Abstract
Although NPM1 gene mutations leading to aberrant cytoplasmic expression of nucleophosmin (NPMc+) are the most frequent genetic lesions in acute myeloid leukemia, there is yet no experimental model demonstrating their oncogenicity in vivo. We report the generation and characterization of a transgenic mouse model expressing the most frequent human NPMc+ mutation driven by the myeloid-specific human MRP8 promoter (hMRP8-NPMc+). In parallel, we generated a similar wild-type NPM trans-genic model (hMRP8-NPM). Interestingly, hMRP8-NPMc+ transgenic mice developed myeloproliferation in bone marrow and spleen, whereas nontransgenic littermates and hMRP8-NPM transgenic mice remained disease free. These findings provide the first in vivo evidence indicating that NPMc+ confers a proliferative advantage in the myeloid lineage. No spontaneous acute myeloid leukemia was found in hMPR8-NPMc+ or hMRP8-NPM mice. This model will also aid in the development of therapeutic regimens that specifically target NPMc+.
Introduction
Nucleophosmin (NPM1) is a nucleolar phosphoprotein and a frequent target of genetic alterations, such as translocations and mutations, in hematopoietic tumors.1
NPM1 gene mutations leading to aberrant cytoplasmic expression of nucleophosmin (NPMc+) have been found in approximately 60% of adult acute myeloid leukemia (AML) with a normal karyotype, making it the most frequent genetic abnormality identified in this disease to date (∼ one-third of cases).2 All NPMc+ mutations consistently result in nucleotide gain at the C-terminus of one NPM1 allele. This results in the loss of at least 1 of 2 tryptophans essential for nucleolar localization and generates an additional nuclear export signal. These alterations determine the aberrant cytoplasmic localization of the protein in leukemic blasts.3 NPMc+ AMLs show a wide morphologic spectrum, multilineage involvement, a unique microRNA and gene expression signature, high frequency of FLT3-internal tandem duplications, and distinctive clinical and prognostic features.4 These unique properties are irrespective of whether it carries a normal karyotype (∼ 85% of cases) or minor secondary chromosomal aberrations (∼ 15% of cases), reinforcing the concept that NPM1 mutation can drive leukemia development.5
The mechanisms by which NPMc+ may exert its oncogenic functions are currently under investigation, and it has been found to play a role in inactivation of the p19Arf tumor suppressor,6,7 to promote RNA Pol II transcription,8 and to stabilize c-Myc oncoprotein.9 In addition, we have reported that NPMc+ can act as a proto-oncogene in vitro in the appropriate genetic milieu.10 Nevertheless, there is no reported in vivo model to assess the leukemogenic potential of NPMc+. Here we generated and characterized transgenic mice expressing the most frequent human NPMc+ mutation (mutation A: TCTG duplication2 ) or its wild-type NPM1 counterpart driven by the myeloid-specific hMRP8 promoter.
Methods
Transgenic mice
Flag-NPMc+ (mutation A2) or Flag-NPM cDNA was amplified from pCMV-Tag2B constructs8 with primers containing BclI ends and subcloned into the compatible BglII site of the hMRP8 cassette.11 Both transgene cassettes were sequenced, released from the parental vector by digestion with ClaI and XbaI, and injected into zygotes from (C57BL/6J × CBA) F1 hybrid mice. Genotyping was carried out by both Southern blotting and polymerase chain reaction (PCR). Southern blot analysis was performed using a 0.6-kb probe amplified from the C-terminus of NPM to the downstream flanking sequence of hMRP8. PCR genotyping was performed using the following primers: NPM874F, (5′-GGTTCTCTTCCCAAAGTGGAAGC-3′); and MRP8R, (5′-GAGGTATTGATGACTTTATTATTCTGCAGG-3′). All animal work was approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
Expression analysis
For reverse-transcribed (RT)–PCR, DNase-treated RNA was reverse transcribed and amplified using the following primers: exon 1F, 5′-ATGTCTCTTGTCAGCTGTCTTTC-3′ + NPM500R, 5′-CAGATATACTTAAGAGTTTCACATC-3′; HPRT1, 5′-CCTGCTGGATTACATTAAAG-CACTG-3′ + HPRT2, 5′-GTCAAGGGCATATCCAACAACAAAC-3′. All PCRs were performed with 40 cycles of (30 seconds at 94°C, 30 seconds at 55°C, 30 seconds at 72°C, followed by 7 minutes of final extension at 72°C). The following antibodies were used for Western blots: polyclonal Flag (2368; Cell Signaling), monoclonal Flag (A8592; Sigma-Aldrich), polyclonal NPMc+ specifically against the mutant (1:500, gift of B. Falini, University of Perugia, Perugia, Italy), and monoclonal NPM recognizing both overexpressed human NPM and endogenous mouse Npm (B0556; Sigma-Aldrich).
