highKIThighFLT3high LMPPs, in line with previously reported findings26 (Figure 1A). The LIN−FLT3high-IL-7R+SCA1lowKITlowB220− (CLP19,30 ) population could be divided into Rag1high and Rag1low cells (Figure 1B), suggesting that this reporter allows for a higher resolution than the Rag2-GFP reporter, which shows uniform expression in the CLP compartment.6 The Rag1high cells could be further subdivided based on the expression of hCD25 into a hCD25+ and a hCD25− fraction. Thus, the reporter gene expression allowed for the identification of 3 distinct populations: low expression of the Rag1-GFP reporter (Rag1low CLPs), high expression of GFP but no expression of hCD25 (Rag1high CLPs), or high expression of both reporter genes (Rag1highλ5Tg+, hereafter referred to as λ5Tg+ CLPs). A similar pattern of reporter gene expression could be observed in LIN−FLT3highIL-7R+SCA1lowKITlowB220+ FrA cells. However, the proportion of cells in each population was different from what could be observed in the B220− cells, with a reduced fraction of Rag1low cells to 7% in the B220+ cells from more than 60% in the B220− cells. By contrast, the fraction of reporter double-positive cells was increased from approximately 5% in the B220− cells to almost 50% in the B220+ fraction. In total, approximately 10% of LIN−FLT3highIL-7R+SCA1lowKITlow cells expressed the λ5Tg reporter.
The combined expression of the Rag1-GFP and λ5 promoter controlled hCD25 gene identifies distinct LIN−FLT3highIL-7R+SCA1lowKITlow early lymphoid progenitor populations. (A) Representative FACS plot showing the expression of the Rag1-GFP reporter gene in LIN−/lowSCA1highKIThigh bone marrow cells. (B) Representative FACS plots where LIN−FLT3highIL-7R+KITlowSCA1low cells are divided into B220− (CLP) and B220+ (FrA). The CLPs and the FrA cells are further subdivided based on expression of the Rag1-GFP reporter (Rag1high or Rag1low) and on expression of the λ5 promoter controlled human CD25 gene (λ5Tg). The dashed line box represents the total percentage of λ5Tg reporter-positive cells in the CLP/FrA compartment. Representative FACS plots shown are from KIT-enriched BM. Numbers indicate percentage of cells in parental gate. LIN includes: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD11C, LY6C, and CD19. (C) Generation of different cell subsets from Rag1low and Rag1high CLPs when cocultured on OP9 stoma cells. Percentages for indicated cells types of total live, GR1, and CD11B− cells within the forward and side scatter gate containing hematopoietic cells (percentage of GR1/CD11B/7AAD− cells) are indicated The data are collected from 2 experiments and represent an average (SEM shown). (D) Representative FACS plots from OP9 stroma cocultures at indicated time points. Gates indicate CD19−hCD25−, CD19−hCD25+, and CD19+ populations, respectively.
The combined expression of the Rag1-GFP and λ5 promoter controlled hCD25 gene identifies distinct LIN−FLT3highIL-7R+SCA1lowKITlow early lymphoid progenitor populations. (A) Representative FACS plot showing the expression of the Rag1-GFP reporter gene in LIN−/lowSCA1highKIThigh bone marrow cells. (B) Representative FACS plots where LIN−FLT3highIL-7R+KITlowSCA1low cells are divided into B220− (CLP) and B220+ (FrA). The CLPs and the FrA cells are further subdivided based on expression of the Rag1-GFP reporter (Rag1high or Rag1low) and on expression of the λ5 promoter controlled human CD25 gene (λ5Tg). The dashed line box represents the total percentage of λ5Tg reporter-positive cells in the CLP/FrA compartment. Representative FACS plots shown are from KIT-enriched BM. Numbers indicate percentage of cells in parental gate. LIN includes: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD11C, LY6C, and CD19. (C) Generation of different cell subsets from Rag1low and Rag1high CLPs when cocultured on OP9 stoma cells. Percentages for indicated cells types of total live, GR1, and CD11B− cells within the forward and side scatter gate containing hematopoietic cells (percentage of GR1/CD11B/7AAD− cells) are indicated The data are collected from 2 experiments and represent an average (SEM shown). (D) Representative FACS plots from OP9 stroma cocultures at indicated time points. Gates indicate CD19−hCD25−, CD19−hCD25+, and CD19+ populations, respectively.
