Abstract
Chronic lymphocytic leukemia (CLL) involves a profound humoral immune defect and tumor-specific humoral tolerance that directly contribute to disease morbidity and mortality. CD154 gene therapy can reverse this immune defect, but attempts to do this pharmacologically have been unsuccessful. The immune-modulatory agent lenalidomide shows clinical activity in CLL, but its mechanism is poorly understood. Here, we demonstrate that lenalidomide induces expression of functional CD154 antigen on CLL cells both in vitro and in vivo. This occurs via enhanced CD154 transcription mediated by a Nuclear Factor of Activated T cells c1 (NFATc1)/Nuclear Factor-κB (NF-κB) complex and also through phosphoinositide-3 (PI3)–kinase pathway-dependent stabilization of CD154 mRNA. Importantly, CD154-positive CLL cells up-regulate BID, DR5, and p73, become sensitized to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–mediated apoptosis, and promote costimulatory activation of normal B cells to produce antibodies. In CLL patients receiving lenalidomide, similar evidence of CD154 activation is observed including BID, DR5, and p73 induction and also development of anti-ROR1 tumor-directed antibodies. Our data demonstrate that lenalidomide promotes CD154 expression on CLL cells with subsequent activation phenotype, and may therefore reverse the humoral immune defect observed in this disease. This study is registered at http://clinicaltrials.gov as NCT00466895.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia, and is characterized by an elevated frequency of infections, secondary malignancy, and autoimmune complications compared with the general population. Current treatment options for CLL are palliative and further exacerbate the immune deficiency seen in this disease. Nonetheless, CLL represents an “immunoresponsive” disease as evidenced by extended disease remission and potential cure with reduced intensity allogeneic stem cell transplantation (reviewed in Gribben1 ). This suggests that strategies that restore immune function have potential to effectively eliminate CLL.
The immune defect in CLL is characterized by both humoral and cellular immune defects. Although detailed studies of normal B cells in CLL patients have not been performed due to the difficulty in isolating these cells, hypogammaglobulinemia is often present at diagnosis and becomes worse with disease progression. A profound cellular immune defect2-4 is present in CLL with significant alterations in genes involved in differentiation, cytoskeleton formation, vesicle trafficking, and cell death.4 Coculture of CLL cells with normal T cells produces the same T-cell defects observed in CLL patients,4 suggesting a direct role of the leukemia cells in contributing to the T cell–dependent cellular immune deficiency. The clinical manifestation of the humoral and cellular immune defects in CLL patients includes hypogammaglobulinemia,5,6 poor response to both polysaccharide-based7-9 and protein-based10 vaccines, and a high predisposition to infections11,12 that represents a leading cause of death.
To date, attempts to reverse the immune defects in CLL have been limited. Most promising has been adenovirus-delivered CD154 gene therapy that in small numbers of patients reversed cellular and humoral tumor tolerance. CD154 is the surface ligand of CD40 and is expressed on activated T cells, natural killer cells, and dendritic cells, but not normal B cells. Activation of T cells promotes increased surface expression of CD154, thereby promoting both activation and antigen presentation in normal and transformed B cells. Congenital mutations in the CD154 gene promote profound cellular and humoral immune deficiency. Although mutations of the CD154 gene have not been described in CLL, these patients have diminished CD154 expression on T cells after CD3 ligation.13 Transduction of murine or human CD154 into primary CLL cells ex vivo with adenovirus gene therapy vectors, followed by systemic reintroduction, has been pursued clinically.14 Surface expression of CD154 on CLL cells after gene therapy treatment promotes expression of costimulatory molecules including CD40, CD80, and CD86 on neighboring bystander CLL cells, thereby making them better costimulants for T-cell activation. As a consequence, increases in interferon-gamma, interleukin-12, and total CD4 T-cell counts were observed after CD154 gene therapy.14 Response to CD154 gene therapy by residual normal B cells was also demonstrated by improvement in both hypogammaglobulinemia and development of antibodies to the CLL tumor-specific antigen ROR1.15 Extending from immune activation, a favorable activation phenotype in the CLL cells occurs after CD154 gene therapy or CD40 ligation. This phenotype includes up-regulation of BID, DR5, and p73, thereby enhancing sensitivity of these tumor cells to both tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) and fludarabine-based therapies.16,17 The CD154 gene therapy approach for CLL represents an exciting proof of concept to reverse the disease-induced immune defect. However, as with other gene therapy approaches, CD154 gene therapy is inefficient, cumbersome, and has not produced durable remissions, perhaps due to the inability to administer therapy over an extended period of time. Identifying an alternative pharmacologic strategy that mimics this approach would represent a major therapeutic advance for CLL and other related lymphoproliferative disorders.
Lenalidomide is an oral therapeutic agent with clinical activity in multiple myeloma,18 lymphoma, myelodysplasia,19 and CLL.20,21 The exact mechanism of action of lenalidomide is uncertain, although it has been reported to promote cellular and innate immune activation,22-25 up-regulation of SPARC,26 and interference with tumor cell microenvironment.27 In addition, lenalidomide increases costimulatory molecules CD40, CD80, and CD86 on CLL cells.28,29 Coculture of CLL cells and autologous T cells with lenalidomide reverses the T-cell immune synapse defect present in this disease.30 Improvement in T-cell function with lenalidomide is dependent upon CLL cell interaction,30 similar to CD154 gene therapy. The subsequent observation of robust in vivo production of polyclonal hypergammaglobulinemia with concurrent disease response in a CLL patient receiving lenalidomide prompted us to examine whether this agent promotes CD154 gene expression in CLL cells. Herein, we demonstrate that lenalidomide promotes expression of functional CD154 on CLL cells through 2 distinct mechanisms of transcriptional activation and posttranscriptional mRNA stabilization. Most importantly, extended duration lenalidomide treatment promotes a CD154 gene therapy phenotype with multiple downstream effects on CLL cells, suggesting novel applications for its clinical use in this disease.
Methods
Cell isolation, culture, and reagents used
Blood was obtained from patients with CLL as described by National Cancer Institute Working Group criteria31 under an Ohio State University Institutional Review Board–approved protocol with informed consent according to the Declaration of Helsinki. The case description and other samples obtained in vivo are derived from a phase 1 clinical trial administering lenalidomide to patients with relapsed and refractory CLL at doses ranging from 2.5 to 7.5 mg daily. All patients enrolled on this trial had received rituximab with exception of the index case. CLL cells were isolated by Ficoll centrifugation using Rosette-Sep reagent (StemCell Technologies) and maintained in culture as previously described.29 Lenalidomide (Revlimid; Celgene) was obtained and extracted as previously described.28 Pokeweed mitogen (PWM), 1,9-dideoxyforskolin, okadaic acid, and transcriptional inhibitor actinomycin D (ActD) were from Sigma-Aldrich.