Histology
Mouse tissue sections were stained with hematoxylin and eosin for histopathologic examination. Pictures of the stained tissue sections were obtained using an Olympus BX41 microscope and a DP20 camera (Olympus). All image acquisition and processing were carried out with Adobe Photoshop 7.0 (Adobe Systems).
Flow cytometry
Flow cytometric analysis of bone marrow (BM) cells and splenocytes was performed with the following monoclonal antibodies conjugated to phycoerythrin, fluorescein isothiocyanate, or allophycocyanin: Mac-1, Gr-1, c-Kit, CD3ϵ, and B220. All data were collected on an LSRII instrument (BD Biosciences) and analyzed by FlowJo software (TreeStar).
Results and discussion
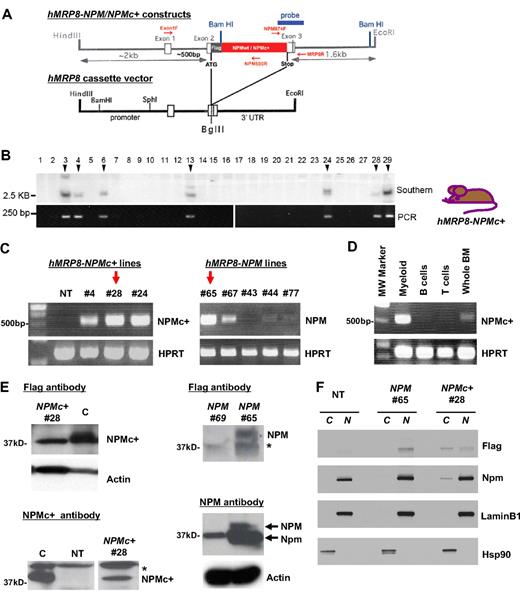
To study the role of NPMc+ mutation in hematopoiesis and leukemogeneis in vivo, we generated NPMc+ transgenic mice expressing the most common cytoplasmic mutation of NPM under control of the human MRP8 promoter. As a control, we also generated transgenic mice expressing wild-type NPM under the same promoter. Both constructs were Flag-tagged at the N-terminus of NPM (Figure 1A). Seven hMRP8-NPMc+ and 8 MRP8-NPM transgenic mice were identified as positive founders (Figure 1B and data not shown).
