Abstract
Mutations in the human nucleophosmin (NPM1) gene are the most frequent genetic alteration in adult acute myeloid leukemias (AMLs) and result in aberrant cytoplasmic translocation of this nucleolar phosphoprotein (NPMc+). However, underlying mechanisms leading to leukemogenesis remain unknown. To address this issue, we took advantage of the zebrafish model organism, which expresses 2 genes orthologous to human NPM1, referred to as npm1a and npm1b. Both genes are ubiquitously expressed, and their knockdown produces a reduction in myeloid cell numbers that is specifically rescued by NPM1 expression. In zebrafish, wild-type human NPM1 is nucleolar while NPMc+ is cytoplasmic, as in human AML, and both interact with endogenous zebrafish Npm1a and Npm1b. Forced NPMc+ expression in zebrafish causes an increase in pu.1+ primitive early myeloid cells. A more marked perturbation of myelopoiesis occurs in p53m/m embryos expressing NPMc+, where mpx+ and csf1r+ cell numbers are also expanded. Importantly, NPMc+ expression results in increased numbers of definitive hematopoietic cells, including erythromyeloid progenitors in the posterior blood island and c-myb/cd41+ cells in the ventral wall of the aorta. These results are likely to be relevant to human NPMc+ AML, where the observed NPMc+ multilineage expression pattern implies transformation of a multipotent stem or progenitor cell.
Introduction
Nucleophosmin (NPM1) is a ubiquitous multifunctional phosphoprotein, which is normally located in the nucleolus but has nucleocytoplasmic shuttling activity. Known functions of NPM1 include roles in ribosome biogenesis and control of centrosome duplication.1 NPM1 also regulates the tumor suppressors p14ARF2,3 and p53,4 and is up-regulated by cellular stresses such as DNA damage.5 Npm1+/− heterozygous mice are viable, and show a hematologic syndrome similar to human myelodysplastic syndrome (MDS), suggesting a role of Npm1 in hematopoiesis.6 Moreover, Npm1+/− mice show genomic instability and a greater propensity to develop hematologic malignancies, defining Npm1 as a haploinsufficient tumor suppressor.7 Heterozygous gain of function mutations in NPM1 are the most frequent genetic abnormalities found in human acute myeloid leukemia (AML), occurring in 30% of all adult cases.8 Such mutations of NPM1 appear to be specific to AML, as they do not occur in other human cancers.8,9 In the absence of FLT3-ITDs, mutated NPM1 in adult AML has been associated with a better prognosis.10 Because of its distinctive biologic and clinical features,11 NPM1-mutated AML has been included as a new provisional entity in the 2008 World Health Organization (WHO) classification of human myeloid neoplasms.12 Consequently, NPM1 mutational analysis is now performed routinely on adults diagnosed with AML.10,11
NPM1 expression is normally detected primarily in the nucleolus, but when mutated it aberrantly relocates to the cytoplasm.13 More than 50 mutations have been described that lead to this aberrant localization, and thus the term NPM1 cytoplasmic–positive (NPMc+) has been used to define cytoplasmic mutants that are found in AML.8 Mechanisms underlying the aberrant localization of NPMc+ have been identified,14,15 and studies have shown that NPMc+ can function as an oncogene in vitro16 and disrupt murine hematopoiesis;17 however, the critical steps leading to leukemic transformation of NPMc+-expressing cells remain unknown.
The zebrafish is a powerful vertebrate disease model system. Several human and murine oncogenes involved in leukemogenesis cause leukemia and disrupt normal hematopoiesis in the zebrafish, providing strong support of the zebrafish as a relevant model system to study mechanisms underlying human leukemia.18-20 Two distinct waves of zebrafish hematopoiesis have been described. Primitive hematopoiesis is characterized by the generation of embryonic erythrocytes in the intermediate cell mass (ICM)21 and a distinct population of primitive macrophages that arise from the anterior lateral plate mesoderm (ALPM).22 Definitive hematopoiesis consists of 2 phases: at first, transient erythromyeloid progenitors (EMPs) form in the posterior blood island (PBI) starting around 24 hours postfertilization (hpf),23 followed by the appearance of hematopoietic stem cells (HSCs) in the ventral wall of the dorsal aorta by 30 hpf, in a region that is thought to be equivalent to the mammalian aorta-gonad-mesonephros (AGM).24 These HSCs then colonize the thymus and the kidney, the hematopoietic organ in adult zebrafish.24
We have used the zebrafish model to study the physiologic role of Npm1 and the perturbation in hematopoiesis induced by NPMc+ expression. We identified 2 zebrafish orthologs of the human NPM1 gene, npm1a and npm1b, and determined that these genes are required for normal hematopoiesis. Furthermore, we have shown for the first time that NPMc+ expression causes expansion of primitive and definitive hematopoietic cells in vivo, establishing a platform to further investigate the mechanisms through which NPMc+ contributes to leukemic transformation.
Methods
Zebrafish, microinjections, and WISH
Wild-type AB stocks of Danio rerio, the transgenic lines Tg(pu.1:EGFP),25 Tg(c-myb:EGFP),26 Tg(cd41:EGFP),27 Tg(gata1:DsRED),28 Tg(lmo2:DsRed),29 and Tg(gata1:EGFP),30 and the mutant line tp53zdf1/zdf1 (tp53M214K/M214K, referred to in the text as p53m/m)31 were maintained as described.32 mRNAs were injected into 1-cell–stage embryos at the indicated amounts. Morpholinos (MOs) targeting the 5′UTR/ATG codon or splice donor sites of npm1a, npm1b, and control morpholinos, were designed by Gene-Tools LLC. Sequences are given in Table 1. MO concentrations were titrated to the lowest dose resulting in phenotypes: 1.6 ng each for npm1a and npm1b ATG MOs, 8 ng and 4 ng for npm1a splice donor MOs (exon2-intron2(2) and exon3-intron3, respectively), and 4 ng for npm1b splice donor MO (exon2-intron2). For experiments in p53-deficient backgrounds, NPM1- or NPMc+-encoding mRNAs were injected into the p53m/m line or with the p53 MO (1.6 ng)33 in the Tg(pu.1:EGFP) line. Observed increases in enhanced green fluorescent protein–positive (EGFP+) cells in the Tg(pu.1:EGFP) line expressing NPMc+ served as an indicator for NPMc+ function.
MO sequences
| Gene . | Target . | MO sequence . |
|---|---|---|
| npm1a | ATG/5′UTR | TAATGTTATCCTCCATTTTTGCGCG |
| npm1a | ATG/5′UTR 5-bp mismatch | TAATcTTATCgTCgATTTTaGCcCG |
| npm1a | Exon2-intron2 splice donor (2)* | AAGAAACATCACATACCATTCTAAC |
| npm1a | Exon3-intron3 splice donor | TTATGACCAAGTCTACTTACACTTG |
| npm1b | ATG/5′ UTR | GACCCATCTGTTCGAGATCCATGTC |
| npm1b | ATG/5′UTR 5-bp mismatch | GAgCCATgTGTTCGAcATCgATcTC |
| npm1b | Exon2-intron2 splice donor | TCAAATATTCATCTTCCTCACCGAC |
| p53 | ATG/5′UTR | GCGCCATTGCTTTGCAAGAATTG |
| Std cont | Human β-globin intron mutation | CCTCTTACCTCAGTTACAATTTATA |
| Gene . | Target . | MO sequence . |
|---|---|---|
| npm1a | ATG/5′UTR | TAATGTTATCCTCCATTTTTGCGCG |
| npm1a | ATG/5′UTR 5-bp mismatch | TAATcTTATCgTCgATTTTaGCcCG |
| npm1a | Exon2-intron2 splice donor (2)* | AAGAAACATCACATACCATTCTAAC |
| npm1a | Exon3-intron3 splice donor | TTATGACCAAGTCTACTTACACTTG |
| npm1b | ATG/5′ UTR | GACCCATCTGTTCGAGATCCATGTC |
| npm1b | ATG/5′UTR 5-bp mismatch | GAgCCATgTGTTCGAcATCgATcTC |
| npm1b | Exon2-intron2 splice donor | TCAAATATTCATCTTCCTCACCGAC |
| p53 | ATG/5′UTR | GCGCCATTGCTTTGCAAGAATTG |
| Std cont | Human β-globin intron mutation | CCTCTTACCTCAGTTACAATTTATA |
Bold letters indicate the ATG codon; lowercase letters, mismatched residues; and Std cont, standard control.
(2) is the 1 working morpholino of 2 tested (supplemental Figure 1F, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Developmental staging of injected zebrafish embryos and uninjected controls was determined by somite number. Embryos were fixed in 4% paraformaldehyde (PFA), and assays for mRNA expression using whole-mount in situ hybridization (WISH) were performed as described.34 Embryos were visualized and imaged with a Nikon SMZ1500 zoom stereomicroscope (Nikon Instruments Inc) using the NIS-Elements software (Nikon Instruments Inc). Some WISH embryos were postfixed in 4% PFA and embedded in 1.5% agar/5% sucrose, equilibrated in 30% sucrose, and cryosectioned (20 μm). All animal protocols were approved by the Dana-Farber Animal Care and Use Committee.