Immunophenotyping studies
Percentage and mean fluorescence intensity of CD154 and CD19 on CLL cells relative to a specific isotype control were determined by flow cytometry (EPICS-XL; Beckman-Coulter) and analyzed using WinMDI (Windows Multiple Document Interface for Flow Cytometry; The Scripps Research Institute). CD19 antibody was from BD Biosciences and CD154 was from Serotec. For acid washing of cells before analysis, cells were exposed to acidic phosphate-buffered saline (pH 4.1) for 3 minutes at room temperature (RT) and then washed twice with normal pH before exposure to antibodies.32
Immunoblotting, immunoprecipitation, and ELISA
Immunoblot assays were performed with the multiple antigen detection immunoblotting method as described.29 Antibodies for Akt, phospho-Akt, c-Rel, Brg-1, nuclear factor of activated T cells c1 (NFATc1), BID, p73, actin, and ROR1 were purchased commercially from Cell Signaling Technology and R&D Systems. Recombinant human ROR1 extracellular domain (ECD) protein, approximately 50 kDa, was expressed in human embryonic kidney (HEK 293F) cells (Invitrogen) and purified by ion exchange and size exclusion chromatography. CLL B cells were washed and cell surface proteins biotinylated using Sulfo-NHS-LC-Biotin (Pierce).33 Lysates were immunoprecipitated overnight at 4°C using either 3 μg of rabbit anti-CD154 or isotype-matched antibody (Santa Cruz Biotechnology) conjugated to rabbit ExactaCruz immunoprecipitation matrix (Santa Cruz Biotechnology). Precipitated materials were boiled, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.2 μm nitrocellulose membranes (Bio-Rad Laboratories), exposed to horseradish peroxidase (HRP)–conjugated streptavidin (Pierce) for 2 hours, and then imaged using a chemiluminescence substrate (SuperSignal; Pierce). An indirect enzyme-linked immunoabsorbance assay (ELISA) protocol was used to detect the presence of anti-ROR1 immunoglobulin in human sera. Polystyrene microtiter plates were incubated overnight at 4°C with 50 μL of ROR1-ECD (at 1 μg/mL in borate buffer), washed 3 times with 0.1% Tween/borate, and incubated in blocking buffer (0.5% bovine serum albumin, 1% goat serum/borate buffer) for 1 hour at RT. Plates were washed 3 times with 0.1% Tween/borate and serial dilutions of human serum in blocking buffer were added to individual wells and incubated for 2 hours at RT. Plates were washed 6 times with 0.1% Tween/borate and incubated with HRP-conjugated goat anti–human immunoglobulin G (IgG; Southern Biotechnology) for 1 hour. Plates were washed 6 times and SureBlue TMB Substrate was added to develop antibodies labeled with HRP. Plates were analyzed using a luminometer. Samples were run in triplicates.
Quantitative real-time PCR
RNA was extracted from 107 cells using TRIzol reagent (Invitrogen). cDNAs were prepared using a SuperScript First-Strand Synthesis System (Invitrogen) as previously described.29 Real-time polymerase chain reaction (PCR) was performed using predesigned TaqMan Gene Expression Assays and ABI Prism 7700 sequence detection system (Applied Biosystems).
ChIP
CD154 luciferase activity
The pGL3-basic CD154-luc reporter constructs (−1280) and the pGL3-basic empty vector have been previously described.35 Transient transfections of CLL B cells were conducted using the Amaxa Nucleofector protocol (Lonza). Luciferase and renilla assays were performed according to the manufacturer's directions (Promega). Luciferase activity values were normalized to transfection efficiency monitored by the cotransfected renilla expression vector.
CD154 functional study
A previously described procedure was followed.35 Effector cells included 5 × 104 autologous T cells (AutoTxr) from the healthy target B-cell donor, lenalidomide-treated CLL B cells (CLLBxr), or vehicle control-treated CLLBxr. Effector cells were irradiated (20 Gy) and placed in culture with 5 × 104 target, purified B cells from the healthy donor, in the absence or presence of PWM (5 μg/mL). Control samples included 5 × 104 target, unirradiated B cells alone, with and without PWM, and 5 × 104 irradiated CLL B cells, in the absence of target B cells. All the experimental conditions were assayed in triplicate. Supernatants were collected after 7 days of coculture and tested for immunoglobulin production by a standard enzyme-linked immunoabsorbance assay (ELISA) protocol. IgG and IgM standards were purchased from Thermo Fisher Scientific Inc, whereas anti-IgG and -IgM antibodies were from Southern Biotech.
Determination of CD154 mRNA half-life
CLL B cells (107) were treated with lenalidomide 0.5μM or vehicle control for 0, 12, 24, and 48 hours. At the end of each time point, cells were treated with 10 μg/mL ActD for varied times. Calculation of mRNA half-lives was carried out by plotting data on a semilogarithmic graph and calculating the first-order decay slope using linear regression. Values of independent experiments were used to calculate the average half-lives (± SD). Alternatively, data of several experiments were plotted on a single graph to calculate the linear regression slope and 95% confidence limits.
IKK-β assay, immunoprecipitation, and immunoblotting
Cell lysates were prepared using a kinase buffer (20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 2mM dithiothreitol, 10mM NaF, 1mM sodium vanadate, and 10mM β-glycerophosphate containing 1% Nonidet P40) plus a protease inhibitors. Cell lysate (500 μg) was incubated with 1 μg of rabbit polyclonal antibody specific for inhibitor of NF-κB kinase subunit β (IKK-β) for 1 hour at 4°C and then with anti–rabbit-specific agarose beads (eBioscience) overnight. Beads were washed 5 times with kinase buffer and then incubated in 20 μL of kinase buffer containing 20μM adenosine triphosphate (ATP), 10 μCi (0.37 MBq) of [γ32P]ATP, and 0.5 μg of glutathione S-transferase (GST)–IκBα (1-54) substrate at 30°C for 30 minutes. The reaction mixture underwent SDS-PAGE (10%). In the radioactivity assay, the gel was dried and phosphorylation of GST-IκBα was measured by autoradiography. To normalize kinase activity, the proteins were transferred to nitrocellulose membrane and probed by Western blotting with anti–phospho-IκBα, total IκBα, and total IKK-β (Santa Cruz Biotechnology). Protein level on the blots was detected by addition of chemiluminescent substrate (SuperSignal; Pierce) followed by quantification by ChemiDoc system with Quantify One software (Bio-Rad Laboratories).
Determination of PP2A activity
Myelin basic protein was labeled with 32P by cyclic adenosine monophosphate–dependent kinase using 32P-ATP according to the protocol provided by New England Biolabs. The protein phosphatase activity of total cell lysate was determined by measuring the released free 32P. Cell lysates were prepared using a low-detergent lysis buffer (1% Nonidet P-40, 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 150mM NaCl, 10% glycerol, 1mM phenylmethylsulphonyl fluoride, 5nM benzamidine, and 10 μg/mL leupeptin). PP2A was immunoprecipitated from the cell lysate using an anti-PP2A antibody (Upstate Biotechnology) and incubated with 32P-myelin basic protein at 30°C for 10 minutes. The reaction was terminated by adding trichloroacetic acid. The mixture was centrifuged and the supernatant was analyzed on a multipurpose liquid scintillation counter (Beckman Coulter). For cell-free assay, purified PP2A, an ABC trimer, was purchased from Upstate Biotechnology and dissolved in 20mM 3-[N-morpholino]propanesulphonic acid (pH 7.4), 0.1M NaCl, 60mM 2-mercaptoethanol, 1mM MgCl2, 1mM ethyleneglycoltetraacetic acid, 0.1mM MnCl2, 1mM dithiothreitol, 10% glycerol, and 0.1 mg/mL serum albumin. Neither Nonidet P40 nor bovine serum albumin was used. In each reaction, 0.5 units of the enzyme were used.
Statistical analyses
All statistical analyses were performed by biostatisticians in the Center for Biostatistics at The Ohio State University. Linear mixed models were used for modeling treatment effects, including patient random effects. Hypothesis testing used contrasts that directly answered primary questions, for example, interaction contrasts were used to directly test the inhibitory or synergistic hypotheses found in the experiments that test mechanisms of action. Random effects associated with these contrasts were included in the random effects portion of the models to avoid biased low estimates of the standard errors of the contrasts. The Holm method was applied to adjust for multiplicity and control the overall family-wise type I error rate at α = .05. SAS/STAT software (Version 9.1.3; SAS Institute Inc) was used for all statistical analyses.
Results
Case reports
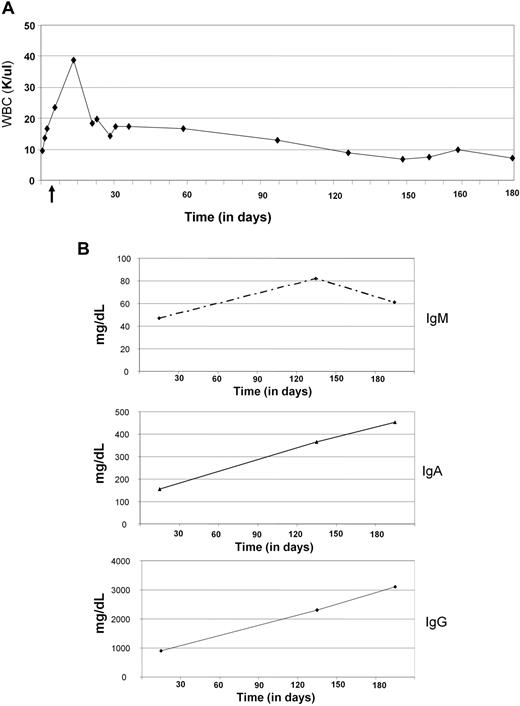
A 78-year-old woman was diagnosed with CLL in June 2007 and ultimately developed symptomatic disease in June 2008. Genetic studies showed IgVH unmutated status together with a complex karyotype including del(17p13.1). A 12-week treatment of subcutaneous alemtuzumab was administered from June 2008 to August 2008. Progressive disease with symptomatic lymphadenopathy was noted 2 months later. A pretreatment bone marrow biopsy had 25% to 30% CLL cells with no plasma cells noted. Lenalidomide was administered in November 2008 as part of an institutional review board–approved phase 1 clinical trial. The lenalidomide dose was escalated from 2.5 to 7.5 mg daily during the first month of therapy. Asymptomatic neutropenia developed during cycle 2 and a dose of 5 mg of lenalidomide daily has been maintained continuously since this time. Over 5 months, a slow reduction in blood lymphocyte count (Figure 1A) and nodal disease was noted. Concomitant with this, a polyclonal IgM, IgA, and IgG expansion was observed (Figure 1B). A bone marrow biopsy at 5 months demonstrated persistent but diminished CLL infiltration (10%) with 5% to 10% polyclonal plasma cells. The patient remains on therapy with continued improvement in her CLL.
Case report. (A) White blood cell (WBC) counts were performed by The Ohio State University Clinical Laboratory using Beckman Coulter LH. WBC count is expressed over time as absolute numbers in thousands of cells per microliter (K/μL). (B) IgM, IgA, and IgG production was detected by The Ohio State University Clinical Laboratory using the immunoturbidimetric method (Synchron LX System). Values are expressed as milligrams per deciliter.
Case report. (A) White blood cell (WBC) counts were performed by The Ohio State University Clinical Laboratory using Beckman Coulter LH. WBC count is expressed over time as absolute numbers in thousands of cells per microliter (K/μL). (B) IgM, IgA, and IgG production was detected by The Ohio State University Clinical Laboratory using the immunoturbidimetric method (Synchron LX System). Values are expressed as milligrams per deciliter.
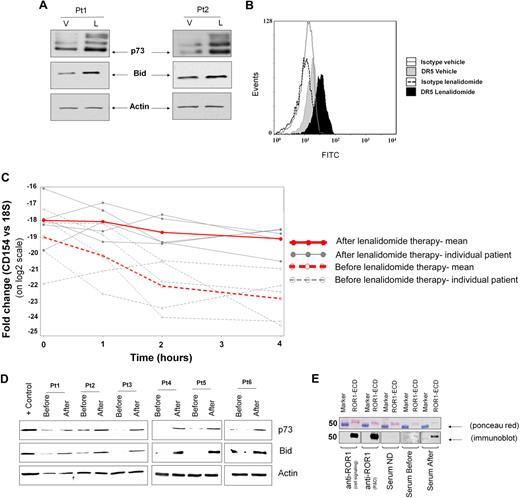
Lenalidomide induces CD154 transmembrane protein expression on CLL cells
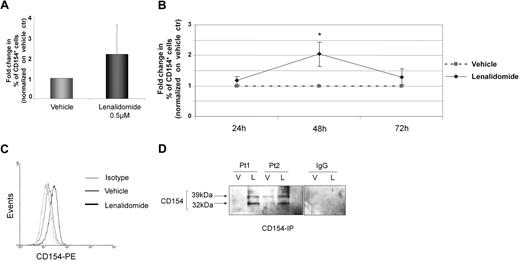
Normal B cells generally do not express CD154, whereas CLL cells have increased CD154 expression at an mRNA level35,36 but rarely express this at the protein level. We hypothesized that lenalidomide was promoting the production of polyclonal IgG and IgM in part through induction of functional CD154 surface expression on CLL cells, similar to that previously reported with CD154 gene therapy.15 Lenalidomide ex vivo treatment for 48 hours induces an increase in surface CD154 expression (P < .001) that was noted in 22 (73%) of the 30 patients examined (Figure 2A). The increase in CD154 was both time dependent (n = 5, P = .006, Figure 2B), with the optimal time of CD154 surface expression being 48 hours, and dose dependent (n = 7, P = .008 linear trend for CD154 expression as the dosage increases; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article), with up-regulation of CD154 in some patients at lenalidomide concentrations as low as 10nM but maximal at approximately 0.5 to 1μM as measured by flow cytometry at the 48-hour time point. Figure 2C demonstrates a representative patient with increased surface CD154 expression on CLL cells. CD154 expression on CLL cells was not from a soluble source, as demonstrated by lack of antigen loss with acid washing (data not shown) and the presence of both the 39-kDa and 32-kDa transmembrane domains of CD154 from lenalidomide-treated cells (n = 5 independent experiments, 2 representative patients are shown), but not vehicle control-treated cells, after biotinylated cell- surface protein pull down (Figure 2D).
Lenalidomide promotes time-dependent CD154 transmembrane protein expression. (A) CD19+ CLL cells were incubated with lenalidomide 0.5μM and vehicle control. CD154 expression was analyzed by flow cytometry after 48 hours of treatment. The graph shows fold change in percentage (%) of CD154+ cells in the lenalidomide-treated group compared with vehicle control (n = 30, P < .001). (B) CD19+ CLL cells were incubated with lenalidomide 0.5μM and vehicle control. CD154 expression was analyzed by flow cytometry after 24, 48, and 72 hours. The graph shows fold change in percentage (%) of CD154+ cells in the lenalidomide-treated group compared with vehicle control (n = 5, P = .006). (C) The panel shows 48-hour results for CD154 expression from a representative experiment. Black line denotes lenalidomide-treated cells and dark gray line denotes vehicle-treated CLL cells. Control IgG1k-PE is shown (light gray line). (D) CLL B cells were treated with vehicle (V) or 0.5μM lenalidomide (L) and after 48 hours, cell-surface proteins were biotinylated, and lysates were prepared. For each condition, 400 μg of protein lysate was subjected to immunoprecipitation using anti-CD154 or anti-IgG1 antibodies. The 39- and 32-kDa bands, corresponding to the 2 transmembrane forms of CD154, are indicated with arrows. Immunoblot shown is representative of 2 patient samples of 5 tested. Error bars represent SD. *Statistical significance.
Lenalidomide promotes time-dependent CD154 transmembrane protein expression. (A) CD19+ CLL cells were incubated with lenalidomide 0.5μM and vehicle control. CD154 expression was analyzed by flow cytometry after 48 hours of treatment. The graph shows fold change in percentage (%) of CD154+ cells in the lenalidomide-treated group compared with vehicle control (n = 30, P < .001). (B) CD19+ CLL cells were incubated with lenalidomide 0.5μM and vehicle control. CD154 expression was analyzed by flow cytometry after 24, 48, and 72 hours. The graph shows fold change in percentage (%) of CD154+ cells in the lenalidomide-treated group compared with vehicle control (n = 5, P = .006). (C) The panel shows 48-hour results for CD154 expression from a representative experiment. Black line denotes lenalidomide-treated cells and dark gray line denotes vehicle-treated CLL cells. Control IgG1k-PE is shown (light gray line). (D) CLL B cells were treated with vehicle (V) or 0.5μM lenalidomide (L) and after 48 hours, cell-surface proteins were biotinylated, and lysates were prepared. For each condition, 400 μg of protein lysate was subjected to immunoprecipitation using anti-CD154 or anti-IgG1 antibodies. The 39- and 32-kDa bands, corresponding to the 2 transmembrane forms of CD154, are indicated with arrows. Immunoblot shown is representative of 2 patient samples of 5 tested. Error bars represent SD. *Statistical significance.
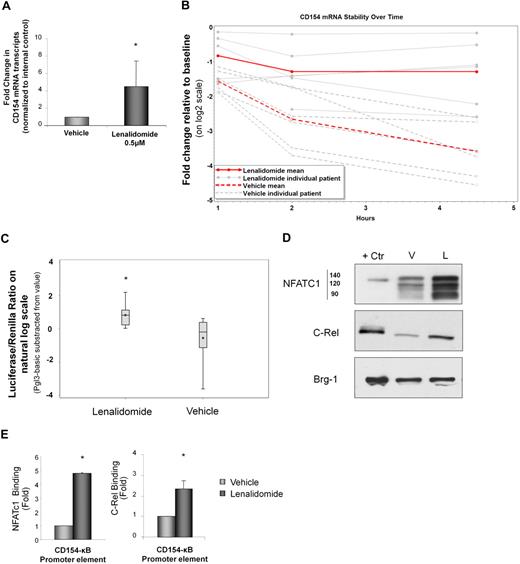
Lenalidomide increases RNA stability and transcription of CD154 in CLL cells
CD154 regulation in T cells has been shown to occur via increased as well as transcript stabilization (reviewed in Vavassori and Covey37 ). In CLL cells, lenalidomide induced CD154 expression at the mRNA level (Figure 3A; P < .001 compared with vehicle control), and this effect was dose dependent (supplemental Figure 2). This enhanced mRNA expression was in part through lenalidomide-induced CD154 mRNA stabilization, as demonstrated by a significant reduction in degradation after actinomycin D treatment (Figure 3B; P < .001 compared with vehicle control). Although increased CD154 gene transcript levels with lenalidomide treatment could be due solely to enhanced mRNA stability, increased transcriptional activation could also contribute. We therefore investigated this using luciferase constructs driven by the CD154 promoter. Lenalidomide significantly increased CD154-driven luciferase activity in primary CLL cells compared with vehicle control (Figure 3C; P = .001). This effect was concomitant with enhanced nuclear expression of c-Rel and NFATc (Figure 3D), and enhanced promoter binding of c-Rel (P < .001) and NFATc1 (P < .001) to the CD154 gene promoter as identified by chromatin immunoprecipitation assay (Figure 3E).
Lenalidomide increases RNA stability and transcription of CD154 in CLL cells. (A) CD154 mRNA level was measured by real-time reverse-transcription–PCR after 48 hours of treatment with lenalidomide 0.5μM or vehicle control (n = 20, P < .001). (B) After 48 hours with lenalidomide (0.5μM) or vehicle control, transcription was inhibited by the addition of ActD and 107 cells were collected for each time point over a 4.5-hour time course. The percentage of RNA remaining from each time point is plotted. Lenalidomide increases CD154 mRNA stability as demonstrated by significantly diminished decay after ActD treatment of CLL cells compared with vehicle control (n = 6, P = .002). (C) CLL B cells were transfected with CD154 luciferase reporter plasmid along with 1 μg of pCMV-renilla reporter vector or pGL3 empty reporter plasmid (pGL3-basic) as a negative control. After 48 hours with lenalidomide 0.5μM or vehicle control, luciferase activity was determined and corrected for transfection efficiency using renilla activity (n = 13, P = .001 for lenalidomide vs vehicle control). (D) Nuclear extracts from lenalidomide- or vehicle control-treated CLL cells were resolved on SDS–polyacrylamide gel electrophoresis and subjected to Western blotting with anti-NFATc1 or anti–c-Rel antibody. Nuclear protein Brg1 was used as an internal loading control. (E) Lenalidomide treatment of CLL B cells enhanced promoter binding of NFATc1 (n = 3, P < .001) and c-Rel (n = 3, P < .001) to the CD154 promoter by chromatin immunoprecipitation (ChIP). PCR to detect the CD154 promoter region (CD154-κB) was performed on the precipitated DNA (n = 3, P = .004). Error bars represent SD. *Statistical significance.
Lenalidomide increases RNA stability and transcription of CD154 in CLL cells. (A) CD154 mRNA level was measured by real-time reverse-transcription–PCR after 48 hours of treatment with lenalidomide 0.5μM or vehicle control (n = 20, P < .001). (B) After 48 hours with lenalidomide (0.5μM) or vehicle control, transcription was inhibited by the addition of ActD and 107 cells were collected for each time point over a 4.5-hour time course. The percentage of RNA remaining from each time point is plotted. Lenalidomide increases CD154 mRNA stability as demonstrated by significantly diminished decay after ActD treatment of CLL cells compared with vehicle control (n = 6, P = .002). (C) CLL B cells were transfected with CD154 luciferase reporter plasmid along with 1 μg of pCMV-renilla reporter vector or pGL3 empty reporter plasmid (pGL3-basic) as a negative control. After 48 hours with lenalidomide 0.5μM or vehicle control, luciferase activity was determined and corrected for transfection efficiency using renilla activity (n = 13, P = .001 for lenalidomide vs vehicle control). (D) Nuclear extracts from lenalidomide- or vehicle control-treated CLL cells were resolved on SDS–polyacrylamide gel electrophoresis and subjected to Western blotting with anti-NFATc1 or anti–c-Rel antibody. Nuclear protein Brg1 was used as an internal loading control. (E) Lenalidomide treatment of CLL B cells enhanced promoter binding of NFATc1 (n = 3, P < .001) and c-Rel (n = 3, P < .001) to the CD154 promoter by chromatin immunoprecipitation (ChIP). PCR to detect the CD154 promoter region (CD154-κB) was performed on the precipitated DNA (n = 3, P = .004). Error bars represent SD. *Statistical significance.
Lenalidomide-induced CD154 on CLL cells is functional
CD154 gene expression on CLL cells has been shown to reverse the T-cell defect and also result in the increased production of immunoglobulin by residual normal B cells.14,38 Given our case observation, we sought to determine whether lenalidomide-induced CD154 expression could cause normal B cells to generate IgM and IgG. To do this, normal B cells were stimulated with pokeweed mitogen and cocultured with CLL cells pretreated with lenalidomide (48 hours) followed by irradiation (Figure 4A-B). Under these conditions, B cells significantly increased their production of IgM (n = 5, P = .006) and IgG (n = 11, P = .001) relative to B cells cocultured with CLL cells pretreated with vehicle only. Furthermore, significant (n = 7, P = .014) normal B-cell proliferation was also observed in this experiment (supplemental Figure 3). These results were consistent with results from autologous T cells, a known positive costimulatory control.35
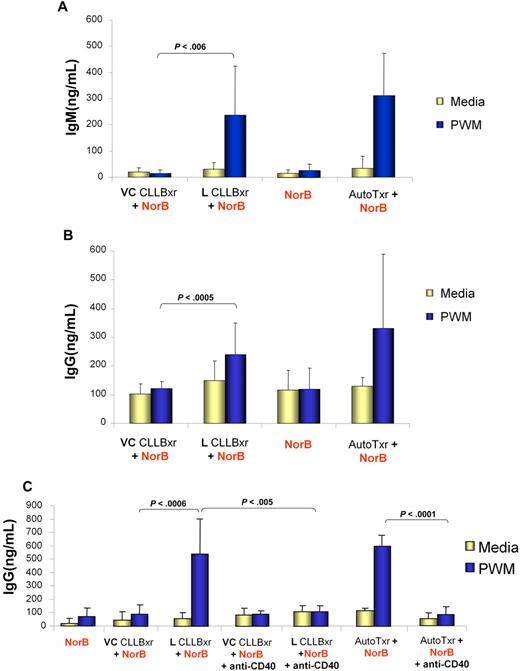
Lenalidomide-induced CD154 expression on CLL cells is functional. Lenalidomide (L)–treated but not vehicle (VC)–treated CLL B cells induce normal, target B cells to produce antibodies in the presence of PWM. A total of 5 × 104 autologous T cells (AutoTxr) from the healthy target B-cell donor, lenalidomide-treated CLL B cells (L CLLBxr), or vehicle control CLL B cells (VC CLLBxr) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM. Control conditions included 5 × 104 target, unirradiated B cells alone, with and without PWM, and 5 × 104 irradiated and unirradiated CLL B cells, in the absence of target B cells (data not shown). IgM (A) and IgG (B) production was assayed by ELISA from supernatants obtained after 7 days of coculture. (C) A total of 5 × 104 autologous T cells (AutoTxr) from the healthy target B-cell donor, lenalidomide-treated CLL B cells (L CLLBxr), or vehicle control CLL B cells (VC CLLBxr) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM and of a CD40 blocking antibody. Control conditions included 5 × 104 targets, unirradiated B cells alone, with and without PWM, and 5 × 104 irradiated and unirradiated CLL B cells, in the absence of target B cells (data not shown). IgM production was assayed by ELISA from supernatants obtained after 7 days of coculture. Error bars represent SD.
Lenalidomide-induced CD154 expression on CLL cells is functional. Lenalidomide (L)–treated but not vehicle (VC)–treated CLL B cells induce normal, target B cells to produce antibodies in the presence of PWM. A total of 5 × 104 autologous T cells (AutoTxr) from the healthy target B-cell donor, lenalidomide-treated CLL B cells (L CLLBxr), or vehicle control CLL B cells (VC CLLBxr) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM. Control conditions included 5 × 104 target, unirradiated B cells alone, with and without PWM, and 5 × 104 irradiated and unirradiated CLL B cells, in the absence of target B cells (data not shown). IgM (A) and IgG (B) production was assayed by ELISA from supernatants obtained after 7 days of coculture. (C) A total of 5 × 104 autologous T cells (AutoTxr) from the healthy target B-cell donor, lenalidomide-treated CLL B cells (L CLLBxr), or vehicle control CLL B cells (VC CLLBxr) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM and of a CD40 blocking antibody. Control conditions included 5 × 104 targets, unirradiated B cells alone, with and without PWM, and 5 × 104 irradiated and unirradiated CLL B cells, in the absence of target B cells (data not shown). IgM production was assayed by ELISA from supernatants obtained after 7 days of coculture. Error bars represent SD.
To confirm that increased antibody production by normal B cells was induced by the expression of CD154 on CLL cells, we first determined whether there was a correlation of CD154 induction on CLL cells with production of IgG and IgM. A significant correlation of increase in CD154 expression with lenalidomide and rise in IgM synthesis was observed (Pearson correlation coefficient; r = 0.94, P = .036). A similar finding correlating increase in CD154 expression on CLL cells and rise in IgG synthesis was observed (r = 0.93, P = .001). To provide additional evidence that the interaction between CD154 (on lenalidomide-treated CLL cells) and CD40 (on normal B cells) has a fundamental role in stimulating normal B cells to secrete immunoglobulin, we repeated these costimulatory studies including a CD40 blocking antibody. Treatment of normal B cells with a blocking CD40 antibody dramatically suppressed the production of IgM by these cells in the presence of lenalidomide-treated CLL cells (Figure 4C, n = 6, P = .001). Collectively, these data demonstrate that the lenalidomide-induced surface expression of CD154 on CLL cells is functional and can promote immunoglobulin production by normal B cells. In addition, the increased IgG production also suggests CD154-mediated class switching is occurring in normal pokeweed mitogen–stimulated B cells after coculture with lenalidomide-treated CLL cells or normal T cells.
Lenalidomide-induced NF-κB activity, CD154 induction, CD154 mRNA stabilization, and costimulatory properties require PI3-kinase pathway activation
The phosphoinositide-3 (PI3)–kinase/AKT axis is a common proximal signaling pathway by which nuclear factor κB (NF-κB) is activated in normal B cells.39 Figure 5A demonstrates that lenalidomide treatment of CLL cells promoted AKT phosphorylation. This kinase also phosphorylates and activates the IKK complex.40 In Figure 5B and C, we demonstrate such activation of IKK, together with increased phosphorylation of the NF-κB inhibitor IκB and its degradation after treatment with lenalidomide. Consistent with this, pretreatment of CLL cells with PI3-kinase signaling pathway inhibitors prior to lenalidomide treatment prevented lenalidomide-induced CD154 mRNA transcription (Figure 5D). Furthermore, pretreatment of CLL cells with PI3-kinase pathway inhibitors also prevented lenalidomide-induced CD154 mRNA stabilization (Figure 5D) and abrogates IgM production of normal B cells cocultured with lenalidomide-treated CLL cells (Figure 5E). Collectively, these data suggest that lenalidomide-mediated activation of the PI3-kinase pathway is essential for downstream NF-κB activation, CD154 mRNA stability, and CLL cell costimulatory activity.
Lenalidomide-induced up-regulation of CD154 is dependent on PI3-kinase. (A) CLL cells were treated with vehicle or 0.5μM lenalidomide for 8 hours. Cell lysates were subjected to immunoblot analysis. (B) IKK-β was immunoprecipitated from cell lysate derived from vehicle or 0.5μM lenalidomide-treated CLL samples and tested for the capability to phosphorylate the GST-IκBα substrate in a kinase buffer containing 20μM ATP and 10 μCi (0.37 MBq) of [γ32P]ATP. The left panel shows a representative experiment of the IKK kinase activity. The graph shows fold change in IKKβ activity in the lenalidomide-treated group compared with vehicle control (n = 8, P = .01). (C) IKK substrate IκB is degraded after lenalidomide treatment. The panel shows 3 representative CLL patients treated with 0.5μM lenalidomide or the vehicle control for 12 hours. (D) After 48 hours with lenalidomide 0.5μM and vehicle control with and without 25μM LY294002, transcription was inhibited by the addition of ActD and 107 cells were collected for each time point over a 4.5-hour time course. The percentage of RNA remaining from each time point is plotted (n = 4, P = .045). (E) A total of 5 × 104 lenalidomide-treated CLL B cells (L) or vehicle control CLL B cells (VC) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM and 25μM LY294002. IgM production was assayed by ELISA from supernatants obtained after 7 days of coculture (n = 8, P = .001). Error bars represent SD.
Lenalidomide-induced up-regulation of CD154 is dependent on PI3-kinase. (A) CLL cells were treated with vehicle or 0.5μM lenalidomide for 8 hours. Cell lysates were subjected to immunoblot analysis. (B) IKK-β was immunoprecipitated from cell lysate derived from vehicle or 0.5μM lenalidomide-treated CLL samples and tested for the capability to phosphorylate the GST-IκBα substrate in a kinase buffer containing 20μM ATP and 10 μCi (0.37 MBq) of [γ32P]ATP. The left panel shows a representative experiment of the IKK kinase activity. The graph shows fold change in IKKβ activity in the lenalidomide-treated group compared with vehicle control (n = 8, P = .01). (C) IKK substrate IκB is degraded after lenalidomide treatment. The panel shows 3 representative CLL patients treated with 0.5μM lenalidomide or the vehicle control for 12 hours. (D) After 48 hours with lenalidomide 0.5μM and vehicle control with and without 25μM LY294002, transcription was inhibited by the addition of ActD and 107 cells were collected for each time point over a 4.5-hour time course. The percentage of RNA remaining from each time point is plotted (n = 4, P = .045). (E) A total of 5 × 104 lenalidomide-treated CLL B cells (L) or vehicle control CLL B cells (VC) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM and 25μM LY294002. IgM production was assayed by ELISA from supernatants obtained after 7 days of coculture (n = 8, P = .001). Error bars represent SD.
Lenalidomide therapy induces a CD154 gene therapy phenotype on CLL cells
In addition to promoting antibody production on normal B cells, CD154 gene therapy results in the increased expression of BID and DR5 concomitant with sensitization to TRAIL.17 In addition, ex vivo CD40 ligation has been shown to activate p73 and enhance fludarabine-mediated killing even in the absence of functional p53.16 Lenalidomide treatment of CLL cells demonstrates similar increases in protein expression of BID, DR5, and p73 (Figure 6A-B). As with preclinical models of CD154 gene therapy,16,17 lenalidomide pretreatment of CLL cells for 5 days enhanced TRAIL- and fludarabine-mediated killing of CLL cells (P < .001 and P = .027, respectively; data not shown). The relevance of this observation in patients was indicated by serial samples from representative CLL patients receiving lenalidomide, in which we observed similar increases in CD154 mRNA stabilization (Figure 6C, n = 5, P = .045 compared with untreated control). BID and p73 induction was also noted in 6 of 9 patients examined (Figure 6D). In addition, our case report patient (Figure 1), the only subject who did not receive rituximab B cell–depleting therapy prior to lenalidomide, had evidence of development of a measurable ROR1 antibody (Figure 6E) after lenalidomide treatment for 6 months. This finding was also confirmed with a ROR1-specific ELISA (data not shown). Together, these findings indicate that lenalidomide reproduces the CLL phenotype of CD154 gene therapy in CLL and can promote an in vivo tumor humoral response to tumor cells.
Lenalidomide promotes CD154 gene therapy phenotype. (A) Whole-cell extracts from 48-hour–treated CLL B cells were analyzed by immunoblot for p73 and BID. Actin was used as internal loading control. (B) Representative histograms show expression of DR5 measured by flow cytometry in CLL B cells cultured without (filled gray) or with (filled black) lenalidomide. The gray- and black-lined open histograms correspond to CLL B cells cultured, respectively, with vehicle or lenalidomide and stained with an isotype monoclonal antibody of irrelevant specificity. (C) Peripheral blood mononuclear cells from before and day 8 of therapy were cultured in the presence of ActD and 107 cells were collected over a 4-hour time course as in Figure 3B (n = 5, P < .05). (D) Whole-cell extracts from patients' peripheral blood mononuclear cells before the dose and on day 8 were analyzed by immunoblot for p73 and BID (representative findings in 6 of 9 patients). (E) Purified recombinant human-ROR1 was run in each lane of an SDS-PAGE gel and then transferred to nitrocellulose membrane. Ponceau red staining was performed to assess both purity and equal loading. The membrane was cut into pieces and probed with rabbit anti–human ROR1 (Cell Signaling), goat anti–human ROR1 (R&D), healthy donor serum (ND), or serum from the index case immediately before lenalidomide (before) and on month 6 of therapy (after). The pieces were incubated with HRP-conjugated anti–rabbit, anti–goat, or anti–human IgG.
Lenalidomide promotes CD154 gene therapy phenotype. (A) Whole-cell extracts from 48-hour–treated CLL B cells were analyzed by immunoblot for p73 and BID. Actin was used as internal loading control. (B) Representative histograms show expression of DR5 measured by flow cytometry in CLL B cells cultured without (filled gray) or with (filled black) lenalidomide. The gray- and black-lined open histograms correspond to CLL B cells cultured, respectively, with vehicle or lenalidomide and stained with an isotype monoclonal antibody of irrelevant specificity. (C) Peripheral blood mononuclear cells from before and day 8 of therapy were cultured in the presence of ActD and 107 cells were collected over a 4-hour time course as in Figure 3B (n = 5, P < .05). (D) Whole-cell extracts from patients' peripheral blood mononuclear cells before the dose and on day 8 were analyzed by immunoblot for p73 and BID (representative findings in 6 of 9 patients). (E) Purified recombinant human-ROR1 was run in each lane of an SDS-PAGE gel and then transferred to nitrocellulose membrane. Ponceau red staining was performed to assess both purity and equal loading. The membrane was cut into pieces and probed with rabbit anti–human ROR1 (Cell Signaling), goat anti–human ROR1 (R&D), healthy donor serum (ND), or serum from the index case immediately before lenalidomide (before) and on month 6 of therapy (after). The pieces were incubated with HRP-conjugated anti–rabbit, anti–goat, or anti–human IgG.
Discussion
Several decades of laboratory research have characterized the profound immune defect that exists in CLL. Investigators have tried a variety of therapeutic approaches to reverse this defect,41-45 the most successful of which has been the introduction of CD154 to CLL cells via gene therapy. Although CD154 gene therapy has generated promising preliminary results, this therapeutic approach is cumbersome, expensive, and not adaptable to extended administration. Herein, we demonstrate the development of polyclonal hypergammaglobulinemia with increasing titers of ROR1 tumor-specific antibodies in a CLL patient receiving lenalidomide, providing in vivo human data that this agent can reverse humoral tumor tolerance. We then show that lenalidomide promotes up-regulation of CD154 antigen on the surface of CLL cells at concentrations readily attainable in the clinic. The increase in CD154 antigen expression on CLL cells with lenalidomide treatment leads to biologic responses similar to those observed with CD154 gene therapy, including increased IgM and IgG production by normal B cells and up-regulation of BIM, DR5, and p73. Consistent with these events, lenalidomide enhances TRAIL- and fludarabine-mediated cytotoxicity. We also show that the CD154 gene therapy phenotype, including increased CD154 transcription and mRNA stabilization, increased immunoglobulin production, and anti-ROR1 antibodies, can be observed in CLL patients receiving lenalidomide. The relevance of these results to the therapy of CLL and related diseases is significant. However, to fully appreciate the benefit of this application, new strategies of lenalidomide administration and combination strategies that build upon previously reported observations with CD154 gene therapy-based approaches14,38 will be required.
Our studies demonstrate CD154 induction is functional in CLL cells, and also detail 2 distinct convergent mechanisms by which lenalidomide increases CD154 gene expression on CLL cells. Lenalidomide enhances mRNA gene transcription with increased binding of the activating c-REL/NFATc1 complex to the CD154 promoter, and also enhances CD154 promoter activity in primary CLL cells similar to that observed in lymphoma.46 In addition, lenalidomide also enhances stabilization of the mRNA transcript as observed in T cells.37 Although CD154 mRNA stabilization is known to lead to increased protein expression in activated T cells,37 this effect has not been previously reported in normal or transformed B cells. In T cells, mRNA transcript stabilization involves a variety of proteins including HnRNP, polypyrimidine tract–binding protein, and HuR. The time course of CD154 surface expression and loss at 72 hours seen with lenalidomide is similar to what has previously been observed with T cells.47 Studies examining the ability of lenalidomide or related drugs to stabilize RNA are limited, although in colon cancer cell lines, thalidomide has been shown to diminish COX2 mRNA stability via shuttling of HuR.48 Further study of the mechanism of CD154 mRNA stabilization by lenalidomide in CLL and the kinetics of this process warrant further study that is ongoing.
Notable to these 2 mechanisms of CD154 up-regulation is the dependence on the PI3-kinase signaling pathway. Our data demonstrate that lenalidomide enhances PI3-kinase pathway activity as measured by increased phosphorylation of AKT and also downstream IKK. Disruption of PI3-kinase signaling with biochemical inhibitors prevents up-regulation of CD154 through both the transcriptional and the mRNA stabilization mechanisms. PI3-kinase inhibition also prevents B-cell costimulatory activity and subsequent increased production of IgM and IgG by normal B cells. How lenalidomide activates the PI3-kinase/AKT pathway in transformed CLL cells is of great interest. In transformed myeloid cells, lenalidomide indirectly inhibits the proapoptotic phosphatase PP2A, which can prevent dephosphorylation of AKT.49 We demonstrate that lenalidomide has similar PP2A inhibitory activity in primary CLL cells, as measured by direct phosphatase activity assays in CLL cells (supplemental Figure 4A) and results in increased AKT phosphorylation (supplemental Figure 4B). The PP2A activating agent, 1,9-dideoxyforskolin, when coincubated with lenalidomide, antagonizes lenalidomide-induced CD154 mRNA and protein (supplemental Figure 4C-D). In addition to PP2A, other mechanisms activating the PI3-kinase/AKT pathway may contribute to CD154 induction. Given the divergent source of activation of PI3-kinase pathway through catalytic domain isoforms, upstream targets directly influenced by lenalidomide might be identified. Studies elucidating the mechanism(s) by which the PI3-kinase pathway is activated by lenalidomide are ongoing.
Our data provide rationale for novel combination treatment strategies with lenalidomide using principles from the previous CD154 gene therapy experience in CLL. The observation that lenalidomide can reverse humoral tolerance to known tumor-specific antigens such as ROR1, which is selectively expressed in CLL and is under investigation as a target for therapeutic monoclonal antibodies,50 is especially intriguing. We hypothesize that the reason our index case was the only one to see recovery of immunoglobulins and development of an anti-ROR1 antibody was due to all of the other subjects having received prior rituximab that depletes normal B cells. A preliminary study of lenalidomide in previously untreated CLL patients by the M. D. Anderson Cancer Center has demonstrated that an overall increase in immunoglobulin levels is observed with extended treatment.51 This finding is supportive of our hypothesis that normal B cells are required to observe this effect. Prospective studies evaluating the relationship of induction of CD154 on CLL cells by lenalidomide in previously untreated patients with subsequent recovery of immunoglobulin levels after lenalidomide treatment are warranted. Given that many CLL therapies deplete normal B cells, application of lenalidomide early in the course of the disease in combination with targeted therapies that spare normal immune cells may be warranted to optimize lenalidomide-induced tumor humoral response. This will be particularly true with anti-CD20 antibodies that deplete both normal and tumor B cells if the humoral B-cell contribution to lenalidomide's tumor mechanism of action in CLL. In addition, these data point to the potential role of this agent to reverse the hypogammaglobulinemia commonly present in CLL that contributes significantly to morbidity from infection. Given the broad activity that lenalidomide has shown across many different B-cell malignancies, it will be vital to explore this agent's ability to reverse humoral tumor tolerance in other diseases and also to enhance vaccine-based approaches. Indeed, given the diverse mechanisms of action of lenalidomide in CLL and other diseases, clinical trials incorporating detailed laboratory correlative studies will be required to attain the full potential of this exciting agent.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Cancer and Leukemia Group B Foundation, D. Warren Brown Foundation, Specialized Center of Research from the Leukemia & Lymphoma Society, and P50-CA140158, 1K12 CA133250, P01 CA95426, and P01 CA101956 from the National Cancer Institute.
L.A. and A.J.J. are Paul Calabresi Scholars. R.B. is a Howard Hughes Medical Institute Research Training Fellow.
National Institutes of Health
Howard Hughes Funding
Authorship
Contribution: R.L. planned the research, performed experiments, analyzed data, drafted the first and subsequent drafts of the paper, and approved the final version of the paper; L.A. planned the research, led the CLL clinical trial, reviewed drafts of the paper, and approved the final version of the paper; Q.L., S.E.M., R.B., C.A.R., L.L.S., A.R., and N.M. were involved in planning components of the research, performed experiments, reviewed drafts, and approved the final version of the paper; K.A.B. planned the research, accrued patients to the CLL clinical trial, reviewed drafts of the paper, and approved the final version of the paper; W.B. wrote the clinical trial, planned the research, reviewed drafts of the paper, and approved the final version of the paper; A.L., X.M., and D.J. were involved in planning components of the research, performed all the statistical analysis, reviewed drafts, and approved the final version of the paper; C.-S.C., R.F., L.V.P., and C.R. were involved in making necessary reagents essential to the hypothesis of this paper, reviewed drafts, and approved the final version of the paper; and A.J.J. and J.C.B. planned every aspect of the proposal, supervised the research, analyzed data, reviewed drafts, obtained funding for the research work, and approved the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Research funding was received from Celgene for the clinical trial described in this paper.
Correspondence: John C. Byrd, B302 Starling-Loving Hall, 320 W 10th Ave, Columbus, OH 43210; e-mail: john.byrd@osumc.edu; or Amy J. Johnson, OSUCCC Bldg, Rm 455C, 410 W 12th Ave, Columbus, OH 43210; e-mail: amy.johnson@osumc.edu.
References
Author notes
A.J.J. and J.C.B. contributed equally to this work.

![Figure 5. Lenalidomide-induced up-regulation of CD154 is dependent on PI3-kinase. (A) CLL cells were treated with vehicle or 0.5μM lenalidomide for 8 hours. Cell lysates were subjected to immunoblot analysis. (B) IKK-β was immunoprecipitated from cell lysate derived from vehicle or 0.5μM lenalidomide-treated CLL samples and tested for the capability to phosphorylate the GST-IκBα substrate in a kinase buffer containing 20μM ATP and 10 μCi (0.37 MBq) of [γ32P]ATP. The left panel shows a representative experiment of the IKK kinase activity. The graph shows fold change in IKKβ activity in the lenalidomide-treated group compared with vehicle control (n = 8, P = .01). (C) IKK substrate IκB is degraded after lenalidomide treatment. The panel shows 3 representative CLL patients treated with 0.5μM lenalidomide or the vehicle control for 12 hours. (D) After 48 hours with lenalidomide 0.5μM and vehicle control with and without 25μM LY294002, transcription was inhibited by the addition of ActD and 107 cells were collected for each time point over a 4.5-hour time course. The percentage of RNA remaining from each time point is plotted (n = 4, P = .045). (E) A total of 5 × 104 lenalidomide-treated CLL B cells (L) or vehicle control CLL B cells (VC) were irradiated (20 Gy) and placed in culture with 5 × 104 targets, purified B cells from the healthy donor, in the absence or presence of PWM and 25μM LY294002. IgM production was assayed by ELISA from supernatants obtained after 7 days of coculture (n = 8, P = .001). Error bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/13/10.1182_blood-2009-09-242438/4/m_zh89990947830005.jpeg?Expires=1769128579&Signature=xxekknI26aL9mw5kcrQl~z94L1j9ElUt8bsglFyDHpgbldQ5HMqVUndK35NKWfPLa61Y2-8jJAQJlLlQiICuDd1ANQBeKCjAN71VqVuni6UofT-vldgtdC8DsiP4Ezle8E2wUhmiO3O24jW7eFzvB5m4jN~JR~-M0x~8XEbsbFa651ic4HTs~hkBHCseEoYbqmxgQ~Pnaexpi997Eqt7OGPbhGBIQb0bPVO-yakSdeZVAmYUFtlbostxUwBppsfZadB9UEe7cPlAnH-3hApm2W7FRGOelEM~yL9SucU2A-Ox89yhjoQqu1oVBdTdEavzVaZRigJmAwMb4-M125-glQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)