Expression of NPMc+ and NPM in hMRP8-NPMc+ and hMRP8-NPM transgenic mice. (A) Schematics of hMRP8-NPMc+ and hMRP8-NPM constructs used to generate NPM transgenic lines. (B) Southern blot analysis using the 3′ probe of BamHI-digested tail DNA from hMRP8-NPMc+ founders. Positive founders were confirmed by PCR as illustrated below the Southern blot panel. (C) Semiquantitative RT-PCR analysis of NPMc+ and NPM mRNA levels from whole BM cells of different founders. Red arrows represent the transgenic lines used for detailed analysis. NT indicates nontransgenic control. (D) Semiquantitative RT-PCR analysis of NPMc+ mRNA levels from sorted populations. Myeloid: Mac-1+Gr-1+ BM cells; B-cell: B220+CD19+ splenocytes; T-cell: CD4+ splenocytes. (E) Western blot of NPMc+ and NPM expression in whole BM cells of NPMc+ 28 (left panels) and NPM 65 (right panels) animals. A monoclonal Flag antibody (top left) and a polyclonal NPMc+-specific antibody (bottom left) were used to detect NPMc+ levels. *Nonspecific band. A polyclonal Flag antibody (top right) and a monoclonal NPM antibody were used to detect both overexpressed human NPM and endogenous mouse Npm levels. C indicates control lysates from HEK293 cells transfected with an NPMc+ expression vector; NPM 69, a negative control (an NPM transgenic mouse that was not found to overexpress Npm at the level of RNA or protein). (F) Western blot analysis of NPMc+ and NPM expression on purified nuclear and cytoplasmic fractions of Mac-1+Gr-1+ sorted granulocytes from BM of NT, NPM 65, and NPMc+ 28 animals. A polyclonal Flag antibody was used to specifically recognize the transgenic wild-type NPM or NPMc+ mutant. A monoclonal NPM antibody was used to detect the endogenous mouse Npm levels. Anti-LaminB1 and anti-Hsp90 assess the purity of the nuclear (N) versus the cytoplasmic (C) fraction, respectively.
Expression of NPMc+ and NPM in hMRP8-NPMc+ and hMRP8-NPM transgenic mice. (A) Schematics of hMRP8-NPMc+ and hMRP8-NPM constructs used to generate NPM transgenic lines. (B) Southern blot analysis using the 3′ probe of BamHI-digested tail DNA from hMRP8-NPMc+ founders. Positive founders were confirmed by PCR as illustrated below the Southern blot panel. (C) Semiquantitative RT-PCR analysis of NPMc+ and NPM mRNA levels from whole BM cells of different founders. Red arrows represent the transgenic lines used for detailed analysis. NT indicates nontransgenic control. (D) Semiquantitative RT-PCR analysis of NPMc+ mRNA levels from sorted populations. Myeloid: Mac-1+Gr-1+ BM cells; B-cell: B220+CD19+ splenocytes; T-cell: CD4+ splenocytes. (E) Western blot of NPMc+ and NPM expression in whole BM cells of NPMc+ 28 (left panels) and NPM 65 (right panels) animals. A monoclonal Flag antibody (top left) and a polyclonal NPMc+-specific antibody (bottom left) were used to detect NPMc+ levels. *Nonspecific band. A polyclonal Flag antibody (top right) and a monoclonal NPM antibody were used to detect both overexpressed human NPM and endogenous mouse Npm levels. C indicates control lysates from HEK293 cells transfected with an NPMc+ expression vector; NPM 69, a negative control (an NPM transgenic mouse that was not found to overexpress Npm at the level of RNA or protein). (F) Western blot analysis of NPMc+ and NPM expression on purified nuclear and cytoplasmic fractions of Mac-1+Gr-1+ sorted granulocytes from BM of NT, NPM 65, and NPMc+ 28 animals. A polyclonal Flag antibody was used to specifically recognize the transgenic wild-type NPM or NPMc+ mutant. A monoclonal NPM antibody was used to detect the endogenous mouse Npm levels. Anti-LaminB1 and anti-Hsp90 assess the purity of the nuclear (N) versus the cytoplasmic (C) fraction, respectively.
NPMc+ transgene mRNA was clearly detected in total BM cells of hMRP8-NPMc+ lines 4, 24, and 28, whereas hMRP8-NPM lines 65 and 67 showed highly detectable NPM transgene mRNA by RT-PCR (Figure 1C). Furthermore, RT-PCR analysis on sorted cells from myeloid and lymphoid compartments in the NPMc+ BM clearly showed the expression of NPMc+ transcripts restricted to myeloid cells (Figure 1D).