Cloning of human and zebrafish NPM1 cDNAs and RNA production
Full-length NPM1 cDNA (wild-type and mutated) were subcloned in the pCS2+ or p3X-FLAG vector (Sigma) from pGEM-T-easy-NPM1 and pGEM-T-easy-NPMmutA.8 Full-length cDNA encoding EGFP-NPM1 (wild-type and mutated) were subcloned in pCS2+ from pEGFP-C1-NPM1 and pEGFP-C1-NPMmutA.8 Full-length cDNA of the 2 zebrafish NPM1 orthologs npm1a and npm1b were reverse transcription–polymerase chain reaction (RT-PCR)–amplified from 24 hpf AB embryos. Primers for full-length npm1a coding region amplification were designed based on the published NCBI sequence (NM_199428), while those for npm1b were based on overlapping expressed sequence tag (EST) sequences and the recently annotated Zv8 sequence BC093285 (Table 2). Full-length npm1a and npm1b amplicons were cloned into pCRII-Topo Dual Promoter vector (Invitrogen), from which WISH probes were made, and then into pCS2+, pEGFP-C1, and pDsRed-Monomer-C1 expression vectors (Clontech). All constructs were verified by sequencing. All mRNAs were transcribed from pCS2+ using the mMessage mMachine kit (Ambion).
Oligonucleotide sequences
| Primer . | Sequence . |
|---|---|
| Npm1a forward BamH1 | 5′-CGGGATCCCGCTTCCTGCACTCGTGTTCGCTGAC-3′ |
| Npm1a reverse XbaI | 5′-GCTCTAGAGCGGCCATTCTGAAACCCACTATTC-3′ |
| Npm1b forward BamHI | 5′-CGGGATCCGCGCAAAATGGAGGATAAC-3′ |
| Npm1b reverse XhoI | 5′-ACTCGAGCGCAGACTTCTGCTGCACT-3′ |
| β-actin forward | 5′-CTGGTCGTTGACAACGGCT-3′ |
| β-actin reverse | 5′-TCCATCACAATACCAGTAGT-3′ |
| Primer . | Sequence . |
|---|---|
| Npm1a forward BamH1 | 5′-CGGGATCCCGCTTCCTGCACTCGTGTTCGCTGAC-3′ |
| Npm1a reverse XbaI | 5′-GCTCTAGAGCGGCCATTCTGAAACCCACTATTC-3′ |
| Npm1b forward BamHI | 5′-CGGGATCCGCGCAAAATGGAGGATAAC-3′ |
| Npm1b reverse XhoI | 5′-ACTCGAGCGCAGACTTCTGCTGCACT-3′ |
| β-actin forward | 5′-CTGGTCGTTGACAACGGCT-3′ |
| β-actin reverse | 5′-TCCATCACAATACCAGTAGT-3′ |
Bold letters indicate restriction enzyme sites.
Comparative analysis and identification of synteny to human 5q35.1-5q35.2
Genes located in 5q35.1-5q35.2 were identified using the National Center for Biotechnology Information (NCBI) Mapviewer.35 Zebrafish orthologs for genes in this region were identified using a “reciprocal best-hit” analysis as described.36 TBLASTN searches were applied for human protein sequences and BLASTX or TBLASTN searches for zebrafish cDNA sequences or proteins, respectively (Ensembl Zv7). Comparative analysis between the human and the 2 zebrafish protein sequences used the Clustal W algorithm.37
RT-PCR, protein extraction, WB, and coimmunoprecipitation studies
Total RNA from zebrafish embryos at various stages of development was extracted using Trizol (Invitrogen) following the manufacturer's instructions. RT-PCR was performed on RNA using the Qiagen One-Step RT-PCR Kit using gene-specific primers. Primers used for npm1a and npm1b amplification spanned the full-length coding sequence. Primers used for β-actin amplification spanned 2 introns. Primer sequences are provided in Table 2. Protein lysates were obtained from single-cell suspensions of approximately 20 zebrafish embryos per condition. Embryos were dissociated with 0.5% trypsin (GIBCO), and protein extraction, Western blot (WB), and coimmunoprecipitation studies were carried out as described.14 Antibodies used were anti-NPM1, clone 376;8 anti–β-tubulin (Abcam); anti-GFP (Roche); and anti-FLAG (Sigma).
Fluorescence analysis in zebrafish embryos and 293T cells
For cytospins (Figure 3A-F), 24-hpf embryos injected with EGFP-NPM1 or EGFP-NPMc+ at the indicated doses were dissociated with 0.5% trypsin (GIBCO). The cell suspension was fixed in 4% PFA. A total of 105 cells were spun onto glass slides and mounted with ProLong Gold Antifade reagent with DAPI (Molecular Probes). For confocal analysis of EGFP+ cells from whole embryos (Figure 3H-I), 24-hpf embryos injected with EGFP-NPM1 (50 ng) or EGFP-NPMc+ (10 ng) were fixed in 4% PFA, deyolked, and flat-mounted on glass slides with ProLong Gold Antifade reagent with DAPI (Molecular Probes). 293T cells (Figure 4) were seeded on glass coverslips and transfected 24 hours later using Fugene 6 (Roche). In cotransfection experiments, plasmids were used at equimolar ratios. At 24 hours after transfection, cells were rinsed in phosphate-buffered saline (PBS), fixed in PFA (10 minutes), and mounted on glass slides with ProLong Gold Antifade reagent with DAPI (Molecular Probes). Whole-mount anti–activated caspase-3 and anti-GFP immunostaining (Figure 6) were performed as follows: embryos fixed in 4% PFA were rinsed in PDT (PBST, 0.3% Triton-X, and 1% DMSO), blocked in CAB (10% HI-FBS, 2% BSA in PBST), and incubated with primary antibodies (rabbit anti–activated caspase-3 [BD Biosciences] and mouse anti-GFP [Molecular Probes]) in CAB (1:500). Embryos were then washed in PDT, blocked in CAB, and incubated with secondary antibodies (anti–mouse AlexaFluor-488–conjugated and anti–rabbit AlexaFluor-568–conjugated; Molecular Probes) in CAB (1:200). Acridine orange stains were performed as previously described.38 Confocal images of 32-hpf Tg(cd41:EGFP), Tg(gata1:dsRed);(c-myb:EGFP), and Tg(lmo2:dsRed);(gata1:EGFP) transgenic embryos injected with NPM1 (50 ng) or NPMc+ (10 ng; Figure 7) were taken from live embryos mounted in 1% low-melting point agarose.
Analysis of zebrafish by flow cytometry
Specific hematopoietic subpopulations from adult dissected kidney marrows were obtained as described.28 Transgenic embryos were dissociated in liberase blendzyme 3 (Roche) for 30 to 60 minutes at 32°C. Embryos were gently dissociated using a pipette tip, passed through a 40-μM filter, and washed 3 times in cold PBS. Tg(pu.1:EGFP) were labeled with annexin V–phycoerythrin (PE) to assess apoptosis.38 DNA content of pu.1:EGFP+ cells was determined by hypotonic propidium iodide (PI) staining of EGFP+ cells sorted on a MoFlo cell sorter (DAKO Cytomation). Fluorescence-activated cell sorter (FACS) analysis of c-myb:EGFP+ cells was conducted in dissected trunks of embryos, gating on the FSCHIGH population as described in Bertrand et al.24 Data from Tg(c-myb:EGFP), Tg(pu.1:EGFP), and double Tg(gata1:EGFP);Tg(lmo2:dsRed) cell suspensions were acquired on a FACSCanto II (BD Biosciences). Dead c-myb:EGFP and pu.1:EGFP cells were excluded by PI staining. Data were analyzed using FlowJo (TreeStar) or Modfit LT (Verity Software).
Results
Identification of 2 zebrafish NPM1 genes
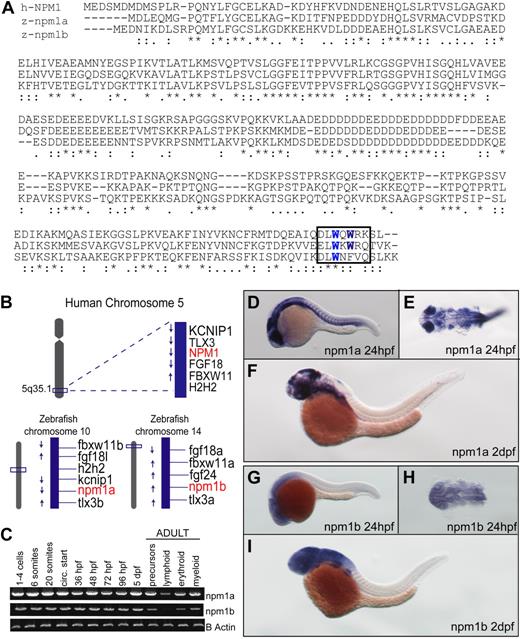
Zebrafish often have 2 copies of genes orthologous to a single mammalian gene as a result of genomic duplication during teleost evolution.39 To identify the zebrafish npm1 gene(s), the human NPM1 amino acid sequence was used in a TBLASTN search using the Ensembl, University of California Santa Cruz (UCSC), and NCBI platforms. A total of 2 loci were identified, including an annotated zebrafish npm1 gene on chromosome 10 whose protein product exhibits 51% identity to NPM1, and multiple overlapping ESTs encoded by a novel locus on chromosome 14, corresponding to a gene with 48% identity to NPM1 (Figure 1A). In silico analysis of the putative proteins encoded by the 2 genes shows that critical NPM1 functional domains are conserved in both Npm1 zebrafish proteins, including the C-terminal tryptophan residues implicated in nucleolar localization14 (Figure 1A boxed area; supplemental Figure 1A-B). Furthermore, the genomic regions surrounding both zebrafish npm1 genes showed high degrees of synteny with human 5q35.1, the locus of human NPM1 (Figure 1B). Thus, each of these genes is orthologous to NPM1; we refer to these genes as npm1a (chromosome 10) and npm1b (chromosome 14). Both zebrafish npm1 genes are expressed maternally and throughout the first 5 days of development (Figure 1C). They are also both expressed in all hematopoietic compartments of the adult kidney marrow (Figure 1C). To investigate the spatial expression pattern of npm1a and npm1b, we performed WISH and found that both npm1a and npm1b are ubiquitously expressed at 24 hpf, with highest levels in the developing brain, eye, and somites (Figure 1D-E and 1G-H, respectively). By 2 days after fertilization (dpf), expression was most prominent in the brain and fin-buds (Figure 1F,I).