We previously reported that hCD25+B220− CLPs develop into almost 100% CD19+ cells after 3 days of incubation on OP9 cells.12 To investigate the developmental relation between the additional progenitor populations identified by the combined reporter gene expression, we cocultured Rag1low and Rag1high CLPs on OP9 stroma cells. At 30, 54, 87, 150, and 240 hours after seeding, we used FACS to investigate the cellular content of the cultures with regard to CD19 and hCD25 (Figure 1C-D). In contrast to Rag1high CLP cells, which generated CD19−hCD25+ as well as low amounts of CD19+ cells already 54 hours after seeding, Raglow CLP cells did not generate detectable amounts of CD19−hCD25+ and CD19+ cells until 150 hours after seeding. The sequential appearance of the different cell types and the differences in kinetics support the idea that Rag1high and Rag1low cells are directly related in such a way that Raglow cells generate Rag1high cells that then generate hCD25+ cells. These data suggest that the LIN−FLT3highIL-7R+SCA1lowKITlow early lymphoid progenitor compartment can be subdivided into 3 developmentally related populations based on the combined reporter gene expression.
Reporter gene expression marks differential developmental potentials
Several lines of investigation have shown that the IL-7R+SCA1lowKITlow population contains progenitor cells with an ability to develop into B-, T-, and NK-lineage cells in vivo.6,12,19,30,31 However, because of the limited expansion potential of these cells, reliable functional readout in vivo demands transplantations of rather large number of cells, which provides limited information about the developmental potential of individual cells. Therefore, to investigate whether the reporter gene expression is linked to differential lineage potentials, we seeded single-sorted cells on OP9 stroma cells or OP9 cells ectopically expressing the notch ligand DELTA1 (OP9DL1 cells) and analyzed the development of the progenitors. All the investigated populations were able to generate B220+CD19+ cells (Figure 2A) on OP9 cells; however, the frequency of developing clones ranged from approximately 45% from Rag1low CLPs to more than 80% from λ5Tg+ CLPs. The Rag1low CLPs contained 12% of progenitors generating CD19− clones. These were not detected in the LMPP cultures, suggesting that they were formed because of the presence of committed progenitors rather than as a result of inefficient B-lymphoid priming by OP9 cells. Changing the culture conditions to allow for the development of NK cells revealed that approximately 60% of the LMPPs generated NK1.1+ cells (Figure 2B); however, the majority of the LMPPs gave rise to clones containing both NK1.1+ and CD19+ cells (Figure 2B). A similar potential for the development of NK cells could be detected in the Rag1low CLPs (Figure 2B). However, the detection of a higher percentage of cells that only generated NK1.1+ cells suggested that NK-restricted progenitors as well as bipotent progenitors can be found in the Rag1low population. The Rag1high populations generated clones with both CD19+ and NK1.1+ cells in 5% to 10% of the cases, suggesting mixed B-/NK-cell potential (Figure 2B). These cells generated only CD19+ cells from a majority of clones, suggesting that NK-cell potential is reduced in Rag1high cells. To investigate the T-lineage potential of the LIN−FLT3highIL-7R+SCA1lowKITlow subpopulations, sorted single cells were cultured on OP9DL1 cells (Figure 2C). In these conditions, more than 70% of Rag1low cells generated clones consisting of cells expressing THY1.2 (Figure 2C), but no CD19+ cells. Among the Rag1highλ5Tg− cells, approximately 50% were able to generate THY1.2+ cells independently of their surface expression of B220. The Rag1high cells, in contrast to the Rag1low cells, did generate a low number of CD19+ clones on OP9DL1 cells, suggesting that they contain a small fraction of B lineage–restricted cells. This fraction was dramatically increased when using λ5Tg+ cells because approximately 50% of these cells generated CD19+ offspring on OP9DL1. Seeding B220−λ5Tg+ cells resulted in 14% developing into THY1.2+ cells, whereas the fraction from B220+ λ5Tg+cells was 7%.