As shown in Figure 1E, NPMc+ protein was detectable in whole BM cells of hMRP8-NPMc+ transgenic mice line 28 by Western blot using anti-Flag, as well as an antibody specific for NPMc+ (gift of B. Falini). Exogenous NPM protein was also easily detectable in whole BM cells of hMRP8-NPM transgenic mice line 65 using an anti-Flag as well as an antibody recognizing both the endogenous mouse Npm and the slightly larger Flag-tagged human NPM. In addition, Western blot analysis using anti-Flag antibody demonstrated that the NPMc+ mutant mainly localizes to the cytoplasmic fraction of granulocytes from hMRP8-NPMc+ transgenic mice, whereas the transgenic wild-type NPM localizes exclusively to the nuclear fraction of granulocytes from hMRP8-NPM line as expected (Figure 1F). Based on the protein expression, NPMc+ 28 and NPM 65 lines were used for detailed analysis.
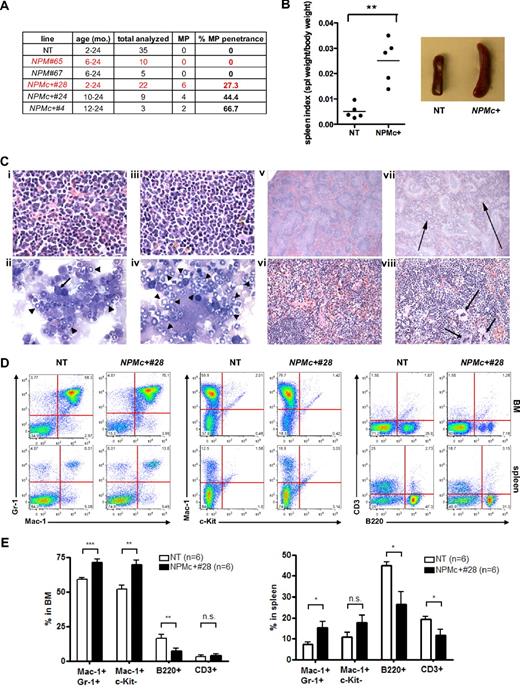
Postmortem pathologic, flow cytometric, and morphologic analyses of NPMc+ mice revealed that 27.3% (6 of 22) of NPMc+ transgenic mice displayed features of myeloproliferation (genetic) as defined by Kogan et al12 in hematopoietic organs (Figure 2A). In contrast, no age-matched nontransgenic (NT) controls, or NPM transgenic mice, showed signs of disease development (Figure 2A and 2 paragraphs below). Mice from 2 to 24 months were analyzed, and signs of myeloproliferation were found in mice from 6 months of age. This myeloproliferation was not found to be fatal, and the animals were not moribund when killed. For each NPMc+ mouse that was killed and studied, an age-matched NT control was killed and studied at the same time. NPMc+ mice showed splenomegaly with an average 5-fold increase in the diseased NPMc+ mice compared with their age-matched NT controls (P > .001, n = 5; Figure 2B). Spleens from NPMc+ mice showed expansion of myeloid cells in the red pulp and a marked extramedullary hematopoiesis (Figure 2Cvii-viii), whereas spleens from NT mice showed a well-defined normal architecture (Figure 2Cv-vi). BM sections from NPMc+ mice demonstrated hypercellularity and accumulation of mature myeloid cells (compare Figure 2Ciii with 2Ci). BM cytospins confirmed an increased fraction of granulocytes (arrowheads) with differentiated morphology and decreased fraction of erythroblasts (arrow; compare Figure 2Civ with 2Cii). Flow cytometric analyses (Figure 2D-E) confirmed the expansion of the differentiated myeloid compartment with an increased percentage of Mac-1+;Gr-1+ cells in BM (58.85% ± 1.276% NT vs 71.28% ± 2.222% NPMc+, P < .001, n = 6) and spleen (7.543% ± 1.078% NT vs 15.45% ± 2.791% NPMc+, P < .025, n = 6) and concomitant reduction of B and T lymphoid fractions. Furthermore, the expanded myeloid cells exhibited a mature immunophenotype in the BM as shown by increased percentage of Mac1+;cKit− cells (52.02% ± 2.764% NT vs 69.50% ± 3.497% NPMc+, P < .003, n = 6).