Identification of 2 putative zebrafish npm1 genes. (A) Clustal W alignment of human NPM1 (top row), zebrafish Npm1a (middle row), and Npm1b (bottom row) proteins. * indicates identical residues; colon (:), highly similar residues; and period (.), similar residues. Critical tryptophan residues necessary for nucleolar localization are highlighted in blue in the boxed area. (B) The genomic loci surrounding human NPM1 on chromosome 5q35.1 (top row) are syntenic with the regions where npm1a (on chromosome 10) and npm1b (on chromosome 14) are located in the zebrafish genome (bottom row left and right, respectively). (C) RT-PCRs showing npm1a (top row) and npm1b (middle row) expression levels. β-actin expression levels are used as a loading control (bottom row). Left: npm1a and npm1b embryonic expression was assessed from whole embryos at the indicated time points. Right: RT-PCRs from precursor, lymphoid, myeloid, and erythroid cells sorted from adult kidney marrow (gating strategy based on forward- and side-scatter plots28 ). (D,F,G,I) npm1a or npm1b WISH assays in 24-hpf or 48-hpf embryos, lateral view, anterior to the left, dorsal upwards. (E,H) Close-up dorsal view (anterior to the left) of the brain and anterior trunk in flat-mounted, deyolked 24-hpf embryos stained with npm1a or npm1b probes. Digoxigenin-labeled RNA probes encoding the full-length npm1a and npm1b sequences were transcribed from linearized cDNA constructs using the DIG RNA labeling kit (Roche) following manufacturer instructions.
Identification of 2 putative zebrafish npm1 genes. (A) Clustal W alignment of human NPM1 (top row), zebrafish Npm1a (middle row), and Npm1b (bottom row) proteins. * indicates identical residues; colon (:), highly similar residues; and period (.), similar residues. Critical tryptophan residues necessary for nucleolar localization are highlighted in blue in the boxed area. (B) The genomic loci surrounding human NPM1 on chromosome 5q35.1 (top row) are syntenic with the regions where npm1a (on chromosome 10) and npm1b (on chromosome 14) are located in the zebrafish genome (bottom row left and right, respectively). (C) RT-PCRs showing npm1a (top row) and npm1b (middle row) expression levels. β-actin expression levels are used as a loading control (bottom row). Left: npm1a and npm1b embryonic expression was assessed from whole embryos at the indicated time points. Right: RT-PCRs from precursor, lymphoid, myeloid, and erythroid cells sorted from adult kidney marrow (gating strategy based on forward- and side-scatter plots28 ). (D,F,G,I) npm1a or npm1b WISH assays in 24-hpf or 48-hpf embryos, lateral view, anterior to the left, dorsal upwards. (E,H) Close-up dorsal view (anterior to the left) of the brain and anterior trunk in flat-mounted, deyolked 24-hpf embryos stained with npm1a or npm1b probes. Digoxigenin-labeled RNA probes encoding the full-length npm1a and npm1b sequences were transcribed from linearized cDNA constructs using the DIG RNA labeling kit (Roche) following manufacturer instructions.
Knockdown of npm1a and npm1b leads to myeloid cell loss
To determine whether the zebrafish npm1 genes had a role in hematopoiesis, each gene was knocked down using antisense MOs. Initial studies were carried out using 5′UTR/ATG MOs. Embryos were analyzed for myeloid peroxidase (mpx) expression by WISH at 2 dpf. Knockdown of each gene individually lead to a significant reduction in the number of mpx-expressing cells (Figure 2A,D,G,M). When npm1a and npm1b were knocked-down simultaneously, the effect on mpx-expressing cell numbers was markedly increased (Figure 2J,M). Embryos injected with both MOs together showed developmental abnormalities, with a smaller head and eyes and a shortened yolk extension (Figure 2J; arrow and arrowhead). We further validated the specificity of this phenotype by testing several MOs directed at splice donor sites of npm1a and npm1b. Combined knockdown of npm1a and npm1b using the splice donor site MOs led to the same myeloid phenotype in these morphants as that induced by the 5′UTR/ATG-targeted double morphants (supplemental Figure 1C-D; quantified in supplemental Figure 1E). Knockdown of each gene was confirmed by RT-PCR and sequencing of the products to demonstrate the introduction of a premature stop codon in aberrantly spliced products (supplemental Figure 1F). Because it is known that some MOs can induce p53-dependent cell death as an off-target effect,40 we knocked-down npm1a and npm1b in the p53m/m line and analyzed mpx expression by WISH at 2 dpf. Compared with control MO-injected p53wt/wt (Figure 2B) and p53m/m (Figure 2H) embryos, injection of npm1(a+b) MO in p53m/m embryos led to a significant reduction of mpx expression (Figure 2K), similar to that observed in p53wt/wt (Figure 2E; quantifications in Figure 2N). These findings show that the loss of myeloid cells seen after knockdown of npm1a and npm1b is not a result of p53-dependent MO-induced toxicity.
Knockdown of npm1a and npm1b leads to loss of myeloid cells. (A-L) WISH for mpx in 48-hpf embryos, lateral view, anterior to the left, dorsal upwards. (A,D,G,J,M) Knockdown of npm1 genes results in loss of mpx-expressing myeloid cells. A significant difference is seen in myeloid cell numbers when each gene is knocked-down individually (npm1a in panel D and npm1b in panel G, quantified in panel M); this effect is at least additive in the double knockdown (npm1a + npm1b in panel J, quantified in panel M). (B,E,H,K,N) mpx expression is reduced upon npm1 knockdown compared with controls in both the p53wt/wt (B,E) and p53m/m (H,K) backgrounds (quantified in panel N). (C,F,I,L,O) Loss of mpx expression by knockdown of npm1 (F) can be rescued completely (I) or partially (L) by NPM1 RNA injection (10 pg). No differential expression of mpx is observed when 10 pg NPM1 or 10 pg control RNA (C) are injected with a control MO (quantified in panel O). npm1a MO indicates 5′UTR MO (1.4 ng); npm1b MO, 5′UTR MO (1.4 ng); npm1(a+b), npm1a MO + npm1b MO (1.4 ng each); control MO, npm1a 5-bp mismatch + npm1b 5-bp mismatch at 1.4 ng each. Error bars represent SEM. ns indicates not significant; *P < .02; **P < .01; ***P < .001 (Student t test). Complete rescue in panel I is defined as mpx+ cell numbers per embryo greater than the lower limit of the 95% confidence interval of the control RNA/control MO-injected embryos. Partial rescue in panel L is defined as mpx+ cells/embryos greater than the upper limit of the 95% confidence interval of the npm1(a+b) MO but below the lower limit of the 95% confidence interval of the control RNA/control MO-injected embryos.
Knockdown of npm1a and npm1b leads to loss of myeloid cells. (A-L) WISH for mpx in 48-hpf embryos, lateral view, anterior to the left, dorsal upwards. (A,D,G,J,M) Knockdown of npm1 genes results in loss of mpx-expressing myeloid cells. A significant difference is seen in myeloid cell numbers when each gene is knocked-down individually (npm1a in panel D and npm1b in panel G, quantified in panel M); this effect is at least additive in the double knockdown (npm1a + npm1b in panel J, quantified in panel M). (B,E,H,K,N) mpx expression is reduced upon npm1 knockdown compared with controls in both the p53wt/wt (B,E) and p53m/m (H,K) backgrounds (quantified in panel N). (C,F,I,L,O) Loss of mpx expression by knockdown of npm1 (F) can be rescued completely (I) or partially (L) by NPM1 RNA injection (10 pg). No differential expression of mpx is observed when 10 pg NPM1 or 10 pg control RNA (C) are injected with a control MO (quantified in panel O). npm1a MO indicates 5′UTR MO (1.4 ng); npm1b MO, 5′UTR MO (1.4 ng); npm1(a+b), npm1a MO + npm1b MO (1.4 ng each); control MO, npm1a 5-bp mismatch + npm1b 5-bp mismatch at 1.4 ng each. Error bars represent SEM. ns indicates not significant; *P < .02; **P < .01; ***P < .001 (Student t test). Complete rescue in panel I is defined as mpx+ cell numbers per embryo greater than the lower limit of the 95% confidence interval of the control RNA/control MO-injected embryos. Partial rescue in panel L is defined as mpx+ cells/embryos greater than the upper limit of the 95% confidence interval of the npm1(a+b) MO but below the lower limit of the 95% confidence interval of the control RNA/control MO-injected embryos.
To functionally validate npm1a and npm1b as true orthologs of human NPM1, we tested whether NPM1 expression could rescue the npm1 knockdown phenotype. We injected 1-cell–stage embryos with NPM1 RNA or control (mCherry-encoding) RNA along with npm1(a+b) MOs or mismatch controls. Although control RNA injection had no effect in the context of control MO or npm1(a+b) double morphants (Figure 2C,F; quantified in Figure 2O), injection of NPM1 mRNA (10 pg) rescued both the myeloid and developmental defects when injected into npm1(a+b) double morphants. Complete rescue was observed in 15% of embryos (Figure 2I), and a partial rescue was seen in 75% of embryos (Figure 2L). Importantly, NPM1 mRNA injection along with the control MO had no effect on the number of mpx cells (quantified in Figure 2O). These results demonstrate that npm1a and npm1b are true orthologs of NPM1.
Mutated human NPM1 is aberrantly expressed in the cytoplasm of zebrafish cells
To test whether NPMc+ was aberrantly localized to the cytoplasm in the zebrafish, as in humans, we injected mRNAs encoding NPMc+ or NPM1 fused to EGFP. The mRNAs were injected into 1-cell–stage embryos at doses up to 100 pg. At the highest dose, embryos appeared normal, and injected and uninjected embryos were morphologically indistinguishable by brightfield microscopic examination at 20 somites. NPM1 protein subcellular localization was assessed using cytospins of cell suspensions obtained from EGFP-NPM1– and EGFP-NPMc+–injected embryos at 24 hpf (Figure 3A-F). At low mRNA concentrations (10 pg), EGFP-NPM1 expression was restricted to nucleoli (Figure 3C), while at higher doses (50 and 100 pg), the protein was found to leak into the nucleoplasm and sometimes the cytoplasm of cells (arrows in Figure 3A-B), possibly because of overexpression artifacts. By contrast, NPMc+ was expressed in the cell cytoplasm at all mRNA concentrations (Figure 3D-F), consistent with observations in human NPMc+ AML cases.8,13 In mammalian cells, NPMc+ cytoplasmic localization is dependent on the presence of a novel nuclear export signal (NES) created by the mutation,14 and thus this function appears conserved in zebrafish.
Mutated human NPM1 is aberrantly expressed in the cytoplasm of zebrafish cells. (A-F) Epifluorescence analysis of EGFP-NPM1 (A-C) and EGFP-NPMc+ (D-F) subcellular localization in cytospins of zebrafish cells after mRNA injection at the indicated amounts. NPM1 or NPMc+ proteins are shown in green, and nuclei are counterstained in blue with DAPI. Cells were visualized with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 numeric aperture (NA) Apochromat oil lens (Carl Zeiss). Images were acquired with Openlab software (Perkin Elmer). (G) WB analysis of protein extracts from NPM1 (top left 3 lanes) and NPMc+ (top right 3 lanes) mRNA-injected embryos. NPM1 expression levels at each mRNA dose are shown along with β-tubulin expression as a loading control (bottom row). The arrows indicate the doses used for subsequent experiments. (H-I) Confocal microscopy of EGFP-NPM1 subcellular localization in zebrafish embryos at 24 hpf injected with 10 pg of EGFP-NPM1 mRNA (H) and 50 pg EGFP-NPMc+ mRNA (I). NPM1 protein is shown in green, while nuclei are counterstained in blue with DAPI. Images of cells were taken in the dorsal trunk region, shown in the insert (H), with a Zeiss LSM 510 META 2-Photon confocal microscope (Carl Zeiss), using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss). Images were acquired with the Zeiss LSM 510 software (Carl Zeiss). The EGFP expression patterns shown were representative of cells throughout the embryo.
Mutated human NPM1 is aberrantly expressed in the cytoplasm of zebrafish cells. (A-F) Epifluorescence analysis of EGFP-NPM1 (A-C) and EGFP-NPMc+ (D-F) subcellular localization in cytospins of zebrafish cells after mRNA injection at the indicated amounts. NPM1 or NPMc+ proteins are shown in green, and nuclei are counterstained in blue with DAPI. Cells were visualized with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 numeric aperture (NA) Apochromat oil lens (Carl Zeiss). Images were acquired with Openlab software (Perkin Elmer). (G) WB analysis of protein extracts from NPM1 (top left 3 lanes) and NPMc+ (top right 3 lanes) mRNA-injected embryos. NPM1 expression levels at each mRNA dose are shown along with β-tubulin expression as a loading control (bottom row). The arrows indicate the doses used for subsequent experiments. (H-I) Confocal microscopy of EGFP-NPM1 subcellular localization in zebrafish embryos at 24 hpf injected with 10 pg of EGFP-NPM1 mRNA (H) and 50 pg EGFP-NPMc+ mRNA (I). NPM1 protein is shown in green, while nuclei are counterstained in blue with DAPI. Images of cells were taken in the dorsal trunk region, shown in the insert (H), with a Zeiss LSM 510 META 2-Photon confocal microscope (Carl Zeiss), using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss). Images were acquired with the Zeiss LSM 510 software (Carl Zeiss). The EGFP expression patterns shown were representative of cells throughout the embryo.
We performed WB analysis to determine protein expression levels in lysates from NPM1 or NPMc+ mRNA–injected embryos, and found that wild-type protein levels were much higher than those of NPMc+ at each mRNA dose (Figure 3G). Thus, we selected mRNA doses that yielded comparable protein levels, injecting 10 pg for NPM1 and 50 pg for NPMc+ (2 black arrows in Figure 3G) in all subsequent assays. We validated these selected doses in vivo by analyzing the EGFP-NPM1 and EGFP-NPMc+ protein subcellular localization in embryos by confocal microscopy (Figure 3H-I), confirming the nucleolar restriction of NPM1 and the cytoplasmic distribution of NPMc+ in the vast majority of cells.
Zebrafish NPM1 orthologs interact with human NPM1 proteins
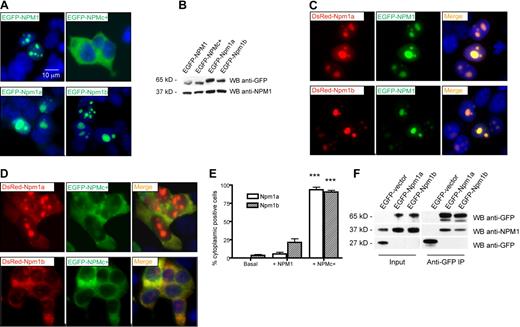
Current evidence supports the hypothesis that the NPMc+ mutant protein heterodimerizes with NPM1 (encoded by the residual normal allele) and may function by shuttling this and other partners out of the nucleus.13,41 To test whether we could study this mechanism in the zebrafish model, we examined the ability of NPMc+ to interact with zebrafish Npm1 proteins. EGFP- or DsRed-tagged Npm1a and Npm1b were expressed with fluorophore-tagged NPM1 or NPMc+ proteins in human 293T cells to assay for colocalization. First, we confirmed that both EGFP-Npm1a and EGFP-Npm1b were expressed in the nucleoli of cells (Figure 4A bottom left and right, respectively), as was NPM1 (compare with EGFP-NPM1 in Figure 4A top left; and to the cytoplasmic localization of EGFP-NPMc+, top right). WB analysis of transfected cells confirmed that all the proteins were expressed at comparable levels (Figure 4B). When coexpressed with EGFP-NPM1, both DsRed-Npm1a and DsRed-Npm1b colocalized with it in the nucleoli of cells (Figure 4C top and bottom rows, respectively). However, when either Npm1a or Npm1b were coexpressed with EGFP-NPMc+, they became aberrantly expressed in the cytoplasm, with or without residual nucleolar expression, in more than 90% of cells (Figure 4D top and bottom rows, respectively; quantification in Figure 4E). These observations indicate that NPMc+ can act in a dominant fashion to redistribute the 2 zebrafish Npm1 proteins to the cytoplasm, similar to human NPM1 (supplemental Figure 2A).14,41
Zebrafish npm1 orthologs interact with human NPM1 protein. (A) Epifluorescent images of 293T cells transfected with pEGFP-C1 expression vector encoding NPM1 (top left), NPMc+ (top right), Npm1a (bottom left), or Npm1b (bottom right). Expressed proteins are shown in green; nuclei are counterstained in blue with DAPI. (B) Anti-GFP WB analysis of protein lysates from transfected cells shown in Figure 2A. Levels of transfected proteins (top row) are shown along with levels of endogenous NPM1 (bottom row). (C) Epifluorescent images of 293T cells cotransfected with pEGFP-C1-NPM1 and pDsRed-monomer-C1 encoding zebrafish Npm1a (top row) or Npm1b (bottom row). Colocalization areas between Npm1a or Npm1b (in red, left column) and NPM1 (in green, middle column) are shown in yellow in the merged images (right column). Nuclei are counterstained in blue with DAPI. (D) Epifluorescent images of 293T cells cotransfected with pEGFP-C1-NPM1c+ and pDsRed-monomer-C1 encoding Npm1a (top row) or Npm1b (bottom row). Colocalization areas between Npm1a or Npm1b (in red, left column) and NPMc+ (in green, middle column) are shown in yellow in the merged images (right column). Nuclei are counterstained in blue with DAPI. All fluorescent images were acquired with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss) and Openlab software (Perkin Elmer). (E) Quantification of the Npm1a and Npm1b delocalization by NPMc+. Bars indicate the percentage of cells with aberrant Npm1a (□) or Npm1b (▧) cytoplasmic expression (with or without residual nucleolar positivity). Error bars indicate SD. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (***P < .001; Student t test). More than 200 cells were counted in 3 different microscopic fields for this analysis. (F) Anti-GFP coimmunoprecipitation assays of 293T cells transfected with the empty pEGFP-C1 vector, pEGFP-C1-Npm1a, or pEGFP-C1-Npm1b. Left panels, input; right panels, anti-GFP coimmunoprecipitation lanes. Top row: anti-GFP WB, detecting EGFP-Npm1a and EGFP-Npm1b fusion proteins. Middle row: anti-NPM1 WB, detecting endogenous NPM1. Bottom row: anti-GFP WB, detecting EGFP expressed by the empty vector. In the anti-GFP immunoprecipitation lanes, a dimmer band under the EGFP-Npm1a and EGFP-Npm1b bands probably represents a degradation product of the fusion protein, as it is also visible in the input lanes at longer exposures.
Zebrafish npm1 orthologs interact with human NPM1 protein. (A) Epifluorescent images of 293T cells transfected with pEGFP-C1 expression vector encoding NPM1 (top left), NPMc+ (top right), Npm1a (bottom left), or Npm1b (bottom right). Expressed proteins are shown in green; nuclei are counterstained in blue with DAPI. (B) Anti-GFP WB analysis of protein lysates from transfected cells shown in Figure 2A. Levels of transfected proteins (top row) are shown along with levels of endogenous NPM1 (bottom row). (C) Epifluorescent images of 293T cells cotransfected with pEGFP-C1-NPM1 and pDsRed-monomer-C1 encoding zebrafish Npm1a (top row) or Npm1b (bottom row). Colocalization areas between Npm1a or Npm1b (in red, left column) and NPM1 (in green, middle column) are shown in yellow in the merged images (right column). Nuclei are counterstained in blue with DAPI. (D) Epifluorescent images of 293T cells cotransfected with pEGFP-C1-NPM1c+ and pDsRed-monomer-C1 encoding Npm1a (top row) or Npm1b (bottom row). Colocalization areas between Npm1a or Npm1b (in red, left column) and NPMc+ (in green, middle column) are shown in yellow in the merged images (right column). Nuclei are counterstained in blue with DAPI. All fluorescent images were acquired with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss) and Openlab software (Perkin Elmer). (E) Quantification of the Npm1a and Npm1b delocalization by NPMc+. Bars indicate the percentage of cells with aberrant Npm1a (□) or Npm1b (▧) cytoplasmic expression (with or without residual nucleolar positivity). Error bars indicate SD. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (***P < .001; Student t test). More than 200 cells were counted in 3 different microscopic fields for this analysis. (F) Anti-GFP coimmunoprecipitation assays of 293T cells transfected with the empty pEGFP-C1 vector, pEGFP-C1-Npm1a, or pEGFP-C1-Npm1b. Left panels, input; right panels, anti-GFP coimmunoprecipitation lanes. Top row: anti-GFP WB, detecting EGFP-Npm1a and EGFP-Npm1b fusion proteins. Middle row: anti-NPM1 WB, detecting endogenous NPM1. Bottom row: anti-GFP WB, detecting EGFP expressed by the empty vector. In the anti-GFP immunoprecipitation lanes, a dimmer band under the EGFP-Npm1a and EGFP-Npm1b bands probably represents a degradation product of the fusion protein, as it is also visible in the input lanes at longer exposures.
To show that human and zebrafish NPM1 proteins can directly bind to each other, we carried out coimmunoprecipitation experiments. Figure 4F shows that an anti-GFP antibody can pull down both EGFP-Npm1a and EGFP-Npm1b along with endogenous NPM1 from transfected 293T cell lysates. In the reciprocal experiment, FLAG-tagged NPM1 and NPMc+ were able to pull down EGFP-tagged Npm1a and Npm1b (supplemental Figure 2B-C). These data demonstrate that NPMc+ can directly interact with zebrafish Npm1 proteins to affect subcellular localization processes, and validates our rationale for testing the consequences of NPMc+ expression in the developing hematopoietic system of zebrafish embryos.
NPMc+ increases the number of primitive myeloid cells in zebrafish embryos
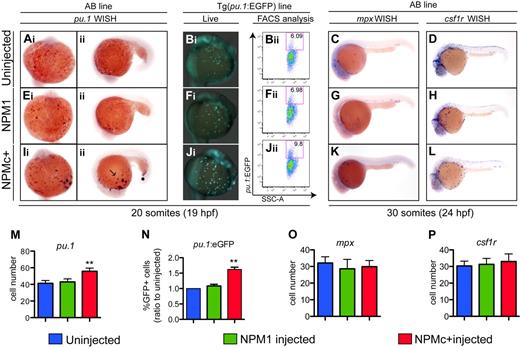
To determine the effects of NPMc+ expression on primitive hematopoiesis, we injected in vitro–transcribed mRNAs encoding NPMc+ (and NPM1 as control) into 1-cell–stage zebrafish embryos, and assayed markers of hematopoietic cell subpopulations by WISH. We found that expression of NPMc+, but not of NPM1, caused an increase in the number of pu.1+ cells at 20 somites compared with controls (Figure 5A,E,I; quantified in Figure 5M). These findings were also confirmed by counting EGFP+ cells in the Tg(pu.1:EGFP) transgenic line25 that expresses EGFP under the control of the pu.1 promoter (Figures 5Bi,Fi,Ji) and by flow cytometric analysis in the same line (Figures 5Bii,Fii,Jii; quantified in Figure 5N). To determine whether other markers of differentiating early myeloid cells were also expanded, we used probes to detect mpx and csf1r expression at 30 somites. Surprisingly, we found that neither of these markers was significantly affected by NPMc+ expression compared with NPM1-injected or uninjected embryos (Figure 5C-D,G-H,K-L; quantified in Figure 5O-P).
NPMc+ increases the number of primitive myeloid cells in zebrafish embryos. (Ai-Aii,Ei-Eii,Ii-Iii,M) WISH assays of AB embryos showing pu.1 expression at 20 somites (19 hpf) in ventral views, anterior to the top (A,E,Ii), and lateral views anterior to the left, dorsal upwards (Aii,Eii,Iii). pu.1-expressing cells are shown as dark purple dots that increase in number with NPMc+ expression (Ii-Iii). (M) pu.1+ cell number quantification shows a statistically significant increase upon NPMc+ expression (20 embryos counted per condition). (B,F,J) pu.1 expression analysis in the Tg(pu.1:EGFP) line at 20 somites (19 hpf). Ventral views, anterior to the top, of the ALPM and yolk of live Tg(pu.1:EGFP) transgenic zebrafish embryos, uninjected (Bi), or injected with NPM1 10 pg (Fi) or NPMc+ 50 pg (Ji) mRNA. Green cells indicate cells expressing EGFP under the control of the pu.1 promoter. NPMc+ expression causes a marked increase in EGFP+ cell numbers that disperse widely across the embryo's yolk (Ji). FACS analysis of Tg(pu.1:EGFP) embryos, uninjected (Bii), or injected with NPM1 10 pg (Fii) or NPMc+ 50 pg (Jii) mRNA. EGFP expression is shown in the y-axis and is increased upon NPMc+ expression (Jii). (N) Quantification of the percentage of EGFP+ cells in 3 independent FACS experiments, each including 50 heterozygous Tg(pu.1:EGFP) embryos per condition, normalized to the percentage of EGFP+ cells of uninjected embryos. (C-D, G-H, K-L, O-P) WISH assays showing lateral views (anterior to the left, dorsal upwards) of 30 somites (24 hpf) embryos stained for mpx (C,G,K) or csf1r expression (D,H,L). No significant change in expression of any of these markers is observed. The number of mpx and csf1r cells is quantified in panels O and P, respectively (20 embryos counted per condition). Note that the csf1r probe also stains xanthophores in the dorsal trunk of the embryo. In all graphs, error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (**P < .005; Student t test). In histograms, blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos.
NPMc+ increases the number of primitive myeloid cells in zebrafish embryos. (Ai-Aii,Ei-Eii,Ii-Iii,M) WISH assays of AB embryos showing pu.1 expression at 20 somites (19 hpf) in ventral views, anterior to the top (A,E,Ii), and lateral views anterior to the left, dorsal upwards (Aii,Eii,Iii). pu.1-expressing cells are shown as dark purple dots that increase in number with NPMc+ expression (Ii-Iii). (M) pu.1+ cell number quantification shows a statistically significant increase upon NPMc+ expression (20 embryos counted per condition). (B,F,J) pu.1 expression analysis in the Tg(pu.1:EGFP) line at 20 somites (19 hpf). Ventral views, anterior to the top, of the ALPM and yolk of live Tg(pu.1:EGFP) transgenic zebrafish embryos, uninjected (Bi), or injected with NPM1 10 pg (Fi) or NPMc+ 50 pg (Ji) mRNA. Green cells indicate cells expressing EGFP under the control of the pu.1 promoter. NPMc+ expression causes a marked increase in EGFP+ cell numbers that disperse widely across the embryo's yolk (Ji). FACS analysis of Tg(pu.1:EGFP) embryos, uninjected (Bii), or injected with NPM1 10 pg (Fii) or NPMc+ 50 pg (Jii) mRNA. EGFP expression is shown in the y-axis and is increased upon NPMc+ expression (Jii). (N) Quantification of the percentage of EGFP+ cells in 3 independent FACS experiments, each including 50 heterozygous Tg(pu.1:EGFP) embryos per condition, normalized to the percentage of EGFP+ cells of uninjected embryos. (C-D, G-H, K-L, O-P) WISH assays showing lateral views (anterior to the left, dorsal upwards) of 30 somites (24 hpf) embryos stained for mpx (C,G,K) or csf1r expression (D,H,L). No significant change in expression of any of these markers is observed. The number of mpx and csf1r cells is quantified in panels O and P, respectively (20 embryos counted per condition). Note that the csf1r probe also stains xanthophores in the dorsal trunk of the embryo. In all graphs, error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (**P < .005; Student t test). In histograms, blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos.
NPMc+ expression causes an increase in the number of primitive mpx+ and csf1r+ cells in the absence of functional p53
We then asked whether we could see a more marked perturbation of primitive myeloid cell development in the absence of p53. Upon NPMc+ expression pu.1 cells were increased in the p53m/m line at the 20-somite stage to a similar extent as observed in the wild-type background, while NPM1 expression had no effect compared with uninjected embryos (Figure 6A,E,I; quantified in Figure 6M). Interestingly, we also observed increased numbers of mpx- and csf1r-expressing cells at the 30-somite stage, providing evidence that a p53-dependent process is required to prevent the increase in mpx+ and csf1r+ early myeloid cells induced by NPMc+ (Figure 6C,G,K and Figure 6D,H,L; quantified in Figure 5N-O, respectively). Furthermore, abnormally located pu.1- and mpx-expressing cells were also observed in the dorsal trunk and head regions in these fish (Figure 6Iii,K arrows). Additional evidence that NPMc+ perturbs the differentiation of myeloid precursors comes from EGFP+ sorted cells from 30-somite Tg(pu.1:EGFP) embryos injected with p53 MO and either NPM1 or NPMc+. Giemsa-stained cytospin slides showed a significant difference in morphology. NPMc+-expressing cells were larger and exhibited more open chromatin, consistent with a more immature phenotype than control and NPM1-expressing cells (Figure 6B,F,J).
NPMc+ expression causes an increase in the number of primitive mpx+ and csf1r+ cells in the absence of functional p53 and causes p53-dependent apoptotic cell death. (Ai-Aii, Ei-Eii, Ii-Iii, M) pu.1 WISH assays in homozygous p53 mutant embryos at 20 somites (19 hpf), uninjected (Ai-Aii), or injected with NPM1 10 pg (Ei-Eii) or NPMc+ 50 pg (Ii-Iii). Embryos are shown in ventral (Ai,Ei,Ii; anterior to the top) and lateral (Aii,Eii,Iii; anterior to the left, dorsal upwards) views. pu.1-expressing cells are indicated by dark purple spots and are increased in embryos injected with NPMc+, 50 pg (Ii-Iii). (M) pu.1+ cell number quantification shows a statistically significant increase upon NPMc+ expression (20 embryos counted per condition). (B,F,J) Giemsa stain of cystospins of EGFP+ cells sorted from Tg(pu.1:EGFP) embryos at 30 somites (24 hpf), injected with p53 MO 1.6 ng alone (B) or in combination with either NPM1 10 pg (F) or NPMc+ 50 pg (J). NPMc+ expression results in larger cells with more immature morphology (J). Images were acquired with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat Oil lens (Carl Zeiss) and Openlab software (Perkin Elmer). (C-D, G-H, K-L, N-O) mpx (C,G,K) and csf1r (D,H,L) WISH are shown in lateral views (anterior to the left, dorsal upwards) of homozygous p53 mutant embryos at 30 somites (24 hpf) uninjected (C-D) or injected with NPM1 10 pg (G-H) or NPMc+ 50 pg (K-L). Note that mpx and csf1r expression is markedly expanded upon NPMc+ expression and p53 loss (K-L). The number of mpx and csf1r cells is quantified in panels N and O, respectively (20 embryos counted per condition). Note that the csf1r probe also stains xanthophores in the dorsal trunk of the embryo. Error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (**P < .005; ***P < .001; Student t test). In histograms, blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos. (P-R) Wild-type p53 embryos are shown either uninjected (Pi-Piii), or expressing NPM1 (Qi-Qiii) or NPMc+ (Ri-Riii). All embryos are at 30 somites (24 hpf) and shown in lateral views of the head and anterior trunk, anterior to the left and dorsal upwards. Acridine orange staining (green indicate dead or dying cells) of embryos uninjected (Pi), or injected with NPM1 10 pg (Qi) or NPMc+ 50 pg (Ri) mRNA. An increase in dying cells is observed upon NPMc+ expression (Ri). Activated caspase-3 immunostaining (green indicates cells in apoptosis) of embryos uninjected (Pii), or injected with NPM1 10 pg (Qii) or NPMc+ 50 pg (Rii) mRNA. Increased numbers of apoptotic cells are observed in NPMc+-injected embryos (Rii), forming aggregates on the yolk (arrow). Anti–activated caspase-3 and anti-GFP immunostaining in Tg(pu.1:EGFP) transgenic embryos (red spots indicate cells that express the cleaved form of caspase-3, and green spots indicate cells expressing EGFP under the control of the pu.1 promoter), uninjected (Piii), or injected with NPM1 10 pg (Qiii) or NPMc+ 50 pg (Riii) mRNA. Note in panel Riii the increase in activated caspase-3–expressing cells that surround and lie adjacent to EGFP+ cells, but do not colocalize with them. Homozygous mutant p53 embryos are shown uninjected (Piv-Pv), or expressing either NPM1 (Qiv-Qv) or NPMc+ (Riv-Rv). All embryos are at 30 somites (24 hpf) shown in lateral views of the head and anterior trunk, anterior to the left and dorsal upwards. Acridine orange staining of embryos uninjected (Piv) or injected with NPM1 10 pg (Qiv) or NPMc+ 50 pg mRNA (Riv) show no difference in the number of dead or dying cells. Anti–activated caspase-3 staining of embryos uninjected (Pv) or injected with either NPM1 10 pg (Qv) or NPMc+ 50 pg (Rv) mRNA show no difference in the number of apoptotic cells. Embryos were equilibrated in glycerol and visualized with a Nikon SMZ1500 zoom stereomicroscope (Nikon) using a 488 nm filter for the EGFP signal and 568 nm filter for the red signal. Images were acquired with NIS-Elements software (Nikon).
NPMc+ expression causes an increase in the number of primitive mpx+ and csf1r+ cells in the absence of functional p53 and causes p53-dependent apoptotic cell death. (Ai-Aii, Ei-Eii, Ii-Iii, M) pu.1 WISH assays in homozygous p53 mutant embryos at 20 somites (19 hpf), uninjected (Ai-Aii), or injected with NPM1 10 pg (Ei-Eii) or NPMc+ 50 pg (Ii-Iii). Embryos are shown in ventral (Ai,Ei,Ii; anterior to the top) and lateral (Aii,Eii,Iii; anterior to the left, dorsal upwards) views. pu.1-expressing cells are indicated by dark purple spots and are increased in embryos injected with NPMc+, 50 pg (Ii-Iii). (M) pu.1+ cell number quantification shows a statistically significant increase upon NPMc+ expression (20 embryos counted per condition). (B,F,J) Giemsa stain of cystospins of EGFP+ cells sorted from Tg(pu.1:EGFP) embryos at 30 somites (24 hpf), injected with p53 MO 1.6 ng alone (B) or in combination with either NPM1 10 pg (F) or NPMc+ 50 pg (J). NPMc+ expression results in larger cells with more immature morphology (J). Images were acquired with a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat Oil lens (Carl Zeiss) and Openlab software (Perkin Elmer). (C-D, G-H, K-L, N-O) mpx (C,G,K) and csf1r (D,H,L) WISH are shown in lateral views (anterior to the left, dorsal upwards) of homozygous p53 mutant embryos at 30 somites (24 hpf) uninjected (C-D) or injected with NPM1 10 pg (G-H) or NPMc+ 50 pg (K-L). Note that mpx and csf1r expression is markedly expanded upon NPMc+ expression and p53 loss (K-L). The number of mpx and csf1r cells is quantified in panels N and O, respectively (20 embryos counted per condition). Note that the csf1r probe also stains xanthophores in the dorsal trunk of the embryo. Error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (**P < .005; ***P < .001; Student t test). In histograms, blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos. (P-R) Wild-type p53 embryos are shown either uninjected (Pi-Piii), or expressing NPM1 (Qi-Qiii) or NPMc+ (Ri-Riii). All embryos are at 30 somites (24 hpf) and shown in lateral views of the head and anterior trunk, anterior to the left and dorsal upwards. Acridine orange staining (green indicate dead or dying cells) of embryos uninjected (Pi), or injected with NPM1 10 pg (Qi) or NPMc+ 50 pg (Ri) mRNA. An increase in dying cells is observed upon NPMc+ expression (Ri). Activated caspase-3 immunostaining (green indicates cells in apoptosis) of embryos uninjected (Pii), or injected with NPM1 10 pg (Qii) or NPMc+ 50 pg (Rii) mRNA. Increased numbers of apoptotic cells are observed in NPMc+-injected embryos (Rii), forming aggregates on the yolk (arrow). Anti–activated caspase-3 and anti-GFP immunostaining in Tg(pu.1:EGFP) transgenic embryos (red spots indicate cells that express the cleaved form of caspase-3, and green spots indicate cells expressing EGFP under the control of the pu.1 promoter), uninjected (Piii), or injected with NPM1 10 pg (Qiii) or NPMc+ 50 pg (Riii) mRNA. Note in panel Riii the increase in activated caspase-3–expressing cells that surround and lie adjacent to EGFP+ cells, but do not colocalize with them. Homozygous mutant p53 embryos are shown uninjected (Piv-Pv), or expressing either NPM1 (Qiv-Qv) or NPMc+ (Riv-Rv). All embryos are at 30 somites (24 hpf) shown in lateral views of the head and anterior trunk, anterior to the left and dorsal upwards. Acridine orange staining of embryos uninjected (Piv) or injected with NPM1 10 pg (Qiv) or NPMc+ 50 pg mRNA (Riv) show no difference in the number of dead or dying cells. Anti–activated caspase-3 staining of embryos uninjected (Pv) or injected with either NPM1 10 pg (Qv) or NPMc+ 50 pg (Rv) mRNA show no difference in the number of apoptotic cells. Embryos were equilibrated in glycerol and visualized with a Nikon SMZ1500 zoom stereomicroscope (Nikon) using a 488 nm filter for the EGFP signal and 568 nm filter for the red signal. Images were acquired with NIS-Elements software (Nikon).
NPMc+ expression causes apoptotic cell death in zebrafish embryos
To determine the mechanism whereby NPMc+ expression resulted in an increase in pu.1-expressing early myeloid cells and, later, in mpx+ and csf1r+ cells only when p53 was not functional, we used the supravital dye acridine orange to assess cell death due to NPMc+ at 30 somites. NPM1 expression has no effect in hematopoiesis, and therefore served as a control. NPMc+ caused a dramatic increase in cell death detected throughout the embryo, particularly in the eye and brain (Figure 6Ri). In contrast, NPM1-injected embryos (Figure 6Qi) only showed a slight increase in the number of dead cells compared with control uninjected embryos (Figure 6Pi). To further dissect the mechanism of cell death, we also assayed injected embryos for activated caspase-3, which is expressed in apoptotic cells. NPMc+-injected embryos (Figure 6Rii), but not NPM1-injected or uninjected embryos (Figure 6Qii and 6Pii, respectively) showed increased activated caspase-3 staining in the brain and eye, and in cells in the ALPM region (Figure 6Rii arrow). This region corresponds to the location of developing myeloid precursors and suggests that pu.1+ cells could be undergoing apoptosis. To test this hypothesis, we injected NPM1 or NPMc+ in the Tg(pu.1:EGFP) transgenic line, and analyzed embryos for the coexpression of activated caspase-3 (in red) and EGFP (in green). NPMc+-injected embryos showed an increase in both signals compared with uninjected and NPM1-injected embryos (Figure 6Piii,Qiii,Riii); however, colocalization of red and green in the same cell was not detected. These findings were confirmed by FACS annexin-V analysis in Tg(pu.1:EGFP) transgenic embryos: NPMc+-injected embryos showed an increase in apoptosis as a whole (supplemental Figure 3A-C) that was not markedly evident in pu.1:EGFP+ cells (supplemental Figure 3D-F). Furthermore, the cell-cycle profile in sorted EGFP+ cells from Tg(pu.1:EGFP) animals showed no difference between control and NPMc+-injected embryos (supplemental Figure 3G-I). Given the marked increase in apoptotic cells evident in NPMc+-injected embryos, we examined the role of p53 in NPMc+-induced cell death. Both wild-type and mutated NPM1 proteins were expressed in the p53m/m line, and acridine orange and anti–activated caspase-3 assays were performed. These assays showed a marked reduction in cell death and apoptosis in p53m/m (Figure 6Riv-v, respectively) compared with p53wt/wt NPMc+-injected embryos (Figure 6Ri-ii), suggesting that the apoptotic phenotype elicited by NPMc+ is p53-dependent.
NPMc+ increases the number of definitive hematopoietic cells in zebrafish embryos
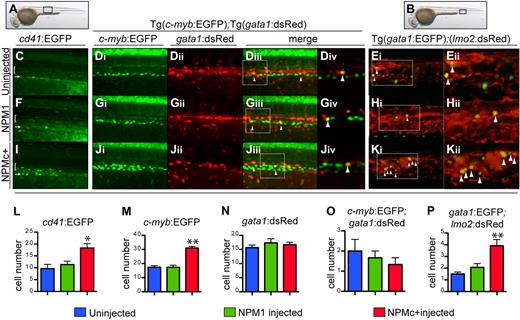
To investigate whether NPMc+ expression affects definitive hematopoietic cell numbers, we examined the expression of the integrin cd41 and the transcription factors c-myb and runx1, which label definitive HSCs starting between 30 and 36 hpf.24 First, we used Tg(cd41:EGFP) transgenic fish to analyze expression of GFPLow cells in the AGM region (Figure 7A black box).24,42 Confocal microscopy showed that NPMc+ expression caused an increase in numbers of cd41-EGFPLow–expressing cells between the dorsal aorta and the cardinal vein (brackets in Figure 7C,F,I; quantified in Figure 7L), the expected region for developing HSCs, and an increase in EGFP+ cells in the pronephric ducts, ventral to the AGM24 (arrows in Figure 7C,F,I). Next, we analyzed c-myb expression by WISH, and found that NPMc+, but not NPM1, expression causes an increase in the numbers of c-myb–expressing cells at 32 hpf (supplemental Figure 4Ai,Ei,Ii; quantified in supplemental Figure 4M). Histologic cross-sections of these embryos confirmed c-myb–expressing cells were correctly localized between the ventral wall of the dorsal aorta and the cardinal vein (ie, the AGM region), and NPMc+-injected embryos showed ventral and lateral expansion of these cells within this domain (supplemental Figure 4Aii-iii,Eii-iii,Iii-iii). Importantly, NPMc+ expression caused the same increase in the number of c-myb–expressing cells in the AGM of p53m/m embryos (supplemental Figure 4D,H,L; quantified in supplemental Figure 4P). Next, we injected NPM constructs into double-transgenic embryos Tg(c-myb:EGFP);(gata1:DsRED). Z-stack images obtained by confocal microscopy in the AGM region (Figure 7A black box) confirmed that NPMc+ expression increased the number of EGFP+ cells expressed from the c-myb promoter relative to controls at 32 hpf (Figure 7Di,Gi,Ji). This observed increase in c-myb+ cells was confirmed by FACS analysis (supplemental Figure 4B,F,J; quantified in supplemental Figure 4N). As c-myb is also expressed in the ICM at this stage,43 it was important to exclude the possibility that the observed increase in c-myb–expressing cells did not result from developmentally delayed erythrocytes that can also express c-myb. We found that the number of gata1:DsRed+ cells was not increased in NPMc+-injected embryos (Figure 7Dii,Gii,Jii; quantified in Figure 7N; also shown by gata1 WISH; supplemental Figure 5C,F,I). Importantly, we demonstrated that the number of cells coexpressing c-myb and gata1 (yellow cells) was not increased in embryos expressing NPMc+ (arrowheads in Figure 7Diii,Giii,Jiii; magnified single Z-sections showing boxed yellow cells in Figure 7Div,Giv,Jiv; quantified in Figure 7O). Therefore, the increase in c-myb was not due to increased numbers of erythroid precursors. To confirm that NPMc+ causes an increase in HSCs in the AGM, we also analyzed runx1 expression, but surprisingly found that the numbers of runx1+ cells were unchanged in NPMc+-injected embryos relative to controls at both the 28- and 32-hpf time points (supplemental Figure 5A-B,D-E,G-H).
NPMc+ increases the number of definitive hematopoietic cells in zebrafish embryos. (A-B) Brightfield images of a 32-hpf embryo (lateral view, anterior to the left, dorsal upwards) where a black box shows the region of the AGM (A) or of the PBI (B), where subsequent images were taken. (C,F,I,L) Confocal images taken from the AGM region of live 32-hpf Tg(cd41:EGFP) transgenic zebrafish embryos (black box in panel A), uninjected (C), or injected with NPM1 10 pg (F) or NPMc+ 50 pg (I) mRNA. Green cells indicate expression of EGFP under the control of the cd41 promoter. The bracket highlights cd41low EGFP+ cells in the AGM region, while the arrow points at the renal EGFP+ population located in the region of the pronephric ducts. EGFP+ cells in the AGM field were counted and showed increased numbers upon NPMc+ expression (L). Images were captured on a Yokogawa spinning disk confocal microscope using a 10×/0.3 NA Plan-fluor phase objective. (Di-iv, Gi-iv, Ji-iv, M-O) Confocal images taken from the AGM region of live 32-hpf Tg(c-myb:EGFP);(gata1:DsRed) transgenic zebrafish embryos (black box in panel A), uninjected (Di-Div), or injected with NPM1 10 pg (Gi-Giv) or NPMc+ 50 pg (Ji-Jiv) mRNA. Green indicates cells expressing EGFP under the control of the c-myb promoter; red indicates cells expressing DsRed under the control of the gata1 promoter; yellow indicates cells coexpressing both genes (highlighted by arrowheads). The white boxes in panels Diii, Giii, and Jiii indicate an area, magnified in a single Z-stack slice (Div,Giv,Jiv), showing how single slices were used to quantify the number of yellow cells. NPMc+ expression causes an increase in EGFP+ cell numbers in the field of analysis (quantified in panel M) without affecting numbers of cells expressing DsRed under the control of the gata1 promoter (quantified in panel N) or double-positive cells (quantified in panel O by counting yellow cells in 3 different Z-stack slices in each of 4 embryos). Images were taken as for Tg(cd41:EGFP) embryos. (E,H,K,P) Confocal images taken from the PBI region of live 32-hpf Tg(gata1:EGFP);(lmo2:DsRed) transgenic zebrafish embryos (black box in panel B), uninjected (E), or injected with NPM1 10 pg (H) or NPMc+ 50 pg (K) mRNA. Green indicates cells expressing EGFP under the control of the gata1 promoter; red indicates cells expressing DsRed under the control of the lmo2 promoter; yellow indicates cells coexpressing both genes (highlighted by arrowheads). NPMc+ expression causes an increase in cells expressing both genes (K), quantified in panel P by counting yellow cells in 3 different Z-stack slices in each of 4 embryos. Images are single Z-stack slices, and the white boxes in panels Ei, Hi, and Ki indicates an area magnified in panels Eii, Hii, and Kii. Images were acquired on a Yokogawa spinning disk confocal microscope using a 20×/0.75 NA Plan-Apo DIC objective. All confocal images were acquired using the Andor IQ software. Analysis of images was performed using ImageJ software. Error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (*P < .05, **P < .005; Student t test). In histograms blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos.
NPMc+ increases the number of definitive hematopoietic cells in zebrafish embryos. (A-B) Brightfield images of a 32-hpf embryo (lateral view, anterior to the left, dorsal upwards) where a black box shows the region of the AGM (A) or of the PBI (B), where subsequent images were taken. (C,F,I,L) Confocal images taken from the AGM region of live 32-hpf Tg(cd41:EGFP) transgenic zebrafish embryos (black box in panel A), uninjected (C), or injected with NPM1 10 pg (F) or NPMc+ 50 pg (I) mRNA. Green cells indicate expression of EGFP under the control of the cd41 promoter. The bracket highlights cd41low EGFP+ cells in the AGM region, while the arrow points at the renal EGFP+ population located in the region of the pronephric ducts. EGFP+ cells in the AGM field were counted and showed increased numbers upon NPMc+ expression (L). Images were captured on a Yokogawa spinning disk confocal microscope using a 10×/0.3 NA Plan-fluor phase objective. (Di-iv, Gi-iv, Ji-iv, M-O) Confocal images taken from the AGM region of live 32-hpf Tg(c-myb:EGFP);(gata1:DsRed) transgenic zebrafish embryos (black box in panel A), uninjected (Di-Div), or injected with NPM1 10 pg (Gi-Giv) or NPMc+ 50 pg (Ji-Jiv) mRNA. Green indicates cells expressing EGFP under the control of the c-myb promoter; red indicates cells expressing DsRed under the control of the gata1 promoter; yellow indicates cells coexpressing both genes (highlighted by arrowheads). The white boxes in panels Diii, Giii, and Jiii indicate an area, magnified in a single Z-stack slice (Div,Giv,Jiv), showing how single slices were used to quantify the number of yellow cells. NPMc+ expression causes an increase in EGFP+ cell numbers in the field of analysis (quantified in panel M) without affecting numbers of cells expressing DsRed under the control of the gata1 promoter (quantified in panel N) or double-positive cells (quantified in panel O by counting yellow cells in 3 different Z-stack slices in each of 4 embryos). Images were taken as for Tg(cd41:EGFP) embryos. (E,H,K,P) Confocal images taken from the PBI region of live 32-hpf Tg(gata1:EGFP);(lmo2:DsRed) transgenic zebrafish embryos (black box in panel B), uninjected (E), or injected with NPM1 10 pg (H) or NPMc+ 50 pg (K) mRNA. Green indicates cells expressing EGFP under the control of the gata1 promoter; red indicates cells expressing DsRed under the control of the lmo2 promoter; yellow indicates cells coexpressing both genes (highlighted by arrowheads). NPMc+ expression causes an increase in cells expressing both genes (K), quantified in panel P by counting yellow cells in 3 different Z-stack slices in each of 4 embryos. Images are single Z-stack slices, and the white boxes in panels Ei, Hi, and Ki indicates an area magnified in panels Eii, Hii, and Kii. Images were acquired on a Yokogawa spinning disk confocal microscope using a 20×/0.75 NA Plan-Apo DIC objective. All confocal images were acquired using the Andor IQ software. Analysis of images was performed using ImageJ software. Error bars represent SEM. *Statistically significant differences between the NPMc+-injected and both NPM1-injected and uninjected embryos (*P < .05, **P < .005; Student t test). In histograms blue indicates uninjected control embryos; green, NPM1-injected embryos; and red, NPMc+-injected embryos.
We then addressed whether expression of NPMc+ affected the EMP population, which can be identified by gata1/lmo2BRIGHT coexpressing cells in the PBI.23 Confocal microscopic analysis of the PBI region (Figure 7B black box) in double-transgenic Tg(gata1:EGFP);(lmo2:DsRed) embryos injected with NPMc+ showed a marked increase in numbers of gata1-lmo2 double-positive cells relative to controls (Figure 7E,H,K; quantified in Figure 7P). We confirmed these findings by FACS analysis and found similar increases in the numbers of EMPs upon expression of NPMc+ (supplemental Figure 4C,G,K; quantified in supplemental Figure 4O), again underscoring the finding that NPMc+ perturbs definitive hematopoiesis in the zebrafish embryo.
Discussion
In this report, we have used the zebrafish embryo as a model to examine the role of NPMc+ in hematopoiesis. We identified 2 npm1 genes orthologous to human NPM1 and have shown that loss of both zebrafish npm1 genes results in a defect in myelopoiesis, which was partially redundant between the 2 genes. This myeloid phenotype is consistent with findings reported in an Npm1 mouse knockout model,6 and could be rescued by forced NPM1 expression, indicating functional conservation between human and zebrafish NPM1. Zebrafish embryos injected with human NPMc+-encoding mRNA demonstrated abnormal cellular localization of the mutated protein in the cell cytoplasm, similar to human NPMc+ AML cells, while normal human NPM1 was localized exclusively in the nucleolus. We also demonstrated that both NPM1 and NPMc+ could bind to the zebrafish Npm1 orthologs and regulate their subcellular localization.
We demonstrated that NPMc+ expression perturbs primitive myelopoiesis by examining cells expressing the myeloid markers pu.1, mpx, and csf1r. While pu.1 is mostly expressed in myeloid precursors at the 20-somite stage,25 mpx is a marker of primitive and definitive granulocytes, and at 30 somites is also expressed in differentiating macrophages.22 csf1r is the zebrafish ortholog of the human macrophage colony-stimulating factor receptor. It is expressed in precursor and mature macrophages on the yolk sac, and in xanthophores of the skin.44 Interestingly, NPMc+ expression resulted in a striking increase in the number of early pu.1-expressing myeloid cells, but only caused a corresponding increase in the numbers of mpx- or csf1r-expressing myeloid cells later in development in the absence of functional p53. Although our data also showed that NPMc+ expression elicited a generalized p53-dependent apoptotic response, we were unable to demonstrate that the p53-dependent apoptotic cell death was occurring specifically in pu.1-expressing myeloid cells upon NPMc+ expression. Technical limitations using reporter zebrafish lines expressing EGFP under the control of the pu.1 promoter may preclude this analysis, because the EGFP signal is known to be degraded early in the course of apoptosis, and therefore a myeloid apoptotic cell might not simultaneously express both EGFP and cleaved caspase-3 or annexin V. Alternatively, apoptosis could occur within a pu.1−, mpx+, and csf1r+ myeloid cell population at a later stage or a p53-dependent, nonapoptotic cell death process such as senescence could explain these observations. Taken together, these data suggest that NPMc+ expression perturbs primitive hematopoiesis by promoting the early expansion of pu.1+ myeloid cells, and this phenotype is even more pronounced in a p53-deficient background where mpx+ and csf1r+ cells are also expanded. The correlation between p53 mutational status and the extent of the phenotype elicited by NPMc+ is intriguing, because p53 mutations rarely occur in NPMc+ AMLs.11 However, it has been shown that NPMc+ binds to and inactivates the tumor suppressor p14ARF in mammalian cells, thus inhibiting the p53 response to oncogene activation.13 Zebrafish and other teleosts lack a homolog of mammalian p14ARF,45 and thus our finding that a p53-dependent apoptotic response is activated by NPMc+ expression may reflect an oncogene-associated cell stress response, which is not evident in human NPMc+ AML cells due to inhibition of p14ARF by NPMc+. These data are also consistent with the myeloproliferative phenotype elicited by NPMc+ expression in a transgenic mouse model,17 and may be relevant to human AML. Other oncogenes involved in human AML have been shown to disrupt primitive hematopoiesis in zebrafish.46,47 Moreover, the phenotype induced by the RUNX1:ETO oncoprotein in zebrafish primitive hematopoiesis has been successfully used in a large-scale screen for chemical modifiers,48 again underscoring the relevance and strengths of the zebrafish system.
Although the transient nature of NPMc+ protein expression after mRNA injection prevents the study of zebrafish hematopoiesis beyond 32 hpf, our data show that definitive hematopoiesis is perturbed by NPMc+, which has important implications for the role of this protein in leukemogenesis. We found that c-myb– and cd41:EGFP-expressing cells were increased in the ventral wall of the aorta of NPMc+-injected embryos, suggesting that presumptive HSC numbers are increased due to its expression. However, the number of cells expressing runx1 remains normal at this time. Runx1 acts upstream of c-myb in the development of normal HSCs,49 suggesting that NPMc+ may expand a c-myb+/cd41+ subpopulation of HSCs in which runx1 expression has already been down-regulated. Alternatively, NPMc+ may specifically down-regulate the expression of runx1 in these cells. We also showed that NPMc+ expression increases the numbers of gata1+/lmo2BRIGHT double-positive cells in the PBI, indicating expansion of EMPs, the first multipotent, definitive hematopoietic progenitors to develop in both mice and zebrafish. Unlike normal HSCs, EMPs express both c-myb and cd41 but only very low levels of runx1, suggesting these could be the main cells targeted by NPMc+ at this time point. However, given the correct anatomical location of c-myb-EGFP– and cd41-EGFP–expressing cells, we favor the hypothesis that both EMPs and HSCs are expanded by NPMc+. Importantly, it has been recently reported that NPMc+ is expressed in the leukemia-initiating cell fraction of human NPMc+ AMLs.50,51 In addition, mutated NPMc+ can be found in granulocytic, monocytic, erythroid, and megakaryocytic cells in the bone marrow of patients with NPMc+ AML.52 These data suggest that a multipotent stem or progenitor cell is the cell of origin for NPMc+ leukemias. Thus, the increase in definitive EMPs and HSCs promoted by NPMc+ expression in zebrafish is likely related to the role of NPMc+ in myeloid leukemogenesis, and provides a tractable in vivo system for study of the mechanisms through which hematopoietic development is perturbed in the presence of NPMc+.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jeff Davies for critical review of the manuscript.
N.B. has been supported by a Princess Borghese and Anna Bulgari Fellowship from The American-Italian Cancer Foundation, and is currently a Leukemia & Lymphoma Society Special Fellow. E.M.P. is the recipient of a Clinical Research Training Fellowship from Leukemia & Lymphoma Research UK and was formerly supported by National Institutes of Health grant 5T32-CA009382-26.
National Institutes of Health
Authorship
Contribution: N.B. and E.M.P. designed and performed research and wrote the manuscript; C.G. contributed important new reagents and analyzed and interpreted data; A.B.J. and J.-S.L. performed research; B.F. designed research and provided important reagents; and J.P.K. and A.T.L. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: B.F. has applied for a patent on the clinical use of NPM1 mutants. The remaining authors declare no competing financial interests.
Correspondence: Niccolò Bolli and A. Thomas Look, Dana-Farber Cancer Institute, Mayer Bldg, Rm M630, 44 Binney St, Boston, MA 02115; e-mail: niccolo_bolli@dfci.harvard.edu, thomas_look@dfci.harvard.edu.
References
Author notes
N.B. and E.M.P. contributed equally to this article.