Reporter gene expression allows for the identification of functionally distinct populations. (A-C) Cloning frequency of seeded single cells (left), and FACS plots from representative clones generated from single Rag1low or λ5Tg+ CLPs (right). Single cells were sorted directly into the cultures based on the expression of surface markers and reporter genes. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. (A) Data from cocultures of single-sorted cells on OP9 stroma cells under B cell–promoting conditions. Bars represent total frequency of clones and frequency of clones containing CD19+ B cells (B). (B) Data from cocultures of single-sorted cells on OP9 stroma cells under conditions promoting NK-cell development. Bars represent the total frequency of clones as well as the frequency of clones containing only CD19+ B-lineage cells (B), NK1.1+ and CD19+ cells (B/NK), or only NK1.1+ cells (NK). (C) Data from cocultures of single-sorted cells on OP9DL1 stroma cells to stimulate the development of THY1.2+ T-lineage cells. Bars represent total cloning frequency, the frequency of clones with only CD19+ cells (B), THY1.2 and CD19+ cells (B/T), or only THY1.2+ (T). Data in graphs represent average (range) from 2 independent experiments analyzing approximately 96 seeded cells.
Reporter gene expression allows for the identification of functionally distinct populations. (A-C) Cloning frequency of seeded single cells (left), and FACS plots from representative clones generated from single Rag1low or λ5Tg+ CLPs (right). Single cells were sorted directly into the cultures based on the expression of surface markers and reporter genes. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. (A) Data from cocultures of single-sorted cells on OP9 stroma cells under B cell–promoting conditions. Bars represent total frequency of clones and frequency of clones containing CD19+ B cells (B). (B) Data from cocultures of single-sorted cells on OP9 stroma cells under conditions promoting NK-cell development. Bars represent the total frequency of clones as well as the frequency of clones containing only CD19+ B-lineage cells (B), NK1.1+ and CD19+ cells (B/NK), or only NK1.1+ cells (NK). (C) Data from cocultures of single-sorted cells on OP9DL1 stroma cells to stimulate the development of THY1.2+ T-lineage cells. Bars represent total cloning frequency, the frequency of clones with only CD19+ cells (B), THY1.2 and CD19+ cells (B/T), or only THY1.2+ (T). Data in graphs represent average (range) from 2 independent experiments analyzing approximately 96 seeded cells.
Even though the CLP compartment is restricted in its ability to generate myeloid cells in vivo,19 in vitro myeloid potential in a fraction of the cells has been reported.6,12 To investigate how the LIN−FLT3highIL-7R+SCA1lowKITlow subpopulations respond to incubation with myeloid cytokines, we seeded single cells into Terasaki plates and investigated the cell growth after 5 days of incubation. Whereas the LMPPs generated large clones with the majority of the cells giving rise to progeny covering more than 10% of the well (Figure 3A), neither B220+ nor B220− Rag1high cells generated colonies where the cultured cells covered more than 10% of the surface of the well. Instead, the majority of the clones generated from the Rag1high cells were composed of 1 to 10 cells. A few larger colonies covering more than 50% of the well were generated using the Rag1low cells (P < .05 compared with Rag1high cells), suggesting that this population contains a limited number of progenitors with an ability to respond robustly to myeloid stimulatory conditions. Even though the Rag1low cells did not generate colonies of a size comparable with those generated from the LMPP under these conditions, they still developed a flattened extended morphology atypical for lymphoid cells (supplemental Figure 2A). In contrast, the cells generated from Rag1high cell (irrespectively of λ5Tg expression) mostly displayed a rounded morphology indicative of lymphoid progenitors (supplemental Figure 2B-C). To investigate this further, we analyzed the expression of lymphoid and myeloid genes in a set of randomly collected clones by nested RT-PCR (Figure 3B). This demonstrated that a majority of the colonies generated from the Rag1low cells expressed either one of or both the myeloid genes Myeloperoxidase and Granulocyte Colony-Stimulating Factor Receptor. Among the colonies generated from reporter positive cells, we were unable to detect transcription of these myeloid associated genes in the absolute majority of the samples, even though B lineage–associated genes were easily detected. Rag1low cells also generated a few clones expressing the T lineage–associated genes Ptα and CD3ϵ, whereas the expression of these genes was undetectable in the clones generated from the reporter-positive cells. Taken together, these data demonstrate that the LIN−FLT3highIL-7R+SCA1lowKITlow early lymphoid progenitor compartment can be subdivided into 3 functionally distinct subpopulations.