hMRP8-NPMc+ transgenic mice develop a nonreactive myeloproliferation with expansion of mature granulocytes/monocytes. (A) Table showing the total number of mice analyzed from different transgenic lines, their age range, and myeloproliferation (MP) penetrance involving either BM or spleen. (B) Composite data from age-matched littermates of indicated genotypes demonstrating splenomegaly in NPMc+ transgenic mice (mean ± SEM; NT, n = 5; NPMc+ 28, n = 5). **P < .01. Spleen masses are 160 ± 60.8 mg for NT (n = 5) versus 567 ± 284 mg for NPMc+ 28 (n = 5; P = .014). (C) Histopathologic sections of BM and spleen from representative NT and NPMc+ transgenic mice. (i) NT BM (hematoxylin and eosin, original magnification, ×1000). (ii) NT BM cytospin (original magnification, ×1000).  represents an erythroblast;
represents an erythroblast;  , granulocytes. (iii) NPMc+ BM (hematoxylin and eosin, original magnification, ×1000). (iv) NPMc+ BM cytospin (original magnification, ×1000).
, granulocytes. (iii) NPMc+ BM (hematoxylin and eosin, original magnification, ×1000). (iv) NPMc+ BM cytospin (original magnification, ×1000).  represent granulocytes. (v) NT spleen (hematoxylin and eosin, original magnification, ×40). (vi) NT spleen (hematoxylin and eosin, original magnification, ×400). (vii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×40).
represent granulocytes. (v) NT spleen (hematoxylin and eosin, original magnification, ×40). (vi) NT spleen (hematoxylin and eosin, original magnification, ×400). (vii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×40).  represents areas of expanded red pulp with extramedullary hematopoiesis. (viii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×400).
represents areas of expanded red pulp with extramedullary hematopoiesis. (viii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×400).  represents extramedullary hematopoiesis. (D) Flow cytometric analysis of single-cell suspensions of BM and spleen from representative NT and NPMc+ transgenic mice demonstrates an increase in granulocytic/monocytic (Mac-1+;Gr-1+) and mature myeloid (Mac-1+;cKit−) cells with a corresponding decrease in the amount of B (B220+) and T (CD3+) lymphoid cell populations. (E) Quantification of granulocytic/monocytic (Mac-1+;Gr-1+), mature myeloid (Mac-1+/cKit+), B cells (B2200+), and T cells (CD3+) in BM and spleen of age-matched NT and NPMc+ transgenic mice analyzed as in panel D (mean ± SEM; NT, n = 6; NPMc+ 28, n = 6). *P < .05. **P < .01. ***P < .001. n.s. indicates not statistically significant.
represents extramedullary hematopoiesis. (D) Flow cytometric analysis of single-cell suspensions of BM and spleen from representative NT and NPMc+ transgenic mice demonstrates an increase in granulocytic/monocytic (Mac-1+;Gr-1+) and mature myeloid (Mac-1+;cKit−) cells with a corresponding decrease in the amount of B (B220+) and T (CD3+) lymphoid cell populations. (E) Quantification of granulocytic/monocytic (Mac-1+;Gr-1+), mature myeloid (Mac-1+/cKit+), B cells (B2200+), and T cells (CD3+) in BM and spleen of age-matched NT and NPMc+ transgenic mice analyzed as in panel D (mean ± SEM; NT, n = 6; NPMc+ 28, n = 6). *P < .05. **P < .01. ***P < .001. n.s. indicates not statistically significant.
hMRP8-NPMc+ transgenic mice develop a nonreactive myeloproliferation with expansion of mature granulocytes/monocytes. (A) Table showing the total number of mice analyzed from different transgenic lines, their age range, and myeloproliferation (MP) penetrance involving either BM or spleen. (B) Composite data from age-matched littermates of indicated genotypes demonstrating splenomegaly in NPMc+ transgenic mice (mean ± SEM; NT, n = 5; NPMc+ 28, n = 5). **P < .01. Spleen masses are 160 ± 60.8 mg for NT (n = 5) versus 567 ± 284 mg for NPMc+ 28 (n = 5; P = .014). (C) Histopathologic sections of BM and spleen from representative NT and NPMc+ transgenic mice. (i) NT BM (hematoxylin and eosin, original magnification, ×1000). (ii) NT BM cytospin (original magnification, ×1000).  represents an erythroblast;
represents an erythroblast;  , granulocytes. (iii) NPMc+ BM (hematoxylin and eosin, original magnification, ×1000). (iv) NPMc+ BM cytospin (original magnification, ×1000).
, granulocytes. (iii) NPMc+ BM (hematoxylin and eosin, original magnification, ×1000). (iv) NPMc+ BM cytospin (original magnification, ×1000).  represent granulocytes. (v) NT spleen (hematoxylin and eosin, original magnification, ×40). (vi) NT spleen (hematoxylin and eosin, original magnification, ×400). (vii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×40).
represent granulocytes. (v) NT spleen (hematoxylin and eosin, original magnification, ×40). (vi) NT spleen (hematoxylin and eosin, original magnification, ×400). (vii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×40).  represents areas of expanded red pulp with extramedullary hematopoiesis. (viii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×400).
represents areas of expanded red pulp with extramedullary hematopoiesis. (viii) NPMc+ spleen (hematoxylin and eosin, original magnification, ×400).  represents extramedullary hematopoiesis. (D) Flow cytometric analysis of single-cell suspensions of BM and spleen from representative NT and NPMc+ transgenic mice demonstrates an increase in granulocytic/monocytic (Mac-1+;Gr-1+) and mature myeloid (Mac-1+;cKit−) cells with a corresponding decrease in the amount of B (B220+) and T (CD3+) lymphoid cell populations. (E) Quantification of granulocytic/monocytic (Mac-1+;Gr-1+), mature myeloid (Mac-1+/cKit+), B cells (B2200+), and T cells (CD3+) in BM and spleen of age-matched NT and NPMc+ transgenic mice analyzed as in panel D (mean ± SEM; NT, n = 6; NPMc+ 28, n = 6). *P < .05. **P < .01. ***P < .001. n.s. indicates not statistically significant.
represents extramedullary hematopoiesis. (D) Flow cytometric analysis of single-cell suspensions of BM and spleen from representative NT and NPMc+ transgenic mice demonstrates an increase in granulocytic/monocytic (Mac-1+;Gr-1+) and mature myeloid (Mac-1+;cKit−) cells with a corresponding decrease in the amount of B (B220+) and T (CD3+) lymphoid cell populations. (E) Quantification of granulocytic/monocytic (Mac-1+;Gr-1+), mature myeloid (Mac-1+/cKit+), B cells (B2200+), and T cells (CD3+) in BM and spleen of age-matched NT and NPMc+ transgenic mice analyzed as in panel D (mean ± SEM; NT, n = 6; NPMc+ 28, n = 6). *P < .05. **P < .01. ***P < .001. n.s. indicates not statistically significant.
Myeloproliferation was confirmed in 2 additional independent NPMc+ transgenic founders and their progeny (NPMc+ line 24 [4/9], NPMc+ line 4 [2/3]; Figure 2A, supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). However, none of the age-matched NT mice (n = 35) or NPM transgenic lines (NPM line 65, n = 10 and NPM line 67, n = 5) displayed features of myeloproliferation (Figure 2A, supplemental Figure 1C).
Total white blood cell counts and differential counts from NPMc+ transgenic mice and their wild-type NT littermates were analyzed periodically, and no significant difference was observed (data not shown). This finding is consistent with the fact that myeloproliferation does not necessarily confer peripheral blood changes seen in myeloproliferative diseases.12 In addition, long-term follow-up of 66 NPMc+ and 44 NPM transgenic mice from different lines failed to develop spontaneous leukemia over the course of 24 months. This suggests that, although NPMc+ expression under control of the hMRP8 promoter can drive myeloproliferation, it is insufficient to trigger full-blown leukemia. Thus, a diagnosis of myeloproliferation (genetic) described by Kogan et al12 is fulfilled in our MRP8-NPMc+ mice: (1) increased nonlymphoid hematopoietic cells in spleen and/or BM (as determined by flow cytometry and histology) without increased counts in the peripheral blood; (2) disorder is not a nonlymphoid leukemia; and (3) disorder is not a myeloid dysplasia.
However, MRP8 is also highly expressed in nondifferentiating and differentiating keratinocytes.13,14 Interestingly, 2 of 14 NPMc+ transgenic mice from a low NPMc+ expressing line, NPMc+ 29, presented with a tumor in their upper left legs. Histopathologic analysis revealed areas of squamous differentiation forming large cystic spaces filled with laminar keratin (supplemental Figure 1D). NPMc+ expression was clearly detected in the tumors of the affected mice (supplemental Figure 1D).
In conclusion, we present the first in vivo evidence that NPMc+ confers a proliferative advantage in the mature granulocytic/monocytic lineage of transgenic mice expressing NPMc+ under the myeloid-specific hMRP8 promoter. The absence of an overt block in myeloid differentiation and megakaryocytic proliferation found in our NPMc+ transgenic mice is in line with the morphologic findings observed in BM biopsies from NPMc+ AML patients.15 In addition, the low penetrance of disease observed is in agreement with our previous in vitro data showing that NPMc+ mutant is able to exert oncogenic cooperative activities, but also to elicit, on its own, failsafe mechanisms that oppose cell growth.10
Importantly, the nonreactive myeloproliferation found in hMRP8-NPMc+ transgenic mice does not progress to overt leukemia. This may be because the NPMc+ transgenic model does not exactly reproduce the NPM1-mutated human AML expression pattern (eg, the cell type in which NPMc+ is expressed), with perhaps the need for cooperating mutations. However, in evaluating the hMRP8-NPMc+ model presented in this study, it is also important to consider that expression levels of NPMc+ under the hMRP8 promoter may not be high enough to counter the function of the abundant endogenous Npm proteins, or expressed early enough in the myeloid lineage to elicit its full leukemogenicity. In support of the former hypothesis, it is important to note that, in patients with NPMc+ AML, the product of these heterozygous mutations is found to accumulate almost exclusively in the cytoplasm. The presence of a residual fraction of NPMc+ in the nucleus of BM granulocytes from hMRP8-NPMc+ transgenic mice (Figure 1F) could be the result of the excess in wild-type NPM16 and may provide an explanation for the reason why animals develop myeloproliferation but not acute myeloid leukemia.
Our findings nevertheless demonstrate, for the first time, in vivo that NPMc+ is not a bystander but contributes actively toward leukemogenesis by conferring a proliferative advantage. Ultimately, our hMRP8-NPMc+ and hMRP8-NPM transgenic mice provide a useful tool for determining additional genetic events that collaborate with NPMc+ to induce AML and to develop a targeted therapeutic approach to eradicate NPMc+ leukemias.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Beth Israel Deaconess Medical Center Flow Cytometry facility for help with cell sorting; Dana-Farber Cancer Center Histopathology Facility and Ainara Egia for histology and immunohistrochemistry assistance; Silvia Grisendi, Benjamin H. Lee, and Brunangelo Falini for reagents and critical advice; William J. Haveman and Steven Feng for assistance in management of mouse colonies and bleeding; and members of the Pandolfi laboratory for helpful discussions.
This work was supported by the National Institutes of Health (grants CA-71692 and CA-74031; P.P.P.). P.S. was supported in part by the Associazione Umbra Leucemie e Linfomi.
National Institutes of Health
Authorship
Contribution: K.C. and P.S. designed and performed experiments, analyzed data, and wrote the paper; K.I. performed experiments; J.G.C. analyzed data and wrote the paper; J.T.-F. and J.L.K. provided expert hematopathologic analysis; and P.P.P. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pier Paolo Pandolfi, Cancer Genetics Program, Beth Israel Deaconess Cancer Center and Department of Medicine, Beth Israel Deaconess Medical Center, 330 Brookline Ave, CLS401, Boston, MA 02215; e-mail: ppandolf@bidmc.harvard.edu.
References
Author notes
K.C. and P.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal