Abstract
Acute myeloid leukemia (AML) may follow a JAK2-positive myeloproliferative neoplasm (MPN), although the mechanisms of disease evolution, often involving loss of mutant JAK2, remain obscure. We studied 16 patients with JAK2-mutant (7 of 16) or JAK2 wild-type (9 of 16) AML after a JAK2-mutant MPN. Primary myelofibrosis or myelofibrotic transformation preceded all 7 JAK2-mutant but only 1 of 9 JAK2 wild-type AMLs (P = .001), implying that JAK2-mutant AML is preceded by mutation(s) that give rise to a “myelofibrosis” phenotype. Loss of the JAK2 mutation by mitotic recombination, gene conversion, or deletion was excluded in all wild-type AMLs. A search for additional mutations identified alterations of RUNX1, WT1, TP53, CBL, NRAS, and TET2, without significant differences between JAK2-mutant and wild-type leukemias. In 4 patients, mutations in TP53, CBL, or TET2 were present in JAK2 wild-type leukemic blasts but absent from the JAK2-mutant MPN. By contrast in a chronic-phase patient, clones harboring mutations in JAK2 or MPL represented the progeny of a shared TET2-mutant ancestral clone. These results indicate that different pathogenetic mechanisms underlie transformation to JAK2 wild-type and JAK2-mutant AML, show that TET2 mutations may be present in a clone distinct from that harboring a JAK2 mutation, and emphasize the clonal heterogeneity of the MPNs.
Introduction
The myeloproliferative neoplasms (MPNs) comprise a group of clonal stem cell disorders associated with a high prevalence of mutations in JAK2, overproduction of mature blood elements, and variable rates of transformation to acute myeloid leukemia (AML).1,2 AML transformation is seen in 2% to 5% of patients with essential thrombocythemia (ET) or polycythemia vera (PV)3,4 and 15% to 30% of those with primary myelofibrosis (PMF).5 Although rates of leukemic transformation are increased after genotoxic therapy, AML also occurs in treatment-naive patients,4 indicating that leukemic transformation is part of the natural history of these disorders. Of note, recent studies have shown a role for the JAK2 V617F mutation in promoting the generation of clones bearing additional genetic damage, suggesting that disease evolution may be driven in part by expression of mutant JAK2.6,7
In striking contrast to chronic myeloid leukemia, in which the blastic phase of disease invariably harbors a BCR-ABL1 fusion, AML transformation after a JAK2 V617F-positive MPN commonly lacks the JAK2 mutation. The first hint of this phenomenon came from the observation that the JAK2 V617F mutation was less common in patients with AML transformation compared with patients with PV or PMF.8 Loss of the JAK2 mutation at leukemic transformation was subsequently shown by analysis of paired MPN and AML samples.9,10 There are several models to explain the development of JAK2 wild-type leukemia. One potential explanation would be loss of the mutant JAK2 allele as a consequence of mitotic recombination. This was not detected in 3 patients transforming to JAK2 wild-type AML,9,10 but, given the small number of patients analyzed, it remains possible that mitotic recombination accounts for a proportion of such cases. Moreover, the previous reports were unable to exclude gene conversion or intrachromosomal deletion. A second model invokes the concept of a “pre-JAK2” phase of disease. This idea is supported by circumstantial evidence, including the familial predisposition to MPN,11,12 clonality studies in female patients with MPN,13,14 and growth of V617F-negative endogenous erythroid colonies from V617F-positive patients.15,16 More recently direct evidence has been reported that the JAK2 mutation may be preceded by a 20q deletion17 or a TET2 mutation18 in some patients, but the frequency of this occurrence remains unclear. A third model that could explain the development of JAK2 wild-type AML would be the coexistence of independent JAK2-mutant and JAK2 wild-type clonal expansions in the same patient. This phenomenon has been documented recently, and, in 2 patients informative for X-linked polymorphic loci, the 2 clones were shown to have arisen independently.19
At the time of AML transformation, blood and bone marrow samples contain cells representing the leukemic process as well as others derived from the preceding MPN. Molecular analyses therefore require careful purification of the leukemic blasts and/or analysis of clonally derived progenitor colonies. We have used these approaches to undertake a detailed study of 16 patients with AML after a JAK2-mutant MPN. Our results show that JAK2-mutant AML, but not JAK2 wild-type AML, usually arises from a preceding myelofibrotic phase. Moreover, although TET2 mutations precede JAK2 mutations in some patients, in others the 2 mutations are present in distinct clones.
Methods
Patients and samples
Comprehensive clinical information and samples of blood or bone marrow were obtained from patients with AML after a JAK2-mutant MPN. Patients were diagnosed with ET, PV, PMF, or refractory anemia with ringed sideroblasts and thrombocytosis (RARS-T) according to standard criteria.20 Diagnostic criteria for myelofibrotic transformation of ET/PV were as previously described21 and required at least grade 3 reticulin fibrosis in a bone marrow biopsy together with at least 2 of the following: increase in spleen size of at least 3 cm, unexplained decrease in hemoglobin by at least 20 g/L, immature myeloid or erythroid cells in the blood smear, tear-drop poikilocytes in the blood smear, or B symptoms (night sweats, bone pain, or weight loss of more than 10% in 6 months). Diagnosis of AML transformation required the presence of at least 20% blasts in the blood or bone marrow.20
Samples of leukemic blasts and constitutional material (derived from buccal swabs or T cells) were available from all 16 patients. Samples from the MPN phase of disease (Table 1) and/or individual progenitor colonies (Table 3) were also available from a proportion of patients. Leukemic blasts were purified when necessary by density centrifugation (Lymphoprep; Axis-Shield), collection of mononuclear cells, and magnetic isolation of CD34+ cells (magnetic-activated cell sorting; Miltenyi Biotec). Constitutional material representative of germline DNA was obtained from buccal swabs (Epicentre), or magnetically isolated CD2+ T cells (Dynabeads; Dynal AS). Granulocytes were purified by density centrifugation. Leukemic blasts and granulocytes were at least 90% pure by morphologic criteria. T-cell purity was also 90% or greater, as assessed by visualizing the proportion of cells directly bound to magnetic beads according to the manufacturer's instructions. Culture of progenitor colonies was performed as previously described.22 Individual colonies were lysed in RLT buffer (QIAGEN), and DNA was precipitated by the addition of isopropanol followed by centrifugation. All patients gave written informed consent for the research, which was approved by the Multi-Region Ethics Committee (Fulbourn) for the Causes of Clonal Haematological Disorders project, which permits the use of biologic samples and clinical data from patients with clonal hematologic disorders, and was carried out in accordance with the principles of the Declaration of Helsinki.
Loss of heterozygosity and AML-associated genetic alterations
Genotyping of single nucleotide polymorphisms was performed by pyrosequencing (PV cohort) or direct sequencing (AML cohort). The JAK2 mutation status of leukemic blasts was assessed by direct sequencing; samples in which the mutant allele was not detected were classified as JAK2 wild-type, those with a 1:1 ratio of mutant to wild-type were classified as heterozygous, and those in which the wild-type allele was not detected or was less than 10% of the mutant allele were classified as homozygous. Mutations in N/KRAS (codons 12, 13, and 61), CEBPA (exon 1), RUNX1 (all coding exons), GATA2 (exon 4), NPM (exon 12), WT1 (exons 7 and 9), TP53 (exons 4-8), CBL (exons 8 and 9), and TET2 (all coding exons) were assessed by direct sequencing. FLT3-ITD, RUNX1-RUNX1T1, and CBFB-MYH11 alterations were assessed by polymerase chain reaction (PCR). Relative levels of JAK2, EVI1, and WT1 transcripts were measured in CD34+ leukemic blasts, and nonmobilized peripheral blood CD34+ cells from 4 healthy donors by SYBR green real-time PCR assays by using the ΔΔCt formula with ABL1 as the housekeeping gene. Increased transcript levels were defined as greater than mean plus 2 standard deviations of the normal controls. JAK2 copy number was assessed by real-time PCR, using a previously reported method23 modified to include a second control region on chromosome 1p.
Statistical analysis
Comparisons of diagnostic variables between patients were performed by using the t test for continuous variables and the Fisher exact test (for 2 × 2 tables).
Results
Association between JAK2 mutation status of leukemic blasts and route of leukemic transformation
Sixteen patients with AML and evidence of a preceding JAK2-mutant MPN (15 JAK2 V617F and 1 JAK2 exon 12) were studied (Table 1). To ascertain the JAK2 mutational status of each AML, it was important to acquire a sample of leukemic blasts free from contamination by the preceding MPN. Direct sequencing of leukemic blasts (≥ 90% pure) showed 6 JAK2 V617F-homozygous, 1 JAK2 V617F-heterozygous, and 9 JAK2 wild-type leukemias, the latter group included the single patient with a preceding JAK2 exon 12 mutation (Table 1; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Features of 16 patients with progression to acute leukemia after a JAK2-mutant MPN
| Pt . | Myeloproliferative neoplasm . | At transformation to AML . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis, y/sex . | MPN . | JAK2 mutation status . | Therapy . | Disease duration, y . | Sample from MPN phase . | JAK2 mutation status . | Cytogenetic analysis . | Additional mutations . | Altered gene expression . | ||
| 1 | 63/M | PMF | JAK2 V617F | HC, AN | 2 | BM at diagnosis | Het | 3 copies AML1* | NRAS G12S, RUNX1 D171N | — | |
| 2 | 47/M | ET->MF | JAK2 V617F | HC | 16 | Grans 1 y pre-AML | Hom | inv(3), del(7) | NRAS G12D | ↑WT1, ↑EVI1 | |
| 3 | 47/F | PV->MF | JAK2 V617F | HC, IFN | 10 | Grans 1 y pre-AML | Hom | ND | RUNX1 N119K | ↑WT1 | |
| 4 | 58/M | PMF | JAK2 V617F | none | 1 | None | Hom | ND | RUNX1 G138V | ↑WT1 | |
| 5 | 70/M | Probable PMF | JAK2 V617F | none | NA | NA | Hom | add(8), del(7q) | RUNX1 D171N | ↑WT1 | |
| 6 | 72/M | PMF | JAK2 V617F | HC | 8 | None | Hom | ND | — | ↑WT1, ↑JAK2 | |
| 7 | 67/M | ET->MF | JAK2 V617F | HC | 6 | Grans 1.5 y pre-AML | Hom | der5, t(5;17) | TP53 R248Q | ↑WT1 | |
| 8 | 43/M | PV | JAK2 N542-E543del | BU, P32, HC | 26 | None | Wild-type | del(20q) | TET2 D1242V, RUNX1 H215X | ↑WT1 | |
| 9 | 79/M | PMF | JAK2 V617F | HC | 3 | Mk 2 y pre-AML | Wild-type | ND | CBL L380P | ↑WT1 | |
| 10 | 75/F | PV | JAK2 V617F | HC | 12 | None | Wild-type | ND | FLT3-ITD, RUNX1 Q127X | NS | |
| 11 | 64/F | PV | JAK2 V617F | HC | 14 | Blood 1 y pre-AML | Wild-type | ND | TP53 S90X | NS | |
| 12 | 76/M | RARS-T | JAK2 V617F | HC | 8 | None | Wild-type | Complex† | TP53 V173M | ↑WT1 | |
| 13 | 67/M | PV | JAK2 V617F | BU, P32, HC | 26 | Blood 1 y pre-AML | Wild-type | ND | TP53 C238F | ↑WT1 | |
| 14 | 65/M | PV | JAK2 V617F | HC | 10 | Blood 2 y pre-AML | Wild-type | Complex‡ | — | ↑WT1 | |
| 15 | 58/F | PV | JAK2 V617F | HC | 3 | Grans 2 y pre-AML | Wild-type | add(9) | TET2 P1549X | — | |
| 16 | 76/F | ET | JAK2 V617F | HC, BU | 10 | Blood 0.5 y pre-AML | Wild-type | ND | — | NS | |
| Pt . | Myeloproliferative neoplasm . | At transformation to AML . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis, y/sex . | MPN . | JAK2 mutation status . | Therapy . | Disease duration, y . | Sample from MPN phase . | JAK2 mutation status . | Cytogenetic analysis . | Additional mutations . | Altered gene expression . | ||
| 1 | 63/M | PMF | JAK2 V617F | HC, AN | 2 | BM at diagnosis | Het | 3 copies AML1* | NRAS G12S, RUNX1 D171N | — | |
| 2 | 47/M | ET->MF | JAK2 V617F | HC | 16 | Grans 1 y pre-AML | Hom | inv(3), del(7) | NRAS G12D | ↑WT1, ↑EVI1 | |
| 3 | 47/F | PV->MF | JAK2 V617F | HC, IFN | 10 | Grans 1 y pre-AML | Hom | ND | RUNX1 N119K | ↑WT1 | |
| 4 | 58/M | PMF | JAK2 V617F | none | 1 | None | Hom | ND | RUNX1 G138V | ↑WT1 | |
| 5 | 70/M | Probable PMF | JAK2 V617F | none | NA | NA | Hom | add(8), del(7q) | RUNX1 D171N | ↑WT1 | |
| 6 | 72/M | PMF | JAK2 V617F | HC | 8 | None | Hom | ND | — | ↑WT1, ↑JAK2 | |
| 7 | 67/M | ET->MF | JAK2 V617F | HC | 6 | Grans 1.5 y pre-AML | Hom | der5, t(5;17) | TP53 R248Q | ↑WT1 | |
| 8 | 43/M | PV | JAK2 N542-E543del | BU, P32, HC | 26 | None | Wild-type | del(20q) | TET2 D1242V, RUNX1 H215X | ↑WT1 | |
| 9 | 79/M | PMF | JAK2 V617F | HC | 3 | Mk 2 y pre-AML | Wild-type | ND | CBL L380P | ↑WT1 | |
| 10 | 75/F | PV | JAK2 V617F | HC | 12 | None | Wild-type | ND | FLT3-ITD, RUNX1 Q127X | NS | |
| 11 | 64/F | PV | JAK2 V617F | HC | 14 | Blood 1 y pre-AML | Wild-type | ND | TP53 S90X | NS | |
| 12 | 76/M | RARS-T | JAK2 V617F | HC | 8 | None | Wild-type | Complex† | TP53 V173M | ↑WT1 | |
| 13 | 67/M | PV | JAK2 V617F | BU, P32, HC | 26 | Blood 1 y pre-AML | Wild-type | ND | TP53 C238F | ↑WT1 | |
| 14 | 65/M | PV | JAK2 V617F | HC | 10 | Blood 2 y pre-AML | Wild-type | Complex‡ | — | ↑WT1 | |
| 15 | 58/F | PV | JAK2 V617F | HC | 3 | Grans 2 y pre-AML | Wild-type | add(9) | TET2 P1549X | — | |
| 16 | 76/F | ET | JAK2 V617F | HC, BU | 10 | Blood 0.5 y pre-AML | Wild-type | ND | — | NS | |
Pt indicates patient number; MPN, myeloproliferative neoplasm; PMF, primary myelofibrosis; HC, hydroxycarbamide; AN, anagrelide; BM, bone marrow; het, heterozygous; AML, acute myeloid leukemia; ET, essential thrombocythemia; MF, myelofibrotic transformation; hom, homozygous; PV, polycythemia vera; IFN, interferon-α; ND, not done; NA, not applicable; Grans, granulocytes; BU, busulphan; P32, radioactive phosphorus; Mk, megakaryocytes; NS, no sample available; and RARS-T, refractory anemia with ringed sideroblasts and thrombocytosis.
Detected by fluorescence in situ hybridization; G-banding failed.
43,XY,del(5)(q1q3),−7,−12,−16 [5],43,idem,del(6)(q1q2) [3],46,XY [2].
45-46,XY,del(5)(q1?),add(16)(q2?),−17,−17,−22, +2∼3 mar [10].
Patients progressing to JAK2-mutant or wild-type leukemia did not differ with regard to sex (P = .6), age at MPN diagnosis (P = .3), prior use of cytoreductive therapy (P = .2), or duration of the preceding MPN (JAK2-mutant AML, 7 ± 6 years; JAK2 wild-type AML, 12 ± 9 years; P = .2), although the relatively small sample size means that a genuine association may not have been detected. Patients with JAK2 wild-type leukemia, however, were significantly older at time of progression to AML (JAK2-mutant AML, 67 ± 8 years; JAK2 wild-type AML, 79 ± 10 years; P = .02). These results contrast with a previous study wherein duration of the preceding MPN was shorter in those progressing to JAK2 wild-type AML,10 and await confirmation in a larger patient cohort.
Of the 7 patients with JAK2-mutant AML, 6 had a well-characterized preceding MPN that fulfilled criteria for PMF20 or myelofibrotic transformation of PV or ET.21 The remaining patient with JAK2-mutant leukemia gave no history of overt prior blood disorder at the time of presentation with AML (patient 5, Table 1). However, the presence of splenomegaly palpable to the umbilicus together with a trephine biopsy showing clusters of hyperlobated megakaryocytes and dense reticulin fibrosis (supplemental Figure 2) indicated the presence of a preceding MPN, most likely PMF or myelofibrotic transformation of ET/PV. Therefore, all 7 transformations to JAK2-mutant AML arose either from myelofibrotic transformation of ET/PV or from PMF. In marked contrast, only 1 of 9 patients with JAK2 wild-type AML had prior evidence of PMF or myelofibrotic transformation (P = .001). In the remaining 8 cases leukemic transformation occurred directly from chronic-phase ET, PV, or RARS-T. In these 8 patients, blood counts 1 year before AML transformation all showed normal levels of hemoglobin (≥ 120 g/L), white cells (4-9 × 109/L), and platelets (150-450 × 109/L) in the absence of constitutional symptoms or increasing splenomegaly. Taken together these data show an association between the JAK2 status of leukemic blasts and the route by which leukemic transformation occurs: AML arising in patients with chronic-phase ET, PV, or RARS-T was generally JAK2 wild-type, whereas JAK2-mutant leukemia was preceded by evidence of primary myelofibrosis or myelofibrotic transformation of ET/PV.
Progression to JAK2 wild-type AML does not reflect mitotic recombination, gene conversion, or deletion of the mutant allele
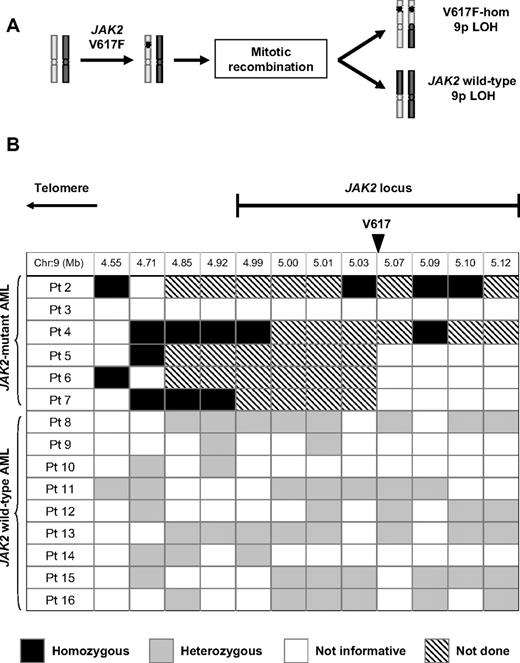
It is unclear why patients with a JAK2-mutant MPN may develop a JAK2 wild-type AML. A proportion of patients with MPN harbor a clone in which the mutant JAK2 allele is duplicated and the wild-type allele is lost.1,2 Such clones result from mitotic recombination in a V617F-heterozygous cell, which would produce V617F-homozygous and JAK2 wild-type daughter cells (Figure 1A). Clonal expansion of the JAK2 wild-type daughter cell could potentially give rise to a subsequent JAK2 wild-type leukemia.
Reversion to wild-type is not a common mechanism of progression from a JAK2-mutant myeloproliferative neoplasm to a JAK2 wild-type leukemia. (A) Mitotic recombination in a JAK2 V617F-heterozygous cell gives rise to both V617F-homozygous and JAK2 wild-type daughter cells, both of which harbor LOH for chromosome 9p. (B) Summary of SNP analysis in patients evolving to acute leukemia after a JAK2-mutant MPN, showing SNP locations relative to the JAK2 locus and genotyping results, whereas all informative JAK2 V617F-homozygous leukemias showed LOH close to JAK2, this was not observed in any of 9 JAK2 wild-type leukemias. Patient 1 is not included in this analysis because leukemic blasts were heterozygous for the JAK2 V617F mutation. SNP indicates single nucleotide polymorphism; hom, homozygous; and LOH, loss of heterozygosity.
Reversion to wild-type is not a common mechanism of progression from a JAK2-mutant myeloproliferative neoplasm to a JAK2 wild-type leukemia. (A) Mitotic recombination in a JAK2 V617F-heterozygous cell gives rise to both V617F-homozygous and JAK2 wild-type daughter cells, both of which harbor LOH for chromosome 9p. (B) Summary of SNP analysis in patients evolving to acute leukemia after a JAK2-mutant MPN, showing SNP locations relative to the JAK2 locus and genotyping results, whereas all informative JAK2 V617F-homozygous leukemias showed LOH close to JAK2, this was not observed in any of 9 JAK2 wild-type leukemias. Patient 1 is not included in this analysis because leukemic blasts were heterozygous for the JAK2 V617F mutation. SNP indicates single nucleotide polymorphism; hom, homozygous; and LOH, loss of heterozygosity.
To search for evidence that mitotically recombined JAK2 wild-type clones may persist in vivo, 1338 individual erythroid colonies from 10 patients with chronic-phase PV were genotyped for both the JAK2 V617F mutation and single nucleotide polymorphisms (SNPs) within or telomeric to the JAK2 locus. All 76 V617F-homozygous colonies showed loss of heterozygosity (LOH) consistent with prior mitotic recombination. LOH was not identified, however, in any of 1195 JAK2-wild type colonies (Table 2), indicating that the mitotically recombined JAK2 wild-type daughter clone does not commonly expand to detectable levels in vivo.
Genotyping of individual erythroid colonies from patients with polycythemia vera for 9p loss of heterozygosity
| Patient . | 9p SNP . | JAK2 wild-type . | JAK2 V617F-het . | JAK2 V617F-hom . | |||
|---|---|---|---|---|---|---|---|
| 9p het . | 9p LOH . | 9p het . | 9p LOH . | 9p het . | 9p LOH . | ||
| a | rs465514 | 160 | 0 | 0 | 0 | 0 | 5 |
| b | rs428111 | 154 | 0 | 19 | 0 | 0 | 5 |
| c | rs428111 | 56 | 0 | 2 | 0 | 0 | 2 |
| d | rs428111 | 78 | 0 | 9 | 0 | 0 | 6 |
| e | rs2230724 | 64 | 0 | 3 | 0 | 0 | 5 |
| f | rs428111 | 58 | 0 | 3 | 0 | 0 | 1 |
| g | rs428111 | 64 | 0 | 5 | 0 | 0 | 1 |
| h | rs7847141 | 119 | 0 | 15 | 0 | 0 | 1 |
| i | rs428111 | 334 | 0 | 2 | 0 | 0 | 49 |
| j | rs428111 | 108 | 0 | 9 | 0 | 0 | 1 |
| Total | 1195 | 0 | 67 | 0 | 0 | 76 | |
| Patient . | 9p SNP . | JAK2 wild-type . | JAK2 V617F-het . | JAK2 V617F-hom . | |||
|---|---|---|---|---|---|---|---|
| 9p het . | 9p LOH . | 9p het . | 9p LOH . | 9p het . | 9p LOH . | ||
| a | rs465514 | 160 | 0 | 0 | 0 | 0 | 5 |
| b | rs428111 | 154 | 0 | 19 | 0 | 0 | 5 |
| c | rs428111 | 56 | 0 | 2 | 0 | 0 | 2 |
| d | rs428111 | 78 | 0 | 9 | 0 | 0 | 6 |
| e | rs2230724 | 64 | 0 | 3 | 0 | 0 | 5 |
| f | rs428111 | 58 | 0 | 3 | 0 | 0 | 1 |
| g | rs428111 | 64 | 0 | 5 | 0 | 0 | 1 |
| h | rs7847141 | 119 | 0 | 15 | 0 | 0 | 1 |
| i | rs428111 | 334 | 0 | 2 | 0 | 0 | 49 |
| j | rs428111 | 108 | 0 | 9 | 0 | 0 | 1 |
| Total | 1195 | 0 | 67 | 0 | 0 | 76 | |
Patients a-j represent a distinct cohort from those transforming to AML (patients 1-16 in Tables 1 and 3). Individual erythroid colonies were genotyped by pyrosequencing for both the JAK2 V617F mutation and an informative SNP on 9p within or telomeric to JAK2. Data are the number of colonies of each individual genotype. All JAK2 wild-type colonies were heterozygous for a 9p SNP.
SNP indicates single nucleotide polymorphism; het-, heterozygous; hom-; homozygous; and LOH, loss of heterozygosity.
The preceding data do not exclude the persistence of a very low level of recombined JAK2 wild-type cells or the presence of such clones in a minority of patients, either of which could still explain the phenomenon of JAK2 wild-type AML. To address this possibility, purified leukemic blasts from patients with AML transformation were investigated for LOH with the use of multiple SNPs within or adjacent to the JAK2 locus. All 5 informative JAK2 V617F-homozygous leukemias showed LOH telomeric to the JAK2 locus, consistent with prior mitotic recombination affecting the JAK2 locus and extending to the telomere, as previously reported.1,2 However, no evidence for LOH was found in any of the 9 JAK2 wild-type leukemias. All JAK2 wild-type leukemias were heterozygous for at least one SNP telomeric to JAK2, 8 of 9 were heterozygous within the JAK2 locus for SNPs telomeric to V617, and 6 of 9 were heterozygous within the JAK2 locus for SNPs centromeric to V617 (Figure 1B; supplemental Figure 1).
Taken together, these data indicate that in PV the JAK2 wild-type daughter cell resulting from mitotic recombination does not commonly expand to detectable levels in vivo. Moreover, after AML transformation, JAK2 wild-type leukemic blasts do not reflect loss of the JAK2 mutation by mitotic recombination, gene conversion, or gene deletion.
Genetic lesions associated with progression to AML include alterations of RUNX1, TP53, NRAS, EVI1, and WT1
Progression from MPN to AML is probably associated with the acquisition of further genetic events. To ascertain whether differences exist between JAK2-mutant and JAK2 wild-type leukemia in the pattern of genetic lesions, patients were screened for aberrations known to be involved in AML pathogenesis. Because BCR-ABL1 copy number and transcript levels may increase in the blastic phase of chronic myeloid leukemia,24 JAK2 copy number and transcript levels were assessed. No increase in JAK2 copy number was observed in any of 7 JAK2-mutant leukemias (Figure 2A). RNA from CD34+ leukemic blasts was available from all 7 JAK2-mutant and from 6 of 9 JAK2 wild-type leukemias. Analysis showed a 12-fold increase in JAK2 transcripts in one JAK2 V617F-homozygous leukemia relative to normal CD34+ cells (patient 6, Figure 2B) despite a normal JAK2 copy number (Figure 2A), suggesting that elevated JAK2 expression may play a role in disease progression in a proportion of patients.
Genetic events associated with progression to acute myeloid leukemia after a JAK2-mutant MPN. (A) Assessment of JAK2 copy number in 7 JAK2-mutant leukemias with the use of a real-time PCR assay; samples from healthy persons and cell lines known to harbor a deletion of 9p (MDA-MB-361 and NB16) or multiple copies of JAK2 (HEL) are shown as controls. (B) Expression of total JAK2 measured by real-time PCR normalized to normal CD34+ cells showing a 12-fold increase in relative JAK2 expression in patient 6. (C) Mutation genotyping from 5 patients progressing to acute leukemia showing that, whereas RUNX1 mutations were acquired at the time of progression to AML, NRAS mutations may be associated with the preceding MPN. Mut indicates mutant; WT, wild-type; grans, purified granulocytes; and BM, bone marrow.
Genetic events associated with progression to acute myeloid leukemia after a JAK2-mutant MPN. (A) Assessment of JAK2 copy number in 7 JAK2-mutant leukemias with the use of a real-time PCR assay; samples from healthy persons and cell lines known to harbor a deletion of 9p (MDA-MB-361 and NB16) or multiple copies of JAK2 (HEL) are shown as controls. (B) Expression of total JAK2 measured by real-time PCR normalized to normal CD34+ cells showing a 12-fold increase in relative JAK2 expression in patient 6. (C) Mutation genotyping from 5 patients progressing to acute leukemia showing that, whereas RUNX1 mutations were acquired at the time of progression to AML, NRAS mutations may be associated with the preceding MPN. Mut indicates mutant; WT, wild-type; grans, purified granulocytes; and BM, bone marrow.
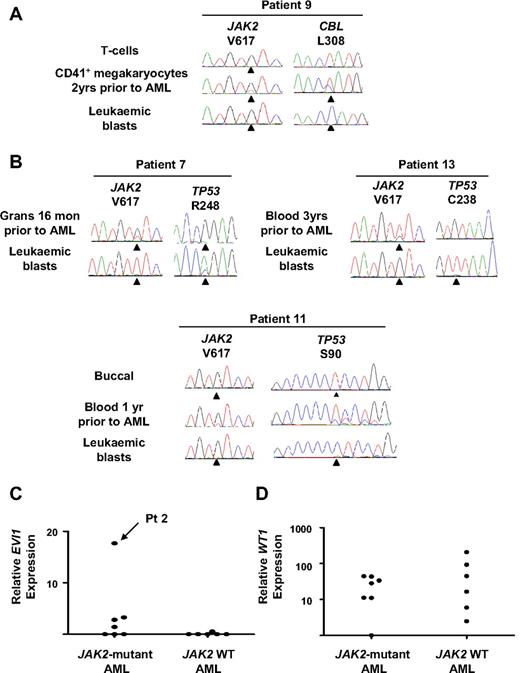
Leukemic blasts were assessed for alterations in known leukemia-associated genes (N/KRAS, CEBPA, RUNX1, GATA2, NPM, WT1, FLT3, CBL, EVI1, and TP53). Two of 7 JAK2-mutant leukemias harbored oncogenic RAS mutations, and in one case the mutation was present 2 years before transformation (patients 1 and 2, Table 1; Figure 2C). Mutations in CBL or FLT3 were identified in single patients with JAK2 wild-type leukemia (patients 9 and 10, respectively, Table 1) and the CBL mutation was also detected in megakaryocytes 2 years before AML transformation (Figure 3A). Mutations in RUNX1 were identified in 2 of 9 JAK2 wild-type and 4 of 7 JAK2-mutant leukemias (Table 1). In all cases RUNX1 mutations were acquired and the wild-type allele was retained. In 4 patients in whom material representative of the preceding MPN was available (patients 1, 3, 5, and 8), RUNX1 mutations were undetectable in either bone marrow, peripheral blood granulocytes, or erythroid colonies (Figure 2C). In patient 5, leukemic blasts harbored mutations in both JAK2 and RUNX1, but erythroid colonies were all RUNX1 wild-type, with or without a JAK2 mutation (JAK2 V617F-homozygous, n = 43; JAK2 wild-type, n = 3; Figure 2C), providing further evidence for an established JAK2 V617F-positive clone in this patient before leukemic transformation.
CBL and TP53 mutations and EVI1 and WT1 expression in patients progressing to acute leukemia. (A) CBL mutation in patient 9 showing detection of the mutation in CD41+ megakaryocytes 2 years before AML transformation and loss of the wild-type CBL allele in leukemic blasts. (B) TP53 mutations in 3 patients progressing to acute leukemia showing the presence of the mutation 1 year before transformation in patient 11. (C) EVI1 expression measured by real-time PCR normalized to normal CD34+ cells showing a 17-fold increase in relative EVI1 expression in patient 2. (D) WT1 expression measured by real-time PCR normalized to normal CD34+ cells showing increased transcript levels (>2 standard deviations above the mean of the normal controls) in 6 of 7 JAK2-mutant and 5 of 6 JAK2 wild-type leukemias. Grans indicates purified granulocytes; and mon, months.
CBL and TP53 mutations and EVI1 and WT1 expression in patients progressing to acute leukemia. (A) CBL mutation in patient 9 showing detection of the mutation in CD41+ megakaryocytes 2 years before AML transformation and loss of the wild-type CBL allele in leukemic blasts. (B) TP53 mutations in 3 patients progressing to acute leukemia showing the presence of the mutation 1 year before transformation in patient 11. (C) EVI1 expression measured by real-time PCR normalized to normal CD34+ cells showing a 17-fold increase in relative EVI1 expression in patient 2. (D) WT1 expression measured by real-time PCR normalized to normal CD34+ cells showing increased transcript levels (>2 standard deviations above the mean of the normal controls) in 6 of 7 JAK2-mutant and 5 of 6 JAK2 wild-type leukemias. Grans indicates purified granulocytes; and mon, months.
Mutations leading to alterations in the DNA binding domain of p53 were detected in 1 of 7 JAK2-mutant and 3 of 9 JAK2-wild-type leukemias (Table 1). In all cases the mutations were acquired and showed complete or partial loss of the wild-type allele in leukemic blasts (Figure 3B). In 2 of these patients, TP53 mutations were undetectable in the preceding MPN (patients 7 and 13), although in a further patient (patient 11) a TP53 mutation could be detected in peripheral blood obtained 1 year before AML transformation (Figure 3B), at which time the patient was well and the full blood count was normal.
A single JAK2-mutant leukemia showed a 17-fold increase in EVI1 transcript levels, consistent with an inversion of chromosome 3 detected at leukemic transformation (patient 2, Table 1), with the remaining AML samples showing either no expression of EVI1 or expression comparable with normal CD34+ cells (Figure 3C). Increased WT1 transcript levels were present in most cases, with only 1 JAK2-mutant and 1 JAK2 wild-type leukemia showing transcript levels comparable with normal CD34+ cells (Figure 3D).
In summary, some leukemia-associated mutations were found only at the time of AML transformation (eg, RUNX1) and were not detected in the preceding MPN, even when this consisted of PMF or myelofibrotic transformation of ET/PV. By contrast other mutations were detectable before leukemia transformation (eg, TP53 and CBL). No consistent differences were observed in the pattern of genetic alterations present in leukemias positive or negative for JAK2 mutations.
Clonal analysis of additional genetic lesions: JAK2 wild-type AML may harbor TET2 mutations that do not precede acquisition of a JAK2 mutation
Clonal analysis was used to explore the phylogenetic relationship between JAK2 wild-type leukemia and a preceding JAK2-mutant MPN. Individual hematopoietic colonies from 4 patients with JAK2 wild-type AML were studied with the use of analysis of X-chromosome inactivation patterns or the detection of additional genetic alterations (Table 3).
Clonal analysis of individual erythroid colonies and purified leukemic blasts from 4 patients progressing to JAK2 wild-type leukemia
| Patient . | Additional mutation . | Erythroid colonies . | JAK2 wild-type leukemic blasts . | |
|---|---|---|---|---|
| JAK2 mutant . | JAK2 wild-type . | |||
| 8 | del(20q) | + | − | − |
| 8 | TET2 | − | + | + |
| 8 | RUNX1 | − | − | + |
| 9 | CBL | − | − | + |
| 12 | TP53 | − | − | + |
| 15 | add(9) | + | − | − |
| 15 | TET2 | − | − | + |
| Patient . | Additional mutation . | Erythroid colonies . | JAK2 wild-type leukemic blasts . | |
|---|---|---|---|---|
| JAK2 mutant . | JAK2 wild-type . | |||
| 8 | del(20q) | + | − | − |
| 8 | TET2 | − | + | + |
| 8 | RUNX1 | − | − | + |
| 9 | CBL | − | − | + |
| 12 | TP53 | − | − | + |
| 15 | add(9) | + | − | − |
| 15 | TET2 | − | − | + |
+ Indicates present; and −, absent.
Samples of RNA from both the JAK2-mutant MPN and JAK2 wild-type AML were available from a single female patient (patient 15, Table 1). In this patient, the same X-chromosome inactivation pattern was observed in JAK2 V617F-positive erythroid colonies, peripheral blood granulocytes obtained 2 years before transformation, and JAK2 wild-type leukemic blasts (supplemental Figure 3A), a result that does not discriminate between a shared versus independent clonal origin for JAK2-mutant MPN and JAK2 wild-type AML.
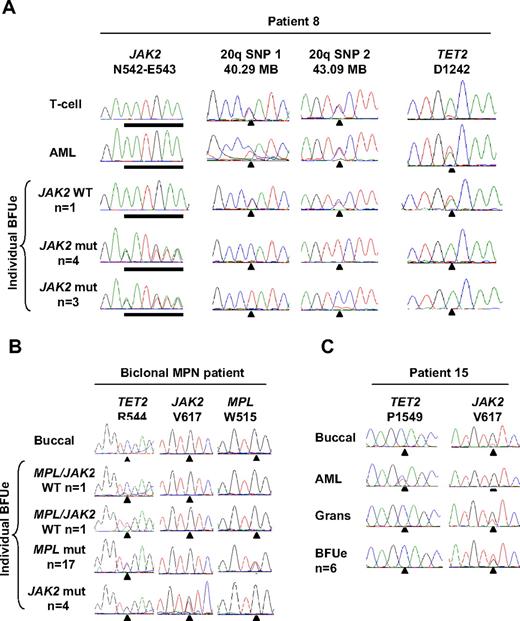
Individual hematopoietic colonies were available from 2 patients in whom cytogenetic abnormalities were detected at transformation to JAK2 wild-type AML (patients 8 and 15, Tables 1 and 3). In patient 15, trisomy 9 was present in 9 of 10 metaphases at AML transformation and was also detected in JAK2-mutant erythroid colonies (as previously described in Beer et al19 ) but was absent from JAK2 wild-type leukemic blasts as shown by a 1:1 allelic ratio of multiple informative SNPs within the JAK2 locus (supplemental Figure 1; data not shown). In patient 8 a deletion of chromosome 20q was present in 9 of 14 metaphases at progression to AML and was also present in JAK2-mutant erythroid colonies as shown by LOH studies but was absent from JAK2 wild-type leukemic blasts (Table 3). Consistent with a recent report,17 different JAK2-mutant erythroid colonies harbored loss of alternative 20q alleles (Figure 4A), indicating the occurrence of at least 2 independent 20q deletions in this patient. In these 2 patients, therefore, cytogenetic aberrations detected at AML transformation reflected cells derived from the preceding JAK2-mutant MPN rather than from the JAK2 wild-type leukemia. Hematopoietic colonies were also available from 2 patients with mutations in CBL or TP53 (patients 9 and 12, Tables 1 and 3). In these patients, CBL or TP53 mutations were present in JAK2 wild-type leukemic blasts but were absent from JAK2-mutant erythroid colonies (Table 3). In summary, in these 4 patients the detection of additional genetic events did not indicate the existence of a shared clone of origin for JAK2-mutant MPN and JAK2 wild-type AML.
Clonal relationship of JAK2 wild-type leukemia to JAK2-mutant MPN with the use of 20q deletion or mutation in TET2 as a clonal marker. (A) Analysis of patient 8 showing a deletion of alternative 20q alleles in the MPN clone but no 20q deletion in the AML clone and a mutation of TET2 in leukemic blasts and a JAK2 wild-type erythroid colony but not in JAK2-mutant erythroid colonies. (B) Analysis of individual erythroid colonies from a patient harboring JAK2 V617F and MPL W515L mutations, showing that mutations in JAK2 and MPL are present in separate clones, with both clones have arisen from a shared TET2-mutant founder clone. (C) Analysis of patient 15 showing a mutation of TET2 in the JAK2 wild-type leukemia but not the JAK2-mutant MPN.
Clonal relationship of JAK2 wild-type leukemia to JAK2-mutant MPN with the use of 20q deletion or mutation in TET2 as a clonal marker. (A) Analysis of patient 8 showing a deletion of alternative 20q alleles in the MPN clone but no 20q deletion in the AML clone and a mutation of TET2 in leukemic blasts and a JAK2 wild-type erythroid colony but not in JAK2-mutant erythroid colonies. (B) Analysis of individual erythroid colonies from a patient harboring JAK2 V617F and MPL W515L mutations, showing that mutations in JAK2 and MPL are present in separate clones, with both clones have arisen from a shared TET2-mutant founder clone. (C) Analysis of patient 15 showing a mutation of TET2 in the JAK2 wild-type leukemia but not the JAK2-mutant MPN.
Mutations in TET2 have recently been described in patients with a chronic-phase JAK2-mutant MPN and may precede acquisition of the JAK2 mutation.18,25 Sequencing of the TET2 coding region identified an acquired mutation (TET2 R544X) in 1 of 3 patients with a biclonal chronic-phase MPN. Analysis of individual erythroid colonies from this patient identified the TET2 mutation in JAK2-mutant, MPL-mutant, and JAK2/MPL wild-type colonies (Figure 4B), showing that the JAK2-mutant and MPL-mutant clones had evolved as subclones from a shared TET2-mutant proliferation. These results show that a TET2-mutant founder clone may give rise to daughter clones bearing different additional genetic alterations.
It is possible, therefore, that a TET2-mutant clone could give rise to both JAK2-mutant MPN and JAK2 wild-type AML within the same patient. Within the AML cohort, 2 patients with JAK2 wild-type leukemia harbored acquired mutations in TET2 (patients 8 and 15, Table 1). To investigate the possibility that a JAK2-mutant MPN and JAK2 wild-type AML may also arise from a shared TET2-mutant founder clone, individual erythroid colonies from both patients were genotyped for the TET2 mutation. In patient 8, a TET2 mutation was present in leukemic blasts and a JAK2 wild-type erythroid colony but was absent from all 7 JAK2-mutant erythroid colonies (Figure 4A; Table 3). Similarly in patient 15, a TET2 mutation was present in leukemic blasts but was not detected in granulocytes obtained 2 years before transformation and was absent from all 6 JAK2-mutant erythroid colonies (Figure 4C; Table 3). Thus, in both patients TET2 mutations were present only in the JAK2 wild-type leukemia and not in the preceding JAK2-mutant MPN. Of note, the presence of a TET2-mutant erythroid colony in a patient with TET2-mutant AML (patient 8) implies that the TET2 mutation was acquired early in disease evolution, before the acquisition of a fully transformed AML phenotype. We considered the possibility that the TET2 and JAK2 mutations were acquired within the same clone, with mitotic recombination or deletion of TET2 leading to loss of the mutant TET2 allele within the JAK2-mutant clone. This possibility was excluded, however, by the detection of a heterozygous SNP within the TET2 locus in JAK2-mutant erythroid colonies and JAK2 wild-type leukemic blasts from both patients (supplemental Figure 3B). Our data therefore indicate that the TET2 mutation did not precede acquisition of the JAK2 mutation in these 2 patients and show that mutations in TET2 and JAK2 may be present in separate clonal expansions.
Discussion
This study used analysis of purified leukemic blasts from 16 patients to address several issues relating to AML transformation after a JAK2-mutant MPN, including the relationship of MPN phenotype to leukemia JAK2 status, the nature of the genetic lesions acquired with progressive disease, and the clonal relationship of JAK2-mutant MPN to JAK2 wild-type AML. Analysis of a pure population of leukemic blasts is critical to establish whether the JAK2 mutation or additional genetic lesions are present within the leukemic clone or in cells representing the preceding MPN. The culture of erythroid colonies from selected patients provided valuable clonal material representing the JAK2-mutant MPN, thus permitting the precise tracking of mutations and cytogenetic aberrations through the MPN and AML phases of disease. The importance of obtaining purified leukemic blasts and clonal MPN-phase material is highlighted by the demonstration in 2 patients that cytogenetic abnormalities found in most cells at AML transformation indicated persistence of the JAK2-mutant MPN and were absent from JAK2 wild-type leukemic blasts.
In this study, a striking correlation was observed between the phenotype of the preceding MPN and the JAK2 status of leukemic blasts. In contrast to JAK2 wild-type AML, evolution to JAK2-mutant leukemia was invariably preceded by myelofibrotic transformation of ET/PV or PMF. It has been suggested that myelofibrotic transformation of ET/PV and at least some cases of PMF may both represent manifestations of accelerated phase disease.1 In support of this notion, PMF and post-ET/PV myelofibrosis are phenotypically indistinguishable,26,27 and PMF is associated with features of advanced disease, including a high prevalence of genomic and epigenetic alterations,28-30 progressive stem cell dysfunction,31,32 and shortened overall survival.5 Our results therefore imply that evolution to JAK2-mutant AML is preceded by the acquisition of genetic changes that give rise to the phenotype of accelerated-phase disease, manifested clinically as PMF or myelofibrotic transformation of ET/PV. Mutant JAK2 may well be responsible for this accumulation of DNA damage. Bcl-XL deamidation in response to DNA damage is inhibited in JAK2 V617F-positive primary cells, thus providing a mechanism for the aberrant survival of DNA-damaged cells.7 Moreover, expression of JAK2 V617F in cell lines and human progenitors is associated with both increased DNA damage and aberrant DNA repair.6
In contrast to JAK2-mutant AML, JAK2 wild-type leukemia usually arose in patients with chronic-phase ET or PV. Although not statistically significant, it is noteworthy that all 3 patients with a history of prior genotoxic therapy (busulphan or P32) progressed to JAK2 wild-type AML (Table 1). It is possible that leukemic transformation in such cases represents therapy-associated damage to a normal stem cell. In the remaining 6 JAK2 wild-type AMLs, the patients had received hydroxycarbamide alone. Concerns have been raised over a possible increased risk of AML in patients treated with hydroxycarbamide, and an association between hydroxycarbamide therapy and alteration of TP53 has been suggested.33,34 Deletion or mutation of TP53 was seen in the current study in 4 patients treated with single-agent hydroxycarbamide, but both these data and previous reports lack a hydroxycarbamide-free control group. It is clear that leukemic transformation does occur in the absence of prior therapy,4,10 and an increased risk with hydroxycarbamide has been difficult to establish, with clinical studies giving conflicting results.4,34-37 Follow-up of patients with sickle cell suggests that hydroxycarbamide is not leukemogenic to normal stem cells,38 although these data do not exclude a mutagenic effect on clonal hematopoiesis (eg, a pre-JAK2 clone). We have previously shown that patients with JAK2 V617F-positive ET are more sensitive to the effects of hydroxycarbamide than are patients without the mutation,39 raising the possibility that hydroxycarbamide might preferentially suppress the JAK2-mutant clone and favor expansion of coexisting JAK2 wild-type clones with subsequent leukemic transformation of the latter. However, at present, the data implicating hydroxycarbamide in progression to AML remain inconclusive.
To gain insight into the mechanisms underlying the development of JAK2 wild-type AML, we studied its clonal relationship to the preceding JAK2-mutant MPN. In all 9 cases examined we were able to exclude the possibility that JAK2 wild-type AML arose from the JAK2 wild-type progeny of mitotic recombination affecting a JAK2 V617F-heterozygous cell (Figure 1A), with analysis of multiple markers within the JAK2 gene also excluding intrachromosomal deletion or gene conversion. These data confirm and considerably extend 3 previously reported cases, in which reversion to wild-type by mitotic recombination was excluded by analysis of a limited number of markers.9,10 Moreover in this study, the persistence of mitotically recombined JAK2 wild-type clones was not detected by analysis of more than 1300 individual erythroid colonies from 10 patients with chronic-phase PV. Taken together, these data indicate that loss of the mutant JAK2 allele by mitotic recombination is unlikely to be a significant mechanism in the genesis of JAK2 wild-type AML.
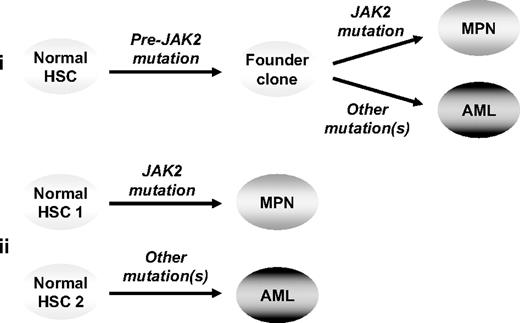
Two models remain to explain the clonal relationship of JAK2 wild-type AML to its preceding JAK2-mutant MPN: (1) the 2 phases of disease are phylogenetically related, having arisen from a shared (pre-JAK2) founder clone, or (2) the MPN and AML are clonally unrelated and reflect transformation of independent stem cells (Figure 5). In the present study, oligoclonal disease was shown in 4 patients transforming to JAK2 wild-type AML, with mutations in TP53, CBL, or TET2 present in a clone distinct from that carrying the JAK2 mutation; however, the phylogenetic relationship of the 2 clones remains unclear. Of note, in 2 of these cases progression to TET2-mutant AML did not reflect transformation of a pre-JAK2 clone carrying the TET2 mutation. It remains theoretically possible, however, that in these 2 patients the JAK2-mutant MPN and TET2-mutant AML represent the phylogenetically related progeny of a shared pre-TET2 clone.
Models to explain progression from a JAK2-mutant MPN to a JAK2 wild-type leukemia. In model 1 (i) the 2 phases of disease are phylogenetically related, having arisen from a shared (pre-JAK2) founder clone, whereas in model 2 (ii) the 2 phases of disease are clonally unrelated, reflecting transformation of independent stem cells. HSC indicates hematopoietic stem cell.
Models to explain progression from a JAK2-mutant MPN to a JAK2 wild-type leukemia. In model 1 (i) the 2 phases of disease are phylogenetically related, having arisen from a shared (pre-JAK2) founder clone, whereas in model 2 (ii) the 2 phases of disease are clonally unrelated, reflecting transformation of independent stem cells. HSC indicates hematopoietic stem cell.
Recent reports have uncovered an increasing degree of clonal heterogeneity in patients with MPN. Mutations in TET2 may precede acquisition of a JAK2 mutation,18 and data presented herein show that genetically dissimilar clones, bearing mutations in JAK2 or MPL, represent the phylogenetically related progeny of a shared TET2-mutant founder clone in a patient with chronic-phase ET. By contrast, we have previously shown that biclonal disease may represent transformation of distinct, unrelated stem cells.19 Further evidence of oligoclonal disease in patients in chronic phase includes deletion of alternative chromosome 20q alleles40 (and present study), the acquisition of multiple JAK2 V617F mutations in individual patients,41 and the detection in patients with CML treated with imatinib of cytogenetically abnormal BCR-ABL1–negative clones.42 Clonal heterogeneity has also been detected in epithelial malignancy, with molecular analysis of multifocal tumors showing that clones with diverse patterns of somatic mutation represent either the phylogenetically related progeny of a shared ancestral clone or transformation of unrelated progenitor cells.43-45 Thus, oligoclonal disease is present in a proportion of patients with myeloid or epithelial malignancy, composed of either phylogenetically related or unrelated clones (akin to models 1 and 2 in Figure 5). Given these observations, it seems plausible that each model may be operating in a different subset of patients with JAK2 wild-type AML.
The presence of clonal heterogeneity in both myeloid and epithelial neoplasia implies that early tumorigenesis is characterized by the proliferation of competing clones harboring diverse patterns of mutation, with clonal dominance arising through a Darwinian process of biologic selection. The preleukemic MPNs offer an experimentally tractable system for the study of early-stage malignancy through the precise tracking of mutations within different clonal expansions. The identification of novel mutations has facilitated the detection of an increasing degree of clonal complexity in patients with MPN. Given that only a limited number of MPN-associated mutations have so far been identified, it is probable that phenomena such as biclonal disease and pre-JAK2 clonal expansions are more prevalent than is currently appreciated.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the patients and staff of the United Kingdom and European MPN clinics who have contributed samples to this study, Dr Wendy Erber and Dr Penny Wright for advice on histology, and the Addenbrooke's Haematology Disorders Sample Bank for processing and managing patient samples.
This work was supported by the UK Medical Research Council, the Leukaemia Research Fund, the Kay Kendall Leukaemia Fund, the NIHR Cambridge Biomedical Research Centre, the Leukemia & Lymphoma Society of America, the Myeloproliferative Disorders Foundation, the Laurette Fugain Association, the Fondation de France, the Ligue Nationale Contre le Cancer, and the Cancéropôle Ile de France.
Authorship
Contribution: P.A.B. designed and performed experiments, analyzed data, and co-wrote the manuscript; F.D., J.-P.L.C., and W.V. performed and analyzed sequencing of TET2; M.A.D. and E.C. performed colony assays; D.B., R.K., M.F.M., C.N.H., and A.M.V. provided patient samples and clinical information; and A.R.G. directed the research and co-wrote the manuscript. All authors had the opportunity to review the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tony Green, Department of Haematology, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 2XY, United Kingdom; e-mail: arg1000@cam.ac.uk.