Rag1-GFPlow cells retain a minimal residual myeloid potential. (A) Percentage of seeded single cells reaching a given clone size as estimated by visual scoring using light microscopy (“In vitro evaluation of myeloid potential”). Cells were sorted based on the expression of surface markers and reporter genes. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A, Each diagram is associated with results from gene expression analysis (B). A set of randomly chosen clones from the different experiments were investigated by multiplex RT-PCR. The dots indicate detected expression of the indicated gene. Each column represents one cell. Data in graphs are collected from 2 independent sorting experiments and represent average (range).
Rag1-GFPlow cells retain a minimal residual myeloid potential. (A) Percentage of seeded single cells reaching a given clone size as estimated by visual scoring using light microscopy (“In vitro evaluation of myeloid potential”). Cells were sorted based on the expression of surface markers and reporter genes. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A, Each diagram is associated with results from gene expression analysis (B). A set of randomly chosen clones from the different experiments were investigated by multiplex RT-PCR. The dots indicate detected expression of the indicated gene. Each column represents one cell. Data in graphs are collected from 2 independent sorting experiments and represent average (range).
The expression of B lineage–associated genes is restricted to Rag1high populations
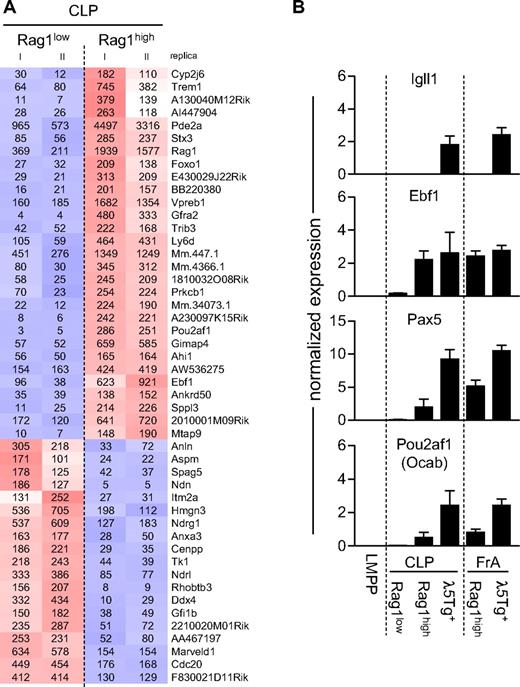
Having established that the subfractionation of the CLP compartment allowed for the prospective isolation of cells with different lineage potentials, we wanted to investigate gene expression patterns in the different populations. Because the gene expression patterns in hCD25+LIN−FLT3highIL-7R+SCA1lowKITlow early lymphoid progenitors has been investigated previously,12 we focused our analysis on the Rag1low and the Rag1high λ5Tg− CLPs. This revealed that, even though the cells displayed highly similar gene expression patterns, specific differences could be detected (Figure 4A). We noted significantly up-regulated expression of B-lineage genes, including Pax5, Vpreb1, Pou2af (OcaB), and CD79a in the Rag1high compared with the Rag1low cells. We could also detect increased expression of Foxo1, in agreement with the fact that Foxo1 is involved in the regulation of the Rag1 gene.32 An increase in the expression of Ebf1 message in the Rag1high cells suggested that high expression of Ebf1 is confined to defined LIN−FLT3highIL-7R+SCA1lowKITlow subpopulations. To verify and more quantitatively investigate the expression patterns of a set of these genes in all the defined populations, we used quantitative PCR to analyze the mRNA expression levels of Igll1, Ebf1, Pax5, and Pou2af1 (Figure 4B). Igll1 message could only be detected in the λ5Tg+ populations, verifying that the reporter gene reflects the expression of the endogenous gene in these early compartments12 (Figure 4B). We could also detect an up-regulation of Pax5 and Pou2af1 in the Rag1high cells, even though the levels did not reach those observed in λ5Tg+ cells. The expression of Ebf1 message was, in agreement with the microarray data, increased in the Rag1high cells relative to the Rag1low cells, and the level was comparable with that observed in the λ5Tg+ cells. Furthermore, the microarray analysis suggested that Rag1high cells expressed higher mRNA levels for the cell surface marker Ly6d than in Rag1low cells. To investigate whether surface expression of this protein can be used as an alternative marker for the identification of Rag1high cells, we stained cells from Rag1-GFP reporter mice with antibodies against LY6D in addition to antibodies needed for the identification of CLP/FrA cells (Figure 5A). This suggested that the majority of the Rag1high cells expressed LY6D. To further investigate the link between LY6D and Rag-1 expression, we subfractionated the LIN-FLT3highIL-7R+SCA1lowKITlow cells based on LY6D expression and analyzed the expression of Rag-1 mRNA by quantitative RT-PCR. This revealed that the Rag-1 expression was increased in the LY6D+ cells compared with their LY6D− counterparts (Figure 5B). To further investigate the usefulness of LY6D as a marker for lineage-restricted progenitor cells, we analyzed the in vitro NK-cell potential of single LY6D+ and LY6D− CLPs. This revealed that, whereas approximately 50% of the LY6D− CLPs were able to generate clones containing NK cells, the LY6D+ cells displayed a dramatically reduced ability to generate NK cells, although they were able to generate B-lineage cells (Figure 5C). This indicates that LY6D might represent a useful marker for subfractionation of early lymphoid progenitor cells.
Alterations in B-lineage gene expression patterns in CLP subpopulations are associated with reporter gene expression. (A) Result of a gene expression microarray analysis comparing the gene expression patterns in Rag1low and Rag1high LIN−FLT3+IL-7R+SCA1lowKITlow CLPs. Cells used for replica arrays originated from independent sorts, and cluster analyses have been done using genes with at least 2.5-fold difference in expressed between either of the cell populations. Red represents high expression; and blue, low expression. Calculated RMA expression values are indicated. (B) Quantitative PCR data where the relative expression of a set of genes is analyzed in purified cells. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. The data are presented as relative expression values normalized to Hprt1 expression levels. Data are average (SD) from cells originating from 3 to 5 independent sorts.
Alterations in B-lineage gene expression patterns in CLP subpopulations are associated with reporter gene expression. (A) Result of a gene expression microarray analysis comparing the gene expression patterns in Rag1low and Rag1high LIN−FLT3+IL-7R+SCA1lowKITlow CLPs. Cells used for replica arrays originated from independent sorts, and cluster analyses have been done using genes with at least 2.5-fold difference in expressed between either of the cell populations. Red represents high expression; and blue, low expression. Calculated RMA expression values are indicated. (B) Quantitative PCR data where the relative expression of a set of genes is analyzed in purified cells. LMPPs were sorted as LIN−/lowSCA1highKIThighFLT3high, whereas CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. The data are presented as relative expression values normalized to Hprt1 expression levels. Data are average (SD) from cells originating from 3 to 5 independent sorts.
The majority of the Rag1high cells express the surface marker LY6D. (A) Representative FACS plots showing Rag1-GFP and LY6D expression in LIN−FLT3+IL-7R+SCA1lowKITlow CLP/FrA cells. Numbers indicate percentage of gated cells. LIN include: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD19, and CD11C. (B) Rag1 quantitative RT-PCR data generated from sorted LY6D+ and LY6D− CLPs. The data are presented as relative expression values normalized to Hprt1 expression levels, and data are collected from 3 quantitative PCR reactions. (C) Distribution of B-, B/NK-, and NK-cell containing clones generated from single LIN−FLT3+IL-7R+SCA1lowKITlow LY6D+/− CLPs as indicated. The imbedded small diagram indicates total cloning frequency. The data are collected from 2 independent experiments, and the error bars represent variation between experiments. For sorting of LY6D+/− CLPs, LIN include: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD19, CD11C, and B220.
The majority of the Rag1high cells express the surface marker LY6D. (A) Representative FACS plots showing Rag1-GFP and LY6D expression in LIN−FLT3+IL-7R+SCA1lowKITlow CLP/FrA cells. Numbers indicate percentage of gated cells. LIN include: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD19, and CD11C. (B) Rag1 quantitative RT-PCR data generated from sorted LY6D+ and LY6D− CLPs. The data are presented as relative expression values normalized to Hprt1 expression levels, and data are collected from 3 quantitative PCR reactions. (C) Distribution of B-, B/NK-, and NK-cell containing clones generated from single LIN−FLT3+IL-7R+SCA1lowKITlow LY6D+/− CLPs as indicated. The imbedded small diagram indicates total cloning frequency. The data are collected from 2 independent experiments, and the error bars represent variation between experiments. For sorting of LY6D+/− CLPs, LIN include: GR1, CD11B, CD3ϵ, TER119, NK1.1, CD19, CD11C, and B220.
Ectopic expression of Ebf1 has been shown to mediate B-lineage restriction in lymphoid progenitor cells; and because the expression level of Ebf1 message was comparable in B cell–committed (Rag1highλ5Tg+ cells) and in B/T potent progenitors (Rag1highλ5Tg− cells), we wanted to investigate the distribution of Ebf1 expressing cells in the different subpopulations. To this end, we used multiplex single-cell RT-PCR (Figure 6) to analyze the expression of Ebf1 message at a single-cell level. This analysis revealed that, whereas more than 97% of the Rag1high CLPs contained detectable Ebf1 message, only 43% of the Rag1low CLPs expressed for Ebf1. One of the Rag1low CLPs expressed detectable levels of Pax5 message, whereas 31% of the Rag1high CLPs and 66% of the Rag1high FrA cells expressed Pax5 message. Among the λ5Tg+ cells, more than 69% of the B220− and 87% of the B220+ cells expressed detectable Pax5 message. We were unable to detect Pou2af1 message in Rag1low cells, whereas 23% of the RaghighB220− cells expressed Pou2af1. 15 of these 20 cells coexpressed Pax5 message, suggesting coordinated gene activation in a subfraction of these cells. Among the λ5Tg+ cells, we detected Pou2af1 expression in 73% of the cells, whereas 37% of the λ5Tg−B220+ FrA cells contained expressed Pou2af1 These data provided molecular support for our functional analysis and suggest that Ebf1 mRNA is not alone sufficient to cause B-lineage restriction in lymphoid progenitors.
Single-cell PCR analysis confirms differential gene expression patterns in CLP and FrA populations. The figure shows the collected result of multiplex single-cell RT-PCR from indicated populations. CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. The dots represent detected expression of the indicated gene. Each horizontal line of dots represents a single investigated cell. Dots indicate that a RT-PCR product could be detected for a given gene in an investigated cell. The values on top of the data panel indicate the number of cells expressing the indicated gene (top portion of the panel) and the percentage of cells expressing the indicated gene of all cells expressing any gene (bottom portion of the panel).
Single-cell PCR analysis confirms differential gene expression patterns in CLP and FrA populations. The figure shows the collected result of multiplex single-cell RT-PCR from indicated populations. CLP (Rag1low, Rag1high, and λ5Tg+) and FrA (Rag1high and λ5Tg+) subpopulations were sorted as shown in Figure 1A. The dots represent detected expression of the indicated gene. Each horizontal line of dots represents a single investigated cell. Dots indicate that a RT-PCR product could be detected for a given gene in an investigated cell. The values on top of the data panel indicate the number of cells expressing the indicated gene (top portion of the panel) and the percentage of cells expressing the indicated gene of all cells expressing any gene (bottom portion of the panel).
