Abstract
Philadelphia chromosome–negative myeloproliferative neoplasms (MPNs) including polycythemia vera, essential thrombocythemia, and primary myelofibrosis show an inherent tendency for transformation into leukemia (MPN-blast phase), which is hypothesized to be accompanied by acquisition of additional genomic lesions. We, therefore, examined chromosomal abnormalities by high-resolution single nucleotide polymorphism (SNP) array in 88 MPN patients, as well as 71 cases with MPN-blast phase, and correlated these findings with their clinical parameters. Frequent genomic alterations were found in MPN after leukemic transformation with up to 3-fold more genomic changes per sample compared with samples in chronic phase (P < .001). We identified commonly altered regions involved in disease progression including not only established targets (ETV6, TP53, and RUNX1) but also new candidate genes on 7q, 16q, 19p, and 21q. Moreover, trisomy 8 or amplification of 8q24 (MYC) was almost exclusively detected in JAK2V617F− cases with MPN-blast phase. Remarkably, copy number–neutral loss of heterozygosity (CNN-LOH) on either 7q or 9p including homozygous JAK2V617F was related to decreased survival after leukemic transformation (P = .01 and P = .016, respectively). Our high-density SNP-array analysis of MPN genomes in the chronic compared with leukemic stage identified novel target genes and provided prognostic insights associated with the evolution to leukemia.
Introduction
Philadelphia chromosome–negative myeloproliferative neoplasms (MPNs) including polycythemia vera (PV), essential thrombocytosis (ET), and primary myelofibrosis (PMF) are defined as clonal hematopoietic stem cell disorders and characterized by increased proliferation of terminally differentiated myeloid cells. The tyrosine kinase JAK2 is directly linked to the pathogenesis of MPN with the identification of JAK2V617F as a recurring gain-of-function mutation.1,2 Almost all cases with PV, and roughly 50% of patients with ET and PMF, carry this specific mutation localized on chromosome 9p24.
The long-term outcome of patients with acute myeloid leukemia (AML) secondary to MPN, myelodysplastic syndrome (MDS), or treatment with cytotoxic agents is relatively poor compared with patients with de novo AML. Patients with de novo and secondary AML have a similar spectrum of cytogenetic abnormalities, but the occurrence of cytogenetic changes associated with unfavorable risk such as 5q−, −7/7q−, trisomy 8, or complex karyotype is higher in secondary AML.3,4 However, so far only a small number of studies with limited number of cases have explored the chromosomal alterations and/or clinical markers associated with acceleration to blast phase of patients with MPN.
Previously, we developed the copy number analyzer for Affymetrix GeneChip (CNAG) program and the new algorithm allele-specific copy number analysis using anonymous references (AsCNAR).5,6 These techniques in combination with high-density single nucleotide polymorphism (SNP) array provide a robust and detailed approach to detect large and small copy number changes, as well as copy number–neutral loss of heterozygosity (CNN-LOH). To obtain a comprehensive profile of genomic alterations associated with leukemic transformation in MPN, we applied this interrogational method and performed a systemic analysis of 159 samples obtained from patients either in chronic phase or blast phase of MPN.
Methods
Patients and clinical samples
In total, samples from 148 patients were analyzed by SNP-array. One hundred fifty-nine samples were obtained, of which 88 (55%) were diagnosed with MPN in chronic phase (23 PV, 32 ET, 33 PMF) and 71 (45%), with MPN in blast phase (19 PV, 19 ET, 33 PMF). Diagnosis was based on the World Health Organization criteria,7 and an overview of patients, including clinical data, is given in Table 1. This study received institutional review board approval from the Cedars-Sinai Medical Center, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Given the relatively high incidence of homozygous JAK2V17F patients diagnosed with ET (3/18 in chronic phase, 2/6 in blast phase), which is usually lower for this disorder,9 we suggest that at least some cases diagnosed with ET may have been incorrect.
Clinical features of MPN/MPN-blast phase cases (unmatched and matched)
| . | Unmatched MPN . | Unmatched MPN-blast phase . | Matched MPN . | Matched MPN-blast phase . |
|---|---|---|---|---|
| All cases, no. (%) | 77 (56) | 60 (44) | 11 (50) | 11 (50) |
| MPN diagnosis no. (%) | ||||
| PV samples | 21 (27) | 17 (28) | 2 (18) | 2 (18) |
| ET samples | 31 (40) | 18 (30) | 1 (09) | 1 (09) |
| PMF samples | 25 (33) | 25 (42) | 8 (73) | 8 (73) |
| Sex, M:F | ||||
| PV samples | 1:2 | 1:1 | 1:1 | 1:1 |
| ET samples | 1:2 | 1:1 | 0:1 | 0:1 |
| PMF samples | 2:1 | 2:1 | 2:1 | 2:1 |
| Mean age at diagnosis, y, ± SD* | ||||
| PV | 57 ± 5 | 68 ± 5 | — | — |
| ET | 59 ± 6 | 69 ± 7 | — | — |
| PMF | 57 ± 6 | 65 ± 9 | 59 ± 9 | 65 ± 7 |
| Mean blast count in bone marrow, ± SD, no. (%)* | ||||
| PV samples | < 5% | 70 ± 20 | — | — |
| ET samples | < 5% | 66 ± 23 | — | — |
| PMF samples | < 5% | 70 ± 21 | < 5% | 66 ± 24 |
| JAK2V617F (+) no. (%) | ||||
| PV samples | 21/21 (100) | 14/17 (82)‡ | 2/2 (100) | 1/2 (50) |
| ET samples | 18/31 (58) | 6/18 (33) | 0/1 (0) | 0/1 (0) |
| PMF samples | 16/25 (64) | 12/25 (48) | 5/8 (62.5) | 4/8 (50) |
| c-MPL mutation positive, no. (%) | ||||
| PV samples | 1/21 (5)† | 0/17 (0) | 0/2 (0) | 0/2 (0) |
| ET samples | 0/31 (0) | 1/18 (6) | 0/1 (0) | 0/1 (0) |
| PMF samples | 3/25 (12) | 2/25 (8) | 1/8 (12.5) | 1/8 (12.5) |
| . | Unmatched MPN . | Unmatched MPN-blast phase . | Matched MPN . | Matched MPN-blast phase . |
|---|---|---|---|---|
| All cases, no. (%) | 77 (56) | 60 (44) | 11 (50) | 11 (50) |
| MPN diagnosis no. (%) | ||||
| PV samples | 21 (27) | 17 (28) | 2 (18) | 2 (18) |
| ET samples | 31 (40) | 18 (30) | 1 (09) | 1 (09) |
| PMF samples | 25 (33) | 25 (42) | 8 (73) | 8 (73) |
| Sex, M:F | ||||
| PV samples | 1:2 | 1:1 | 1:1 | 1:1 |
| ET samples | 1:2 | 1:1 | 0:1 | 0:1 |
| PMF samples | 2:1 | 2:1 | 2:1 | 2:1 |
| Mean age at diagnosis, y, ± SD* | ||||
| PV | 57 ± 5 | 68 ± 5 | — | — |
| ET | 59 ± 6 | 69 ± 7 | — | — |
| PMF | 57 ± 6 | 65 ± 9 | 59 ± 9 | 65 ± 7 |
| Mean blast count in bone marrow, ± SD, no. (%)* | ||||
| PV samples | < 5% | 70 ± 20 | — | — |
| ET samples | < 5% | 66 ± 23 | — | — |
| PMF samples | < 5% | 70 ± 21 | < 5% | 66 ± 24 |
| JAK2V617F (+) no. (%) | ||||
| PV samples | 21/21 (100) | 14/17 (82)‡ | 2/2 (100) | 1/2 (50) |
| ET samples | 18/31 (58) | 6/18 (33) | 0/1 (0) | 0/1 (0) |
| PMF samples | 16/25 (64) | 12/25 (48) | 5/8 (62.5) | 4/8 (50) |
| c-MPL mutation positive, no. (%) | ||||
| PV samples | 1/21 (5)† | 0/17 (0) | 0/2 (0) | 0/2 (0) |
| ET samples | 0/31 (0) | 1/18 (6) | 0/1 (0) | 0/1 (0) |
| PMF samples | 3/25 (12) | 2/25 (8) | 1/8 (12.5) | 1/8 (12.5) |
MPN indicates myeloproliferative neoplasm; PV, polycythemia vera; ET, essential thrombocytosis; M, male; F, female; and PMF, primary myelofibrosis.
Data are available for 27 unmatched MPN (10 PV, 10 ET, and 7 PMF) and 54 unmatched MPN-blast phase (15 PV, 18 ET, and 21 PMF) cases, and 8 matched MPN (PMF) cases.
This c-MPL mutation in a PV patient has already been validated and reported by Kawamata et al.8
Significantly fewer cases with JAK2V617F in blast phase vs chronic phase (P=.045).
Samples were provided by (1) Department of Hematology, Mayo Clinic (n = 35); (2) Brigham and Women's Hospital, Harvard University, School of Medicine (n = 46); (3) Department of Hematology, Archet Hospital (n = 44); (4) MLL Munich Leukemia Laboratory, (n = 14); (5) Division of Hematology-Oncology, Chang Gung Memorial Hospital (n = 14); and (6) Division of Hematology, Sheba Medical Center and Sackler School of Medicine, Tel-Aviv University (n = 6).
SNP-Chip analysis
A total of 159 tumor specimens (MPN and/or MPN-blast phase) were analyzed on GeneChip SNP genotyping microarrays (GeneChip Mapping 50K and/or 250K arrays; Affymetrix) as described previously.5,6 After appropriate normalization of mean array intensities, signal ratios were calculated between tumors and anonymous normal references in an allele-specific manner. Genome-wide determination of allele-specific copy numbers (AsCNs) and detection of CNN-LOH at each SNP were inferred from the observed signal ratios based on the hidden Markov model using CNAG/AsCNAR algorithms (http://www.genome.umin.jp).5,6 For clustering of patient samples with regard to the status of copy number changes, as well as CNN-LOH, CNAG-Graph software (Tokyo University) was used. Size, position, and location of genes were identified with the University of California, Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu)10 and Ensemble Genome Browser (http://www.ensembl.org).11 Germline copy number changes previously described as copy number variants at Database of Genomic Variants (http://projects.tcag.ca/variation)12 and UCSC Genome Browser were excluded. SNP-array data used in this study are available in the Gene Expression Omnibus (GEO) database under accession number GSE19647.13
Comparison of 50K versus 250K SNP-Chip analysis in MPN chronic phase
SNP-array analysis of 46 of our MPN samples (10 PV, 20 ET, 16 PMF; kindly provided by D.G.G. at Brigham and Women's Hospital, Harvard University) has already been reported by our group.8 At that time, only 50K arrays were available, whereas later in this study, the 250K arrays were accessible and used to analyze additionally 42 MPN and 71 MPN-blast phase samples. Because no significant differences in either number of deletions, duplications/amplifications, or CNN-LOH per case were found as analyzed by the 50K compared with 250K array (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article), we combined the analysis of both platforms in our results. Supplemental Table 2 lists all individual samples and the array that was used.
Cytogenetics
Routine cytogenetic analysis with conventional banding techniques was performed in 35 of 88 MPN (10/23 PV, 10/32 ET, 15/33 PMF) and 63 of 71 MPN-blast phase (15/19 PV, 18/19 ET, 30/33 PMF) cases according to standard procedures as previously described.14 No routine fluorescent in situ hybridization (FISH) panel was applied, but in some cases, however, FISH analysis was performed to supplement conventional cytogenetic analysis (supplemental Table 2).
Allele-specific PCR for JAK2V617F mutation
For the detection of JAK2V617F, allele-specific polymerase chain reaction (PCR) was performed according to the previously reported method.15
Direct mutation screening
Primers were designed to amplify and sequence coding exons and splice junctions of the following genes: TET2, c-CBL, TP53, and RUNX1. We screened only the 11 matched samples that showed genomic changes in the particular gene regions. Primer details are available from the corresponding author (N.H.T.).
We evaluated all MPN and MPN-blast phase patients with 1pCNN-LOH for the MPLW515 mutation (exon 10) by direct sequencing. If no mutation was detected in this cohort, we also screened the other coding exons of the c-MPL gene previously shown to be mutated in MPN.8
Validation of acquired genomic copy number changes including CNN-LOH
To confirm the somatic origin of genomic copy number changes, quantitative genomic real-time (QG RT)–PCR was performed on the genomic DNA from the hybridized MPN and matched MPN-blast phase samples according to the calculation method described by Weksberg et al.16 For example, we used primers for the RUNX1 gene (21q22.12; supplemental Figure 1A) as well as TET2 gene (4q24; data not shown) and a random region on chromosome 21q21.1 and 4p15.1, respectively, as a reference in patient 121.
Detection of acquired CNN-LOH was also validated by QG RT-PCR and subsequently by nucleotide sequencing. Three SNP sequences (rs919275, rs10854117, and rs10854117) on chromosome 19p in case 36 at diagnosis of PV, as well as at leukemic transformation, were determined (supplemental Figure 1B). The genomic region of each SNP site was amplified, and products were purified and sequenced (supplemental Figure 1C). In addition, we confirmed loss of CNN-LOH on 9p after leukemic transformation in matched case 120 using SNP sequences rs3858029, rs1360461, and rs10818814 on chromosome 9 (data not shown).
Homozygous deletions of CUTL1 and SH2B2 (case 138) as well as PIG-A (case 121) in both MPN and/or MPN-blast phase samples were also confirmed by QG RT-PCR (supplemental Figure 2). Primers for these experiments will be provided upon request.
Statistical analysis
Wilcoxon rank sum tests were used to assess differences in continuous variables, and categoric variables were assessed using chi-square tests, all with a significance level of α = .05. The methods of estimations included the standard deviation (± SD) of the sampling distribution. Asterisks shown in the figures indicate significant differences of experimental groups in comparison with the corresponding control condition (*P < .05; **P < .001). Survival analysis was performed using the Kaplan-Meier method, and survival curves were compared using the log-rank test.
Results
Lower frequency of JAK2V617F and 9p alterations after leukemic transformation
In the present study, we examined 159 samples (88 MPN and 71 MPN-blast phase) from a total of 148 patients. An overview of the clinical features of matched and unmatched cases including sex, age, leukemic blast infiltration, and mutational status (JAK2V617F, c-MPL) is provided in Table 1. The sex ratio of male and female patients in chronic phase was 1:2 for PV and ET, whereas after transformation, the ratio was balanced with 1:1. For PMF patients, the male-to-female ratio was 2:1 in both MPN chronic and blast phase.
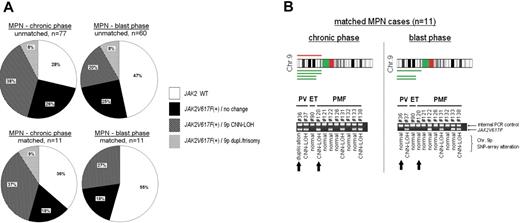
Overall, the incidence of JAK2V617F was almost 20% less in the blast phase compared with the chronic phase for both the matched and unmatched MPN cases (unmatched cases: P < .05; Figure 1A). Cases that were negative for JAK2V617F were also exclusively negative for 9p duplication, trisomy 9, or 9pCNN-LOH in the chronic as well as leukemic stage of MPN. 9pCNN-LOH was noted approximately 3 to 4 times more often than 9p duplication and/or trisomy 9 in JAK2V617F+ MPN cases during either the chronic or blast phase (Figure 1A), but the frequency of 9pCNN-LOH was significantly less in the blast crisis compared with the chronic phase of unmatched PMF and PV patients (supplemental Table 3). In contrast, unmatched ET cases had about the same frequency of 9pCNN-LOH in the chronic phase versus the blast phase of the disease. Furthermore, in the analysis of the 11 matched MPN cases, 7 were positive for JAK2V617F (64%), 4 had 9p CNN-LOH (37%), and 1 had 9p duplication (9%) at first diagnosis (Figure 1A). In comparison, 2 of these patients were JAK2V617F+ with either trisomy 9 or 9pCNN-LOH during their chronic phase (1 PV, 1 PMF), but no longer had detectable JAK2V617F with a normal chromosome 9 after leukemic evolution (Figure 1B).
Frequency of JAK2V617F and associated alterations on chromosome 9. (A) Diagrams represent matched and unmatched MPN cases in chronic versus blast phase. Indicated are frequencies of JAK2V617F and association to 9p duplication (dupl)/trisomy 9 or 9pCNN-LOH. Data and statistical evaluation for underlying MPN subgroups are shown in supplemental Table 3. (B) CNAG software represents duplication (red) and CNN-LOH (green) on 9p detected in 11 patients with matched samples (chronic MPN vs MPN-blast phase). In addition, allele-specific PCR for the detection of JAK2V617F was performed in these samples. Arrows indicate 2 MPN patients who were initially positive for JAK2V617F in association with 9p imbalances; leukemic transformation was accompanied with loss of JAK2V617F and a normal chromosome 9.
Frequency of JAK2V617F and associated alterations on chromosome 9. (A) Diagrams represent matched and unmatched MPN cases in chronic versus blast phase. Indicated are frequencies of JAK2V617F and association to 9p duplication (dupl)/trisomy 9 or 9pCNN-LOH. Data and statistical evaluation for underlying MPN subgroups are shown in supplemental Table 3. (B) CNAG software represents duplication (red) and CNN-LOH (green) on 9p detected in 11 patients with matched samples (chronic MPN vs MPN-blast phase). In addition, allele-specific PCR for the detection of JAK2V617F was performed in these samples. Arrows indicate 2 MPN patients who were initially positive for JAK2V617F in association with 9p imbalances; leukemic transformation was accompanied with loss of JAK2V617F and a normal chromosome 9.
JAK2V617F mutational status had no impact on time to transformation or survival
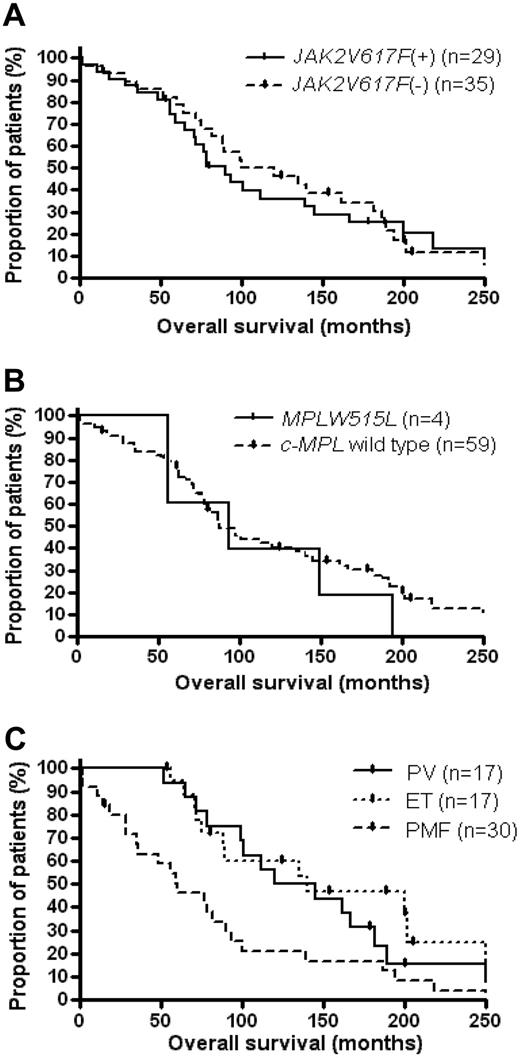
In the evaluation of clinical data for MPN-blast phase patients, no significant correlation was noted between the prevalence of JAK2V617F at transformation and either age, percentage of leukemic blast cells in the marrow, or pretreatment with alkylating agents and/or hydroxyurea (data not shown). Moreover, we found no statistical association between either time to leukemic transformation or overall survival and the JAK2V617F status at transformation in PV, ET, or PMF patients. The overall survival of MPN-blast phase patients with JAK2V617F versus blast phase patients without this mutation is provided in Figure 2A (P = .6). In addition, with respect to the comparably low frequency of MPLW515-positive MPN-blast phase patients (6%), we noted no impact of the c-MPL mutational status on either time to transformation (data not shown) or the overall survival in MPN patients who underwent leukemic transformation (P = .5; Figure 2B).
Overall survival of MPN patients with subsequent transformation to blast crisis. Kaplan-Meier plots of all MPN-blast phase patients from the diagnosis of pre-existing MPN were stratified for (A) the presence or absence of a JAK2V617F mutation at transformation, (B) the presence or absence of a MPLW515L mutation at transformation, and (C) the underlying type of MPN.
Overall survival of MPN patients with subsequent transformation to blast crisis. Kaplan-Meier plots of all MPN-blast phase patients from the diagnosis of pre-existing MPN were stratified for (A) the presence or absence of a JAK2V617F mutation at transformation, (B) the presence or absence of a MPLW515L mutation at transformation, and (C) the underlying type of MPN.
However, regardless of the mutational status of MPN-blast phase patients, we noted that the time from diagnosis of MPN to leukemic transformation was significantly shorter in those with pre-existing PMF (median, 58 months) compared with patients with either prior PV (median, 98 months) or ET (median, 110 months; P = .01). This earlier transformation resulted in a decreased overall survival from the time of diagnosis of the underlying MPN in leukemic patients with preceding PMF patients compared with preceding PV or ET (P = .02; Figure 2C), which is congruent with previously published results.17
Increased number of additional genomic changes after leukemic transformation
Altogether, a relatively low number of genomic alterations was found by SNP-array analysis in the chronic phase of the MPN samples (Figure 3A). In contrast, 2 to 3 times more abnormalities per sample were detected after leukemic evolution in both matched and unmatched cases with MPN (both P < .001; Figure 3A). We found no statistical relationship between the JAK2V617F status and the number of genomic changes in matched as well as unmatched samples (data not shown). However, samples from ET patients had fewer copy number changes than those from either PV or PMF patients in the chronic phase, which was highly significant in the unmatched cases (P < .001; Figure 3A, supplemental Figure 3A). After leukemic transformation, a similar number of SNP-array changes occurred in cases with prior ET compared with those with pre-existing PV and PMF (unmatched cases: P = .59). Statistical evaluation of the matched samples divided into each subentity was not possible because of the small number of cases (Figure 3Aii and supplemental Figure 3B). A subanalysis of the number of either deletions, duplications/amplifications, or CNN-LOH per case, matched and unmatched, is shown in supplemental Figure 3.
Genomic alterations per MPN patient in chronic versus blast phase. (A) Mean of SNP-array alterations per patient in MPN versus MPN-blast phase with (i) unmatched samples and (ii) matched samples (± SD); **P < .001. (B) Mean of SNP-array aberrations compared with cytogenetic alterations per patient in chronic versus blast phase with (i) unmatched samples and (ii) matched samples (± SD).
Genomic alterations per MPN patient in chronic versus blast phase. (A) Mean of SNP-array alterations per patient in MPN versus MPN-blast phase with (i) unmatched samples and (ii) matched samples (± SD); **P < .001. (B) Mean of SNP-array aberrations compared with cytogenetic alterations per patient in chronic versus blast phase with (i) unmatched samples and (ii) matched samples (± SD).
Compared with the cytogenetic data, SNP-array analysis detected more than 2-fold of additional chromosomal changes in the MPN samples of either chronic or blast phase, whereas SNP-array practically captured all cytogenetic abnormalities (Figure 3B).
Candidate genes involved in leukemic transformation of MPN patients
SNP-chip analysis detected several additionally altered regions in patients after leukemic evolution compared with the MPN chronic phase in both unmatched (Figure 4; supplemental Figure 4) and matched (Figure 5A) cases. The altered regions included chromosome 8q (MYC), 12p (ETV6), 17p (TP53), and 21q (RUNX1), which are already known to be involved in leukemogenesis.18-22 Trisomy 8 was detected in 12% of unmatched and 9% of matched cases in MPN-blast phase; interestingly, almost all these samples were negative for JAK2V617F. PMF patient 148, who was also JAK2V617F−, showed amplification of 8q24.21 in blast crisis involving the MYC gene. MPN-blast phase patients with trisomy 8 did not show an inferior outcome compared with cases without this abnormality (P = .11; data not shown).
Overview of gains and losses detected by CNAG software. Indicated are the most common altered regions in unmatched MPN-blast phase patients (n = 60; right-sided cytobands) compared with unmatched MPN patients (n = 77; left-sided cytobands). Each line represents 1 sample with either deletion (blue), duplication/amplification (red), or CNN-LOH (green). Candidate genes of the minimal altered regions are highlighted by arrows.
Overview of gains and losses detected by CNAG software. Indicated are the most common altered regions in unmatched MPN-blast phase patients (n = 60; right-sided cytobands) compared with unmatched MPN patients (n = 77; left-sided cytobands). Each line represents 1 sample with either deletion (blue), duplication/amplification (red), or CNN-LOH (green). Candidate genes of the minimal altered regions are highlighted by arrows.
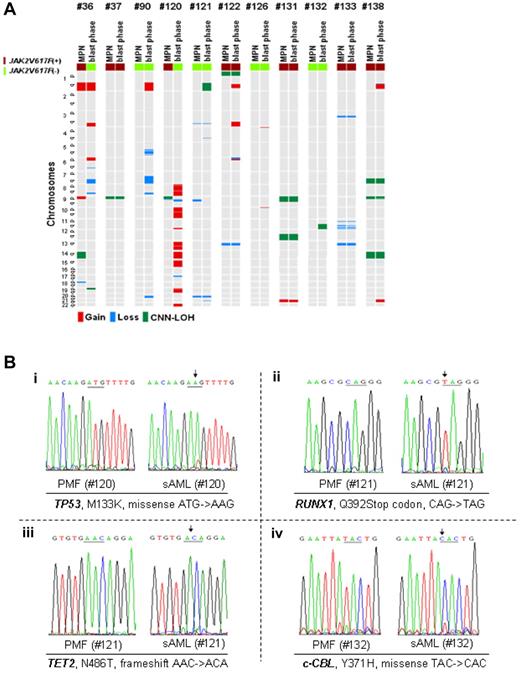
Gains and losses in matched MPN samples and mutational analysis. (A) Most commonly altered genomic regions in MPN samples (left sample column) compared with matched blast phase samples (right sample column) evolved from 11 patients (2 PV, 1 ET, 8 PMF). Each line represents 1 sample with either deletion (blue), duplication/amplification (red), or CNN-LOH (green). (Bi) Hemizygous TP53 mutation detected in MPN-blast phase sample of case 120 associated with acquired 17q deletion, which was not present in the MPN phase of case 120. (ii) Hemizygous RUNX1 mutation detected in MPN-blast phase sample (case 121) associated with acquired deletion at 22q22.1, which was not present in the MPN phase of case 121. (iii) Hemizygous TET2 mutation detected in MPN-blast phase sample of case 121 associated with acquired cryptic deletion on 4q24, which was not present in the MPN phase of case 121. (iv) Homozygous c-CBL mutation detected in MPN-blast phase sample of case 132 associated with acquired 11q CNN-LOH, which was not present in the MPN phase of case 132.
Gains and losses in matched MPN samples and mutational analysis. (A) Most commonly altered genomic regions in MPN samples (left sample column) compared with matched blast phase samples (right sample column) evolved from 11 patients (2 PV, 1 ET, 8 PMF). Each line represents 1 sample with either deletion (blue), duplication/amplification (red), or CNN-LOH (green). (Bi) Hemizygous TP53 mutation detected in MPN-blast phase sample of case 120 associated with acquired 17q deletion, which was not present in the MPN phase of case 120. (ii) Hemizygous RUNX1 mutation detected in MPN-blast phase sample (case 121) associated with acquired deletion at 22q22.1, which was not present in the MPN phase of case 121. (iii) Hemizygous TET2 mutation detected in MPN-blast phase sample of case 121 associated with acquired cryptic deletion on 4q24, which was not present in the MPN phase of case 121. (iv) Homozygous c-CBL mutation detected in MPN-blast phase sample of case 132 associated with acquired 11q CNN-LOH, which was not present in the MPN phase of case 132.
In 20% of unmatched cases in MPN-blast phase, deletions (12%) or CNN-LOH (8%) occurred on chromosome 17 including TP53 at p13.1. Deletions on the short arm of chromosome 17 were detected significantly often in MPN-blast phase patients who received prior treatment with hydroxyurea with or without the addition of alkylating agents (P = .035, Table 2). Supplemental Table 4 indicates pretreatment (hydroxyurea and/or alkylating agents) of 47 MPN-blast phase patients and their individual SNP-array findings. Deletion or CNN-LOH on 17p in unmatched blast phase cases was associated with either complex karyotype or isochromosome 17 (P = .01), and significantly decreased survival (with 17p deletion: P = .012; with 17p CNN-LOH: P = .018). One of the 11 matched MPN samples (case 120) acquired a 17p deletion at diagnosis of blast phase (Figure 5A), resulting in a hemizygous mutant TP53 (M133K; Figure 5Bi).
Pretreatment in 47 MPN-blast phase cases and frequency of 17p and 7q deletions
| Pretreatment | ||||
| Hydroxyurea | − | + | − | + |
| Alkylating agents | − | − | + | + |
| SNP-array alteration (17p vs 7q) | ||||
| No del(17)(p), no del(7(q) | 17 | 16 | 0 | 1 |
| del(7)(q) | 2 | 2 | 2 | 2 |
| del(17)(p) | 0 | 4* | 0 | 1* |
| del(17)(p) and del(7)(q) | 0 | 0 | 0 | 0 |
| Pretreatment | ||||
| Hydroxyurea | − | + | − | + |
| Alkylating agents | − | − | + | + |
| SNP-array alteration (17p vs 7q) | ||||
| No del(17)(p), no del(7(q) | 17 | 16 | 0 | 1 |
| del(7)(q) | 2 | 2 | 2 | 2 |
| del(17)(p) | 0 | 4* | 0 | 1* |
| del(17)(p) and del(7)(q) | 0 | 0 | 0 | 0 |
Numbers of blast-phase patients are presented.
MPN indicates myeloproliferative neoplasm; and SNP, single nucleotide polymorphism.
A total of 5 cases with del(17)(p) pretreated with hydroxyurea (P = .035).
On chromosome 21, SNP-chip analysis revealed either deletions or CNN-LOH in 8% of unmatched cases in MPN-blast phase involving the transcription factor RUNX1 at q22.12. Patient 121 acquired a small deletion of that locus in the leukemic sample (Figure 5A) associated with a mutation of the Runt domain of the RUNX1 gene on the remaining allele (Q392Stop codon; Figure 5Bii).
Deletion or CNN-LOH on 4q24 spanning the TET2 gene was detected in 6% of unmatched blast phase cases and 1% in chronic phase. One TET2 mutation was found by nucleotide sequencing in the matched MPN samples. JAK2V617F− case 121 had no genomicimbalances on 4q at diagnosis of PMF, but acquired a microdeletion (1 Mbp) on 4q24 (TET2) after leukemic evolution 1 year later (Figure 5A). The remaining allele had a TET2 frameshift mutation (N486T; Figure 5Biii), and the mutation was absent in the matched PMF sample.
CNN-LOH involving 11q23.3, which has been shown to be strongly associated with c-CBL mutations,23 had an even lower frequency, with only 2% of unmatched MPN cases in either chronic phase or blast crisis. The JAK2V617F− patient 132 had 11q CNN-LOH with a homozygous c-CBL missense mutation (Y371H) in the MPN-blast phase sample. Both the CNN-LOH and the mutation were absent in the corresponding chronic phase, 2 years before disease progression (Figure 5A-Biv).
Besides these already well-known targets, SNP-array analysis detected commonly altered regions on chromosomes 1, 7, 16, 19, and 21 encompassing potentially new candidate genes involved in MPN transformation. These imbalances were either absent or at least very infrequent in the chronic phase of the disease (Figures 4 and 5A, supplemental Figure 4). Ten percent of unmatched and 18% of matched MPN-blast phase cases had either duplication/amplification or CNN-LOH on 19p. The commonly involved region spanned a small locus (2 Mbp) at 19p13.2, where, among others, the genes PIN1, ICAM1, and CDC37, which have been associated with carcinogenesis, are located.24-26 In addition, the minimal region (1.8 Mbp) of amplifications/duplications/trisomy on chromosome 21 detected in 8% of unmatched and 9% matched MPN-blast phase samples harbored the oncogenic transcription regulator ERG (q22.2).
Complete or partial deletion (−7/7q−), as well as CNN-LOH of the long arm of chromosome 7, was one of the most common abnormalities detected by SNP-array analysis in up to 25% of unmatched and 27% matched samples evolved in the blast phase. SNP-array also revealed 3 unmatched cases (32, 87, and 116) with a heterozygous microdeletion encompassing the 7q22.1 locus, which was not detectable by cytogenetic analysis. Moreover, case 138 with 7qCNN-LOH had a homozygous deletion on 7q22.1 in both the matched MPN and MPN-blast phase samples (supplemental Figure 2A). The minimally deleted region spanned a small region of 0.88 Mbp at 7q22.1 covering only 2 target genes, CUTL1 and SH2B2. The homozygous deletion of these genes in patient 138 was confirmed by QG RT-PCR (supplemental Figure 2B). Deletions of the long arm of chromosome 7 were found more often in MPN-blast phase patients pretreated with hydroxyurea and/or alkylating agents, but the findings were not statistically significant (P = .2; Table 2).
Also worth mentioning, 1 microdeletion encompassing the chromosome X–linked PIG-A gene occurred in male patient 121 at leukemic transformation (supplemental Figure 2C). This patient had a normal chromosome X in his chronic phase of PMF.
CNN-LOH is a marker of poor survival in MPN patients after leukemic evolution
SNP-array technology provides efficient and effective detection of segmental CNN-LOH. In the present study, the most prominent regions for CNN-LOH besides chromosome 9p (JAK2) were on 7q and 17p (TP53) in patients with MPN-blast phase. In marked contrast to CNN-LOH on 9p, CNN-LOH on 7q or 17p almost never occurred in the chronic phase of the disorder in matched and unmatched samples. As mentioned previously, cases with CNN-LOH and/or deletion of 17p were associated with either complex karyotype or isochromosome 17 and decreased survival.
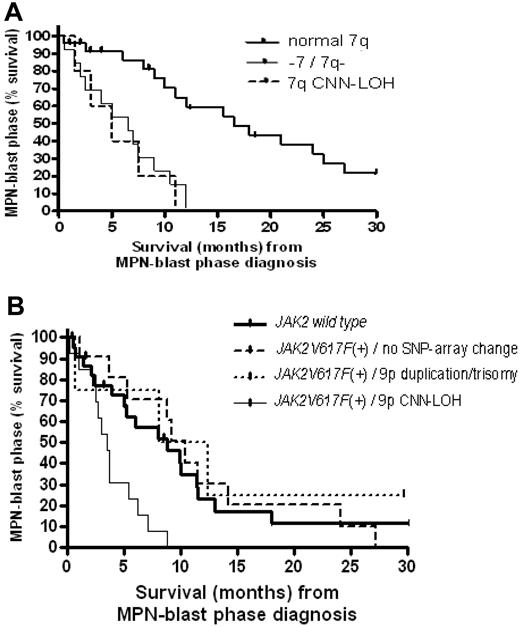
As also expected, survival in the MPN-blast phase was significantly decreased in patients with −7/7q− (median, 3.75 months) compared with those without chromosome 7 alterations (median, 9 months; P = .008). In addition, the unbalanced translocation, der(1;7)(q10;p10), a nonrandom chromosomal abnormality rarely found in AML, was detected by SNP-chip and FISH in 7% of unmatched samples after leukemic evolution and was also associated with an inferior outcome compared with patients without chromosome 7 imbalances (P = .014). Strikingly, survival continued to be significantly decreased in MPN-blast phase, when cases with only 7qCNN-LOH were compared with those with a normal 7q (P = .01; Figure 6A; supplemental Table 5).
Survival analysis in MPN-blast phase. (A) Survival from the time of diagnosis of blast phase in transformed MPN patients with normal chromosome 7 (normal 7q) compared with either monosomy 7 (−7)/deletion of 7q (7q−) or 7qCNN-LOH. (B) Survival from the time of diagnosis of blast phase in transformed MPN patients with homozygous JAK2V617F+ associated with 9pCNN-LOH compared with either heterozygous JAK2V617F+ with 9p duplication/trisomy 9 or no abnormality, or patients without the mutation (JAK2 wild type). Median survival (months) and the case numbers for each group (transformed PV, ET, or PMF) are listed in supplemental Table 5.
Survival analysis in MPN-blast phase. (A) Survival from the time of diagnosis of blast phase in transformed MPN patients with normal chromosome 7 (normal 7q) compared with either monosomy 7 (−7)/deletion of 7q (7q−) or 7qCNN-LOH. (B) Survival from the time of diagnosis of blast phase in transformed MPN patients with homozygous JAK2V617F+ associated with 9pCNN-LOH compared with either heterozygous JAK2V617F+ with 9p duplication/trisomy 9 or no abnormality, or patients without the mutation (JAK2 wild type). Median survival (months) and the case numbers for each group (transformed PV, ET, or PMF) are listed in supplemental Table 5.
The JAK2V617F mutational status in terms of heterozygosity or homozygosity appeared to have no influence on the duration to leukemic evolution. Regardless of the underlying MPN subgroup, no statistical difference in the time to leukemic transformation was found comparing JAK2V617F+ patients with normal chromosome 9 to mutant positive blast phase patients with either 9p duplication/trisomy 9 (P = .28) or 9pCNN-LOH (P = .21). Instead, we found that homozygous JAK2V617F had an impact on survival after MPN transformation. Blast phase patients with 9pCNN-LOH resulting in a homozygous JAK2 mutation had a worse outcome (median, 4 months) compared with JAK2V617F+ MPN-blast phase patients with either 9p duplication/trisomy 9 (median, 7.5 months) or no abnormality on 9p (median, 9 months), as well as patients without JAK2V617F (median, 7 months, P = .016; Figure 6B; supplemental Table 5). Homozygous JAK2V617F in association with CNN-LOH diagnosed at leukemic transformation was independent of known risk factors such as 5q−, −7/7q−, or complex karyotype (P > .05).
Discussion
Oncogenic JAK2 signaling is an important event in MPN.1,2 Recently, we and others showed that homozygosity for JAK2V617F is closely related to chromosome 9pCNN-LOH in MPN patients.1,6,8,9 However, the transformation process of MPN to MPN-blast phase is not well understood.
Recent findings suggested that transition from heterozygosity to homozygosity for JAK2V617F is associated with a hyperproliferative disease profile and may be important for disease progression, at least from PV to secondary myelofibrosis.27 Moreover, Barosi et al showed in a longitudinal prospective study that the presence of a JAK2V617F hematopoietic clone was significantly associated with leukemic transformation in PMF.28 This is in contrast to our present findings showing that not only the mutational status of JAK2V617F, but also 9pCNN-LOH with homozygous JAK2V617F, had no impact on the time to leukemic transformation in patients with MPN-blast phase. In addition, 2 of the 11 matched MPN samples, initially positive for JAK2V617F with either trisomy 9 or 9pCNN-LOH, became negative for these abnormalities after leukemic transformation. Although only tested in unpaired samples, PMF and PV samples also showed a significantly smaller number of both JAK2V617F+ and 9pCNN-LOH in the blast phase compared with the chronic phase. Interestingly, and also contrary to the previously cited studies, Tefferi et al noted a significant association between a low JAK2V617F allelic burden and evolution to blast phase in a large cohort of PMF patients.29 Even though these data are not completely congruent with our findings, the results of Tefferi et al and our results point to the coexistence of a more dominant JAK2V617F-negative clone with a higher propensity to undergo clonal evolution. This is congruent with recent studies indicating that JAK2V617F+ MPN can result in JAK2V617F− MPN-blast phase.30,31 But still, some of our matched cases with JAK2V617F+ had no change in abnormalities including JAK2 mutational status as well as 9pCNN-LOH, allowing the existence of a common pre-JAK2V617F clone. Taken together, the presence of JAK2V617F appears not to be a prerequisite for leukemic transformation of MPN, suggesting that additional genetic events are required for full transformation.
SNP-array analysis was able to capture practically all cytogenetic abnormalities and to uncover additional lesions with potentially important clinical implications. The number of genomic alterations was more than 2 to 3 times greater in the blast phase as in the chronic phase of matched and unmatched cases with MPN. Noticeably, ET patients had fewer alterations in their chronic phase samples compared with the PV and PMF cases, whereas the number was comparable in all 3 MPN subgroups after their transformation. Being aware of the increased number of new genomic changes enables investigators to focus on the identification of causative genes associated with the evolution of MPN to leukemia.
Commonly altered regions in blast crisis samples were detected on chromosomes 8, 12, 17, and 21 encompassing MYC, ETV6, TP53, and RUNX1, respectively, which are already known to be involved in the development of de novo and secondary AML.18-22 Gain of chromosomal material at 8q24.21 was almost exclusively found in JAK2V617F− samples, suggesting that increased activity of MYC might allow selection of clones that do not require the JAK2 gain-of-function mutation. Furthermore, deletion of 17p (TP53) was significantly associated with prior exposure to hydroxyurea as well as a complex karyotype in samples with MPN-blast crisis, which is in accordance with recent results.32,33 Interestingly, not only deletion, but also 17pCNN-LOH, was associated with a complex karyotype, a poor prognostic marker in myeloid malignancies.
In addition, regions on chromosomes 1q, 7q, 16q, 19p, and 21q were frequently altered in the evolution to the leukemic phase and may harbor promising new candidate genes. Abnormalities involving chromosome 7 are frequently detectable in de novo and secondary AML,34-37 and preceding studies have found a critical breakpoint region involving a locus at centromeric band 7q22, whereas the telomeric breakpoint varies from q32 to q36. Interestingly, the minimal deleted region in our cohort was located at 7q22.1 encompassing only 2 promising target genes, SH2B2 (previously named APS) and CUTL1. SH2B2 regulates and enhances JAK2-mediated cellular responses,38 and the CUTL1 gene encodes for a CUT family member of the homeodomain proteins that can repress the expression of developmentally regulated myeloid genes.39 Moreover, genome-wide inspection for minimal regions of duplications/amplifications and CNN-LOH revealed several interesting genes, such as PIN1, ICAM1, and CDC37 on 19p as well as ERG on 21q. Whereas the latter 3 targets have been shown to possess potential progrowth activity in de novo AML and/or MDS,25,26,40 PIN1 is known to be overexpressed in a variety of cancers and may act as an oncogene via promotion of cell cycle progression and proliferation.24
Mutations of the c-CBL gene are tightly associated with 11qCNN-LOH and are commonly diagnosed in patients with chronic myelomonocytic leukemia.23,41,42 Although MPN shares clinical as well as hematologic features with chronic myelomonocytic leukemia, we detected 11qCNN-LOH only in a minority of our study population, suggesting that c-CBL mutations are rare events leading to transformation of chronic MPN to leukemic blast phase.
In contrast to recent findings showing frequent LOH on 4q associated with TET2 mutations in patients diagnosed with MDS/MPN,43 we detected CNN-LOH or deletions at 4q24 (TET2) only in a minority of our patients in the chronic as well as blast phase of MPN. Nevertheless, our study was not sufficient to explore these findings in more detail and make conclusions on tumor suppressor TET2 and its potential role in leukemic transformation.
However, with regard to the variety of detected allelic imbalances, we suggest that no single candidate gene or molecular pathway is sufficient and necessary to cause transformation of chronic MPN to blast phase. Like de novo AML, MPN-blast phase appears to be a heterogeneous disease prone to have evolved multiple mechanisms to provide a proliferative advantage to the abnormal leukemic clone.
CNN-LOH involving chromosomal regions that are also frequently affected by deletions may have prognostic implications similar to the deletions visible by karyotyping. In our study, prognostic evaluation was based mainly on SNP-array data from blast phase samples without the incorporation of SNP-array results from the matched chronic phase. Moreover, we implied the survival and clinical outcome only of MPN patients who underwent leukemic transformation, without comparison with survival and outcome in untransformed chronic phase. However, as expected, blast phase patients with loss of chromosomal material on 7q showed poor survival, because this is known to be predictive for rapid progression and poor response in AML therapy.35-37 MPN-blast phase patients with cytogenetically undetectable 7qCNN-LOH had comparable survival rates to those with −7/7q− in their leukemic cells, which is in accordance with previously published data.44
In addition, 9pCNN-LOH with homozygous JAK2 mutation was also linked to an inferior outcome in MPN-blast crisis in comparison with patients with either heterozygous JAK2V617F or wild-type JAK2. In contrast to LOH on 17p, the prognostic impact of 9pCNN-LOH was independent of established risk factors such as −7/7q−, 5q−, or complex karyotype. Although JAK2V617F in association with 9pCNN-LOH appeared to have no impact on the time to MPN transformation, we suggest that the homozygous driver mutation in combination with additional newly acquired aberrations in terms of a second hit may have an implication on the clinical course of MPN-blast phase patients.
In conclusion, high-density SNP-array technology allowed precise identification of chromosomal aberrations, including CNN-LOH, and complemented conventional cytogenetic techniques in patients with chronic and transformed MPN. Our analysis provided prognostic details to further improve clinical prognosis, as well as novel interesting candidate genes potentially involved in the transformation of MPN.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the members of all cooperating laboratories for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; TH 1438/1-1, N.H.T.), National Science Council (NSC; NSC96-2314-B-182-003, L.-Y.S.), Chang Gung Memorial Hospital (CMRP; CMRPG330303, L.-Y.S.), and the National Institutes of Health (NIH; grant 5R01CA026038-31, H.P.K.). H.P.K. is the holder of the Mark Goodson endowed Chair in Oncology Research at Cedars Sinai Medical Center and is a member of the Jonsson Cancer Center and the Molecular Biology Institute, UCLA, and is supported by the A*STaR award from the National University of Singapore.
National Institutes of Health
Authorship
Contribution: N.H.T and U.O.K. performed the research, analyzed the data, and wrote the paper; D.H.T.L., N.K., G.B.I., T.L., T.W., D.N., M.K.-M., M.K., M.S., L.-Y.S., A.N., and S.D.R. assisted with the research; C.M.-T., R.M., T.H., D.G.G., and A.T. designed and performed the research; and S.O. and H.P.K. directed the overall study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nils H. Thoennissen, Division of Hematology and Oncology, Cedars Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, Los Angeles, CA 90048; e-mail: nils.thoennissen@cshs.org.
References
Author notes
N.H.T. and U.O.K. contributed equally and should be considered as co–first authors.
S.O. and H.P.K. contributed equally and should be considered as co–last authors.