Discussion
We report here that the combined expression of Rag1 and λ5Tg reporter genes allows for the prospective isolation of functionally distinct CLP subpopulations. It was previously suggested that the B220+CD43+ FrA cells do not contain B lineage–committed cells because FrA cells can generate T-lineage cells both in vitro and in vivo at a high frequency.6,31 This has led to the general conclusion that B-lineage commitment does not occur until the CD19+ progenitor stage.6 At the molecular level, this idea is supported by the finding that CD19 is a direct target for Pax5 and therefore could serve as an indicator of functional Pax5 levels and subsequent lineage restriction.9,10 The data presented in this report suggest that only a subpopulation of the FrA cells retain T-cell potential, whereas a significant proportion of the cells are indeed B-lineage committed, even in the absence of detectable CD19 surface expression. The identification of these cells is complicated by the fact that, unless a reporter gene can be used in combination with single-cell differentiation analysis, the experiments will not give sufficient resolution to detect the committed cells. Furthermore, among the FLT3- and IL-7R–expressing cells, B220 surface expression only appears to result in an alteration in the relative percentage of committed cells and not in qualitative differences related to B/T-lineage potential. The finding that B220 expression allows for an enrichment of the Rag1high cells probably explains the findings that these cells display reduced NK and myeloid potential.6 Even though our findings do not exclude that the subpopulations we identify are generated stochastically and then adopt differential cell fates in response to external signals, the kinetics experiments (Figure 1C-D) suggest that the cells develop sequentially from Rag1low cells to Rag1high cells and eventually to cells that express the λ5Tg reporter (Figure 7). The percentage of λ5Tg reporter-positive cells was approximately 10% of the LIN−FLT3highIL-7R+SCA1lowKITlow population; and considering that the burst size for B-cell development from a CLP is estimated to be 1900,30 it would not appear unrealistic that these cells represent a transient commitment step in the major pathway of the generation of the adult B-lymphoid compartment.
Schematic drawing of a novel model for lymphoid lineage restriction events. The figure shows a schematic drawing of the development of lineage-restricted cells within the LIN−KITlowSCA1lowFLT3+IL-7+ compartment in the mouse bone marrow. NK indicates NK-cell potential; and T, competence for development into T lymphocytes.
Schematic drawing of a novel model for lymphoid lineage restriction events. The figure shows a schematic drawing of the development of lineage-restricted cells within the LIN−KITlowSCA1lowFLT3+IL-7+ compartment in the mouse bone marrow. NK indicates NK-cell potential; and T, competence for development into T lymphocytes.
A sequential series of events is also supported by the molecular analysis of gene expression patterns in the different subpopulations with a gradually increased expression of B lineage–associated genes. The low level of Ebf1 message in the Rag1low CLPs may also explain the finding that Ebf1 expression is reduced in CLPs from TCFE2A (E2A)33 and IL-7–deficient mice.20,21,34 These mice display a reduced CLP compartment, and a specific reduction in Rag1high cells would result in decreased amounts of Ebf1 message. This does not contradict the functional role for EBF1 in the stimulation of B-cell development from CLPs but might provide alternative explanations to the disturbed gene expression patterns observed. Whereas Ebf1 levels were up-regulated to the same level as in the committed cells in the transition from Rag1low to Rag1high CLPs, the levels of the target genes Pax5 and Pou2af1 were not induced to the same level as in the committed cells, and the direct target Igll1 was essentially silent (Figure 4B). However, considering that the fractions of message expressing cells are altered, the Pax5 and Pou2af1 mRNA levels may be comparable with those in the λ5Tg+ cells on a per-cell basis. The coordinated expression of Ebf1, Pax5, and Pou2af1 as well as the functional data suggest that, in addition to the λ5Tg+ cells, a small fraction of the Rag1highλ5Tg− is composed of B lineage–restricted cells.
It has been shown that Ebf1 expression in Pax5-deficient cells is sufficient to cause lineage restriction.11 However, our single-cell RT-PCR experiments revealed that all the Rag1high cells expressed Ebf1 message and that the average expression level in the population was comparable with that of λ5Tg+ cells (Figure 4B), shown previously to express levels comparable with those of CD19+ pro-B cells.12 Because these cells retain an in vitro T-cell potential, this would suggest that physiologic levels of Ebf1 message is not sufficient to cause lineage restriction. Expression of Pax5 and Pou2af1 appears to be more restricted and, although probably not totally linked to commitment, a safer sign of lineage restriction. Reporter mice carrying a GFP reporter in the 3′ noncoding region of the Pax5 gene have been shown to express GFP within a subpopulation of the CLPs,35 but the lineage potential of these cells has not been determined. We find it reasonable that, in normal development, EBF1 and PAX5 collaborate to induce B-lineage commitment via a positive feedback loop.36-38 The inability of EBF1 to fully activate target genes in the Rag1high cells could be explained by specific expression of suppressor proteins, lack of cofactors, or by posttranscriptional regulation of the EBF1 protein per se. One potential explanation comes from the findings that NOTCH signaling, shown to favor T-cell fate,39-41 directly reduces the functional activity of EBF1.42 Furthermore, it has been reported that Notch signaling destabilizes the transcription factor TCFE2A43,44 known to collaborate with EBF1 in early progenitor cells.36,45 Thus, it is possible that active NOTCH signaling can primarily target genes, such as λ5, whose functional activity is highly dependent on the synergy between EBF1 and TCFE2A,45,46 whereas the induction of other target genes, such as Pou2af1, Rag1,22 and Pax5,36,38 is less dependent of the coordinated activity of the 2 transcription factors and thus not as sensitive to NOTCH signaling. These data all support the idea that EBF1 plays a crucial role in B-lymphoid commitment and that this process is achieved in CD19− FLT3highIL-7R+ cells.
We also report that Rag-1 expression is correlated to the expression of LY6D and that expression of this surface marker is linked to a reduced ability of progenitor cells to develop into NK cells in vitro. This is in line with a recent report where this surface marker has been used to identify lineage-restricted subpopulations of the LIN−FLT3highIL-7R+SCA1lowKITlow progenitor compartment.47 This work suggests that the LY6D+ cells lack in vivo T-cell potential and thus would represent a pure B lineage–committed population. This is not fully in compliance with our in vitro differentiation data because, even though Rag1high cells express LY6D, they display a robust T-cell potential, even at a single-cell level. This could reflect that the in vitro differentiation analysis reveals lineage potentials that are not developed in vivo or a difference in the lineage readout in the transplantation setting per se. Thus, the combined use of LY6D and the Rag1 reporter may allow for an even more stringent subfractionation of these early cells, allowing a more detailed insight into B- versus T-cell fate decisions.
In conclusion, we propose that the initial stages of lymphoid development are associated with transcription of lymphoid-associated genes in the LMPP compartment.14,15 This priming is dependent of the transcription factors IKAROS17,18 and TCFE2A48 and do not appear to involve lineage-restricted genes but rather genes expressed in both B- and T-lineage cells. B-lineage genes are expressed in a CLP-resembling FLT3highIL-7R+ population, composed mainly of B lineage–committed cells that are generated through the gradual loss of first NK potential and then T-cell potential (Figure 7). The expression of these B-lineage genes is critically dependent on functional EBF1 protein,22 whereas the stable commitment of these cells is dependent on the action of PAX59,10,49 and IKAROS.50
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Inga-Lill Mårtensson and Fritz Melchers for the gift of λ5 transgenic mice and expert advice regarding their usage, J.C. Zuniga-Pflücker for providing the OP9 and OP9DL1 stromal cell lines and expert advice regarding their usage, Liselotte Lenner and Gerd Sten for advice and assistance, and Elinore M. Mercer for critical reading of the manuscript.
This work was supported by the Swedish Cancer Society, the Swedish Research Council, the Swedish Childhood Cancer Foundation, the Swedish Foundation for Strategic Research, and the Faculty of Medicine at Linköping University.
Authorship
Contribution: R.M. and M.S. devised conceptual idea; R.M., S.Z., E.W., and P.T. have performed the majority of the in vitro differentiation experiments and single-cell PCRs; D.B. performed cell sorting experiments; R.M., S.Z., E.W., P.T., N.S., D.B., and M.S. have all been involved in the design of the experiments; N.S. has provided crucial reagents; and all authors have read and given comments to the manuscript written by R.M., S.Z., and M.S.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mikael Sigvardsson, Faculty of Health Sciences, Linköping University Lab1, Level 13, Linköping, Sweden; e-mail: miksi@ibk.liu.se.
References
Author notes
R.M. and S.Z. contributed equally to this study.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal