Abstract
Studies of childhood leukemia and the potential etiologic role of genetic variation in folate metabolism have produced conflicting findings and have often been based on small numbers. We investigated the association between polymorphisms in key folate metabolism enzymes (MTHFR 677 C>T, MTHFR 1298 A>C, SHMT1 1420 C>T, MTR 2756 A>G, TS 1494del6, and TS 28bp repeat) in 939 cases of childhood acute lymphoblastic leukemia (ALL) and 89 cases of acute myeloid leukemia (AML) recruited into the United Kingdom Childhood Cancer Study. We also examined the maternal genotypes of 752 of these cases. Data from 824 noncancer controls recruited were used for comparison. No evidence of an association with MTHFR 677 was observed for ALL or AML, either in children or their mothers. However, in children an increased risk of ALL (odds ratio [OR] = 1.88; 95% confidence interval [CI], 1.16-3.07; P = .010) and AML (OR = 2.74; 95% CI, 1.07-7.01; P = .036) was observed with the MTR 2756 GG genotype; the association was most pronounced for cases with the MLL translocation (OR = 4.90; 95% CI, 1.30-18.45; P = .019). These data suggest that genetic variation in methionine synthase could mediate risk of childhood leukemia, either via effects on DNA methylation or via effects on fetal growth and development.
Introduction
Leukemia accounts for approximately one-third of all malignancies diagnosed in childhood, with approximately 57 000 cases reported worldwide each year. The major morphologic subtypes of leukemia, acute lymphoblastic (ALL) with a B-cell precursor phenotype and acute myeloid leukemia (AML), are characterized by gross chromosomal abnormalities,1,2 several of which have been shown to originate in utero.3-6 Although there has been much speculation about the nature of the potential agents that could cause such alterations, there is, as yet, no consistent evidence to support a link with either specific exposures or modifiers of exposure.7 Folate levels along with genetic regulation of folate metabolism have been the focus of many investigations,8-19 predicated on the notion that they may influence the creation and/or expansion of the preleukemic clone via DNA hypomethylation of key regulatory genes as well as uracil misincorporation into DNA leading to double-strand breaks and chromosomal aberrations.20,21
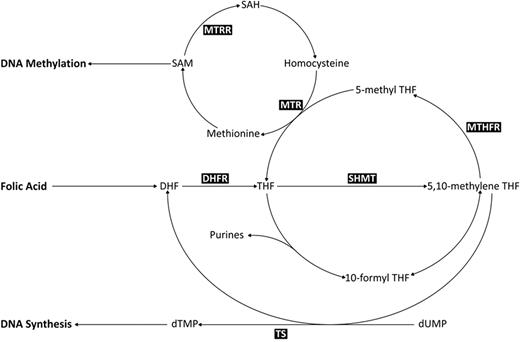
A critical component of the folate metabolic pathway is methylene tetrahydrofolate reductase (MTHFR), which controls the balance between DNA methylation and synthesis via the irreversible conversion of 5,10-methylenetetrahydrofolate (5,10-MeTHF), required for DNA synthesis, to 5-methyltetrahydrofolate (5-MeTHF), a methyl donor for conversion of homocysteine to S-adenosylmethionine (SAM; Figure 1). Two common polymorphisms in MTHFR (677 C>T and 1298 A>C), which result in decreased catalytic activity22,23 and subsequent availability of 5,10-MeTHF and SAM, have been extensively studied in relation to childhood leukemia, but findings have been inconsistent. However, MTHFR is only 1 of more than 30 different enzymes involved in this pathway, and functional polymorphisms in other key enzymes, such as methionine synthase (MTR), thymidylate synthase (TS), and serine hydroxymethyltransferase 1 (SHMT1), have been shown to moderate the risk of hematological malignancies.24-26 These polymorphisms include MTR 2756 (A>G), which moderates the flux of single-carbon moieties for DNA methylation processes27 ; SHMT1 1420 (C>T), which reduces circulating folate levels, thus shunting 5,10-MeTHF toward DNA synthesis28 ; a 6-bp deletion (1494del6) in the 3′-untranslated region of TS that influences RNA levels29 ; and a polymorphic tandem 28-bp repeat sequence within the promoter enhancer region of TS where the triple repeat increases gene expression levels and reduces DNA damage.30
Metabolic folate pathway. Metabolites: 5-MeTHF indicates 5-methyltetrahydrofolate; 10-formyl THF, 10-formyltetrahydrofolate; SAM, adenosylmethionine; SAH, S-adenosylhomocysteine; DHF, dihydrofolate; THF, tetrahydrofolate; dTMP, deoxythymidine monophosphate; and dUMP, deoxyuridine monophosphate. Enzymes: MTR indicates methionine synthase; SHMT, serine hydroxymethyltransferase; MTHFR, 5,10-methylenetetrahydrofolate reductase; and TS, thymidylate synthase.
Metabolic folate pathway. Metabolites: 5-MeTHF indicates 5-methyltetrahydrofolate; 10-formyl THF, 10-formyltetrahydrofolate; SAM, adenosylmethionine; SAH, S-adenosylhomocysteine; DHF, dihydrofolate; THF, tetrahydrofolate; dTMP, deoxythymidine monophosphate; and dUMP, deoxyuridine monophosphate. Enzymes: MTR indicates methionine synthase; SHMT, serine hydroxymethyltransferase; MTHFR, 5,10-methylenetetrahydrofolate reductase; and TS, thymidylate synthase.
With a view to providing further insight into the association between childhood leukemia and folate metabolism, we analyzed polymorphisms in MTHFR, MTR, SHMT1, and TS in more than 1000 cases of acute leukemia and their mothers recruited as part of the United Kingdom Childhood Cancer Study (UKCCS).
Methods
Study population
Cases were children 0 to 14 years of age diagnosed with leukemia between 1991 and 1996 recruited into the UKCCS.31 Samples taken at the time of diagnosis underwent immunophenotype and cytogenetic analysis.31,32 Specific chromosomal abnormalities, including MLL lesions, TEL-AML1 translocations, and hyperdiploidy, were identified by a combination of banded karyotyping, reverse-transcribed polymerase chain reaction, and fluorescence in situ hybridization where appropriate.32 In addition, peripheral blood samples were taken in remission from which DNA was extracted for this and other genetic studies.31,32 In total, DNA was available for 1028 white cases (55.2% male) of which 939 (91.3%) were ALLs and 89 (8.7%) were AMLs. Of the 939 ALLs, 765 (81.5%) were B-lineage (738 precursor B-cell and 27 pro-B cell) and 87 (9.3%) were T-lineage in origin. The remaining 87 (9.3%) were not entered into clinical treatment trials, and details of their immunophenotype were not recorded. With respect to common cytogenetic groups, genotype data were available for 103 cases with a TEL-AML1 translocation (52.4% male), 316 cases with hyperdiploidy (56.7% male), and 34 cases with an MLL lesion (35.3% male). Maternal DNA was available for 752 of the leukemia cases, which included 685 ALLs (573 B-lineage and 54 T-lineage) and 58 AMLs. DNA was amplified using DNA polymerase Phi-29, which has been previously validated for use in genetic epidemiology studies.33,34 As part of routine quality control procedures, we also compared preamplified and postamplified DNA genotyping results on a random sample set.
DNA was obtained from peripheral blood samples taken from 824 noncancer white controls (54.1% male) selected from population registers as part of a United Kingdom–based case-control study. DNA from this control series has been included in several genetic association studies.25,35 Both the UKCCS and the case-control study from which the controls were obtained were carried out with approval from the United Kingdom multiregional ethics committee and in compliance with the Declaration of Helsinki.
Genotyping
Genotyping was carried out using TaqMan Assays-by-Design supplied by Applied Biosystems with probes and primer sets for MTHFR 677C>T (rs1801133), MTHFR 1298A>C (rs1801131), SHMT1 1420C>T (rs1979277), MTR 2756A>G (rs1805087), and TS 1494del6 (rs16430) polymorphisms and the protocol for the TS 28-bp repeat identical to those previously published.26,36 Case samples were genotyped for all 6 polymorphisms, whereas analysis of mother samples was restricted to MTHFR 677C>T, MTHFR 1298A>C, and MTR 2756A>G. TaqMan genotyping assays for MTHFR were verified by running 96 Coriell samples of known genotypes (http://snp500cancer.nci.nih.gov). All other TaqMan assays were verified by direct sequencing or using standard restriction fragment length polymorphism analysis. For added quality assurance, 5% of control samples were selected at random for repeat analysis, 4 independent control samples were analyzed on each 96-well plate, 30 duplicate DNAs were randomly distributed across the entire plate series, and 3 duplicate plates were included in each genotype analysis.
Statistical analysis
Estimates of the odds ratios (OR) for having leukemia were obtained for each polymorphism using univariate logistic regression models (Genmod procedure).37 Genotypes were considered as classes in the regression models so there was no predetermined expectation of a particular dose-response relationship between the number of variant alleles and the risk of having leukemia. Bivariate gene-gene interactions were assessed by adding multiplicative interaction terms between pairs of genes, one at a time, to a multiple logistic regression model that included all of the genes as covariates. Only persons with nonmissing genotype data for all 6 polymorphic sites were included in the multiple regression. To be included in a specific analysis, persons must have had nonmissing genotype information for all single nucleotide polymorphisms included as covariates in the regression model being assessed. Associations in the distributions of gene polymorphisms in pairs of genes were assessed separately for cases and controls using a series of χ2 tests.
Results
Genotype distributions for leukemia cases and controls are shown in Table 1. The control frequencies for MTHFR 677C>T, MTHFR 1298A>C, SHMT1 1420C>T, MTR 2756A>G, TS 1494del6, or TS 28-bp repeat were all in Hardy-Weinberg equilibrium (data not shown) and are similar to those reported in other white populations10,11,14-18,24,36
MTHFR 677C>T, MTHFR 1298A>C, MTR 2756A>G, SHMT1 1420C>T, TS 1494del6, and TS 28-bp repeat genotype frequencies, ORs and 95% CIs in acute leukemia cases and controls
| . | Controls n . | ALL . | AML . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total ALL . | B-lineage . | T-lineage . | |||||||
| n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | ||
| Total* | 824 | 939 | 765 | 87 | 89 | ||||
| MTHFR (677 C>T) | |||||||||
| CC | 359 (47.2) | 374 (46.4) | 1.00 | 302 (46.0) | 1.00 | 37 (50.7) | 1.00 | 47 (59.5) | 1.00 |
| CT | 317 (41.7) | 341 (42.4) | 1.03 (0.84-1.27) | 275 (41.8) | 1.03 (0.83-1.29) | 29 (39.7) | 0.89 (0.53-1.48) | 21 (26.6) | 0.51 (0.30-0.87) |
| TT | 84 (11.1) | 90 (11.2) | 1.03 (0.74-1.43) | 80 (12.2) | 1.13 (0.80-1.59) | 7 (9.6) | 0.81 (0.35-1.88) | 11 (13.9) | 1.00 (0.50-2.01) |
| CC versus CT/TT | 401 (52.8) | 431 (53.6) | 1.03 (0.85-1.26) | 355 (54.0) | 1.05 (0.85-1.30) | 36 (49.3) | 0.87 (0.54-1.41) | 32 (40.5) | 0.61 (0.38-0.98) |
| MTHFR (1298A>C) | |||||||||
| AA | 350 (46.1) | 408 (51.9) | 1.00 | 343 (53.2) | 1.00 | 33 (48.5) | 1.00 | 41 (52.6) | 1.00 |
| AC | 332 (43.7) | 305 (38.8) | 0.79 (0.64-0.97)a | 243 (37.7) | 0.75 (0.60-0.93)b | 32 (47.1) | 1.02 (0.61-1.70) | 26 (33.3) | 0.67 (0.40-1.12) |
| CC | 77 (10.2) | 73 (9.3) | 0.81 (0.57-1.15) | 59 (9.1) | 0.78 (0.54-1.13) | 3 (4.4) | 0.41 (0.12-1.38) | 11 (14.1) | 1.22 (0.60-2.48) |
| AA versus AC/CC | 409 (53.9) | 378 (48.1) | 0.79 (0.65-0.97)c | 302 (46.8) | 0.75 (0.61-0.93)d | 35 (51.5) | 0.91 (0.55-1.49) | 37 (47.4) | 0.77 (0.48-1.23) |
| SHMT1 (1420C>T) | |||||||||
| CC | 351 (46.1) | 401 (49.0) | 1.00 | 330 (49.0) | 1.00 | 35 (47.9) | 1.00 | 41 (52.6) | 1.00 |
| CT | 318 (41.8) | 320 (39.1) | 0.88 (0.71-1.09) | 259 (38.5) | 0.87 (0.69-1.08) | 33 (45.2) | 1.04 (0.63-1.71) | 28 (35.9) | 0.75 (0.46-1.25) |
| TT | 92 (12.1) | 97 (11.9) | 0.92 (0.67-1.27) | 84 (12.5) | 0.97 (0.70-1.35) | 5 (6.9) | 0.55 (0.21-1.43) | 9 (11.5) | 0.84 (0.39-1.79) |
| CC versus CT/TT | 410 (53.9) | 417 (51.0) | 0.89 (0.73-1.09) | 343 (51.0) | 0.89 (0.72-1.10) | 38 (52.1) | 0.93 (0.57-1.50) | 37 (47.4) | 0.77 (0.48-1.23) |
| TS 28bp repeat† | |||||||||
| 2R/2R | 181 (24.0) | 193 (25.4) | 1.00 | 165 (26.6) | 1.00 | 13 (18.1) | 1.00 | 15 (18.5) | 1.00 |
| 2R/3R | 368 (48.8) | 344 (45.3) | 0.88 (0.68-1.13) | 274 (44.2) | 0.82 (0.63-1.06) | 33 (45.8) | 1.25 (0.64-2.43) | 40 (49.4) | 1.31 (0.71-2.44) |
| 3R/3R | 205 (27.2) | 222 (29.3) | 1.02 (0.77-1.34) | 181 (29.2) | 0.97 (0.72-1.30) | 26 (36.1) | 1.77 (0.88-3.54) | 26 (32.1) | 1.53 (0.79-2.98) |
| 2R2R versus others | 575 (76.0) | 570 (74.6) | 0.93 (0.74-1.17) | 457 (73.4) | 0.87 (0.68-1.11) | 59 (81.9) | 1.43 (0.77-2.66) | 66 (81.5) | 1.38 (0.77-2.49) |
| TS (1494 del6) | |||||||||
| 6-bp+/6-bp+ | 373 (49.0) | 423 (49.5) | 1.00 | 353 (50.4) | 1.00 | 36 (46.8) | 1.00 | 41 (48.8) | 1.00 |
| 6-bp+/6-bp− | 331 (43.4) | 336 (39.3) | 0.90 (0.73-1.10) | 268 (38.3) | 0.86 (0.69-1.06) | 32 (41.6) | 1.00 (0.61-1.65) | 30 (35.7) | 0.82 (0.5-1.35) |
| 6-bp−/6-bp− | 58 (7.6) | 96 (11.2) | 1.46 (1.02-2.08)e | 79 (11.3) | 1.44 (1.00-2.08)f | 9 (11.7) | 1.61 (0.74-3.51) | 13 (15.5) | 2.04 (1.03-4.03)g |
| 6-bp+/6-bp+ versus 6-bp+/6-bp−/6-bp−/6-bp− | 389 (51.0) | 432 (50.5) | 0.98 (0.81-1.19) | 347 (49.6) | 0.94 (0.77-1.16) | 41 (53.2) | 1.09 (0.68-1.75) | 43 (51.2) | 1.02 (0.64-1.58) |
| MTR 2756 (A>G) | |||||||||
| AA | 510 (67.2) | 531 (61.0) | 1.00 | 431 (60.6) | 1.00 | 48 (61.6) | 1.00 | 43 (55.8) | 1.00 |
| AG | 223 (29.4) | 288 (33.1) | 1.24 (1.00-1.53)h | 240 (33.8) | 1.27 (1.02-1.59)i | 26 (33.3) | 1.24 (0.75-2.05) | 28 (36.4) | 1.49 (0.90-2.46) |
| GG | 26 (3.4) | 51 (5.9) | 1.88 (1.16-3.07)j | 40 (5.6) | 1.82 (1.09-3.03)k | 4 (5.1) | 1.63 (0.55-4.88) | 6 (7.8) | 2.74 (1.07-7.01)l |
| AA versus AG/GG | 249 (32.8) | 339 (39.0) | 1.31 (1.07-1.60)m | 280 (39.4) | 1.33 (1.07-1.65)n | 30 (38.4) | 1.28 (0.79-2.07) | 34 (44.2) | 1.62 (1.01-2.60)o |
| . | Controls n . | ALL . | AML . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total ALL . | B-lineage . | T-lineage . | |||||||
| n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | ||
| Total* | 824 | 939 | 765 | 87 | 89 | ||||
| MTHFR (677 C>T) | |||||||||
| CC | 359 (47.2) | 374 (46.4) | 1.00 | 302 (46.0) | 1.00 | 37 (50.7) | 1.00 | 47 (59.5) | 1.00 |
| CT | 317 (41.7) | 341 (42.4) | 1.03 (0.84-1.27) | 275 (41.8) | 1.03 (0.83-1.29) | 29 (39.7) | 0.89 (0.53-1.48) | 21 (26.6) | 0.51 (0.30-0.87) |
| TT | 84 (11.1) | 90 (11.2) | 1.03 (0.74-1.43) | 80 (12.2) | 1.13 (0.80-1.59) | 7 (9.6) | 0.81 (0.35-1.88) | 11 (13.9) | 1.00 (0.50-2.01) |
| CC versus CT/TT | 401 (52.8) | 431 (53.6) | 1.03 (0.85-1.26) | 355 (54.0) | 1.05 (0.85-1.30) | 36 (49.3) | 0.87 (0.54-1.41) | 32 (40.5) | 0.61 (0.38-0.98) |
| MTHFR (1298A>C) | |||||||||
| AA | 350 (46.1) | 408 (51.9) | 1.00 | 343 (53.2) | 1.00 | 33 (48.5) | 1.00 | 41 (52.6) | 1.00 |
| AC | 332 (43.7) | 305 (38.8) | 0.79 (0.64-0.97)a | 243 (37.7) | 0.75 (0.60-0.93)b | 32 (47.1) | 1.02 (0.61-1.70) | 26 (33.3) | 0.67 (0.40-1.12) |
| CC | 77 (10.2) | 73 (9.3) | 0.81 (0.57-1.15) | 59 (9.1) | 0.78 (0.54-1.13) | 3 (4.4) | 0.41 (0.12-1.38) | 11 (14.1) | 1.22 (0.60-2.48) |
| AA versus AC/CC | 409 (53.9) | 378 (48.1) | 0.79 (0.65-0.97)c | 302 (46.8) | 0.75 (0.61-0.93)d | 35 (51.5) | 0.91 (0.55-1.49) | 37 (47.4) | 0.77 (0.48-1.23) |
| SHMT1 (1420C>T) | |||||||||
| CC | 351 (46.1) | 401 (49.0) | 1.00 | 330 (49.0) | 1.00 | 35 (47.9) | 1.00 | 41 (52.6) | 1.00 |
| CT | 318 (41.8) | 320 (39.1) | 0.88 (0.71-1.09) | 259 (38.5) | 0.87 (0.69-1.08) | 33 (45.2) | 1.04 (0.63-1.71) | 28 (35.9) | 0.75 (0.46-1.25) |
| TT | 92 (12.1) | 97 (11.9) | 0.92 (0.67-1.27) | 84 (12.5) | 0.97 (0.70-1.35) | 5 (6.9) | 0.55 (0.21-1.43) | 9 (11.5) | 0.84 (0.39-1.79) |
| CC versus CT/TT | 410 (53.9) | 417 (51.0) | 0.89 (0.73-1.09) | 343 (51.0) | 0.89 (0.72-1.10) | 38 (52.1) | 0.93 (0.57-1.50) | 37 (47.4) | 0.77 (0.48-1.23) |
| TS 28bp repeat† | |||||||||
| 2R/2R | 181 (24.0) | 193 (25.4) | 1.00 | 165 (26.6) | 1.00 | 13 (18.1) | 1.00 | 15 (18.5) | 1.00 |
| 2R/3R | 368 (48.8) | 344 (45.3) | 0.88 (0.68-1.13) | 274 (44.2) | 0.82 (0.63-1.06) | 33 (45.8) | 1.25 (0.64-2.43) | 40 (49.4) | 1.31 (0.71-2.44) |
| 3R/3R | 205 (27.2) | 222 (29.3) | 1.02 (0.77-1.34) | 181 (29.2) | 0.97 (0.72-1.30) | 26 (36.1) | 1.77 (0.88-3.54) | 26 (32.1) | 1.53 (0.79-2.98) |
| 2R2R versus others | 575 (76.0) | 570 (74.6) | 0.93 (0.74-1.17) | 457 (73.4) | 0.87 (0.68-1.11) | 59 (81.9) | 1.43 (0.77-2.66) | 66 (81.5) | 1.38 (0.77-2.49) |
| TS (1494 del6) | |||||||||
| 6-bp+/6-bp+ | 373 (49.0) | 423 (49.5) | 1.00 | 353 (50.4) | 1.00 | 36 (46.8) | 1.00 | 41 (48.8) | 1.00 |
| 6-bp+/6-bp− | 331 (43.4) | 336 (39.3) | 0.90 (0.73-1.10) | 268 (38.3) | 0.86 (0.69-1.06) | 32 (41.6) | 1.00 (0.61-1.65) | 30 (35.7) | 0.82 (0.5-1.35) |
| 6-bp−/6-bp− | 58 (7.6) | 96 (11.2) | 1.46 (1.02-2.08)e | 79 (11.3) | 1.44 (1.00-2.08)f | 9 (11.7) | 1.61 (0.74-3.51) | 13 (15.5) | 2.04 (1.03-4.03)g |
| 6-bp+/6-bp+ versus 6-bp+/6-bp−/6-bp−/6-bp− | 389 (51.0) | 432 (50.5) | 0.98 (0.81-1.19) | 347 (49.6) | 0.94 (0.77-1.16) | 41 (53.2) | 1.09 (0.68-1.75) | 43 (51.2) | 1.02 (0.64-1.58) |
| MTR 2756 (A>G) | |||||||||
| AA | 510 (67.2) | 531 (61.0) | 1.00 | 431 (60.6) | 1.00 | 48 (61.6) | 1.00 | 43 (55.8) | 1.00 |
| AG | 223 (29.4) | 288 (33.1) | 1.24 (1.00-1.53)h | 240 (33.8) | 1.27 (1.02-1.59)i | 26 (33.3) | 1.24 (0.75-2.05) | 28 (36.4) | 1.49 (0.90-2.46) |
| GG | 26 (3.4) | 51 (5.9) | 1.88 (1.16-3.07)j | 40 (5.6) | 1.82 (1.09-3.03)k | 4 (5.1) | 1.63 (0.55-4.88) | 6 (7.8) | 2.74 (1.07-7.01)l |
| AA versus AG/GG | 249 (32.8) | 339 (39.0) | 1.31 (1.07-1.60)m | 280 (39.4) | 1.33 (1.07-1.65)n | 30 (38.4) | 1.28 (0.79-2.07) | 34 (44.2) | 1.62 (1.01-2.60)o |
ALL indicates acute lymphoblastic leukemia; AML, acute myeloid leukemia; and OR, odds ratio.
Total includes persons for whom a result was not available, and varied between single nucleotide polymorphisms.
Includes 2 cases with 1R/1R, one case with 2R/4R, and one case with 3R/4R, and 2 controls with 3R/4R genotypes.
aP = .03, bP = .01, cP = .02, dP = .01, eP = .03, fP = .05, gP = .04, hP = .05, iP = .03, jP = .01, kP = .02, lP = .04, mP = .01, nP = .01, oP = .05.
No statistically significant case-control differences in the distribution of the MTHFR 677C>T, SHMT1 1420C>T, or TS 28-bp repeat polymorphisms were observed for ALL or AML (Table 1). However, a dose-response relationship between the number of copies of the MTR 2756 G-allele and increased risk of ALL, specifically that of B-lineage ALL, as well as AML was observed (Table 1). Specifically, heterozygosity (AG) was associated with a 1.24-fold increased risk of ALL (95% confidence interval [CI], 1.00-1.53; P = .05), and homozygosity for the variant allele (GG) with a 1.88-fold increased risk of ALL (95% CI, 1.16-3.07; P = .01) and 2.74-fold increased risk of AML (95% CI, 1.07-7.01; P = .036). Findings were similar for B- and T-lineage ALL. In addition, homozygosity for the TS 1494del6 polymorphism (6-bp−/6-bp−) was associated with an increased risk of ALL (OR = 1.46; 95% CI, 1.02-2.08; P = .04), B-lineage ALL (OR = 1.44; 95% CI, 1.00-2.08; P = .05), and AML (OR = 2.04; 95% CI, 1.03-4.03; P = .04; Table 1). There was also limited evidence to suggest that the MTHFR 1298 variant C allele was associated with total ALL (OR = 0.79; 95% CI, 0.65-0.97) and B-lineage ALL (OR = 0.75, 955 CI, 0.61-0.93; Table 1). When data for all polymorphisms were included in a multiple logistic regression model, similar trends were observed as those shown in Table 1 (data not shown).
When data were stratified by sex, no differences between boys and girls were observed with respect to MTHFR 677C>T, SHMT1 1420C>T, MTR 2756A>G, TS 1494del6, or TS 28-bp repeat polymorphisms (data not shown). However, there was some evidence that homozygosity for the MTHFR 1298 A>C polymorphism (CC) was associated with a decreased risk of ALL in girls (OR = 0.51; 95% CI, 0.30-0.89; P = .02) and B-lineage ALL (OR = 0.48; 95% CI, 0.27-0.87; P = .02), but not in boys (ALL OR = 0.96; 95% CI, 0.65-1.43; B-cell ALL OR = 0.95; 95% CI, 0.62-1.46; data not shown).
Genotype data were also stratified according to the presence of specific chromosomal abnormalities, including MLL and TEL-AML1 lesions, as well as hyperdiploidy (Table 2). Homozygosity for the MTR 2756 polymorphism (GG) was strongly associated with MLL-positive leukemia (OR = 4.90; 95% CI, 1.30-18.45; P = .02). Similar findings were also observed when genotypes from MLL-positive leukemias were compared with those for all other leukemias combined (MTR 2756 AG OR = 2.21 95% CI, 1.01-4.84; MTR 2756 GG OR = 2.60; 95% CI, 0.71-9.49) and those from cases with a normal cytogenetic profile (MTR 2756 AG OR = 2.16; 95% CI, 0.90-5.19; MTR 2756 GG OR = 3.94; 95% CI, 0.78-19.88). There was also evidence to suggest an association, although not statistically significant at the conventional 5% level, with the TS 28-bp repeat polymorphism, 3R/3R (OR = 3.53; 95% CI, 0.98-12.71). Furthermore, homozygosity for the TS 6-bp deletion polymorphism (6-bp−/6-bp−) was related to hyperdiploidy (OR = 1.69; 95% CI, 1.07-2.68; P = .02). No significant associations were observed between TEL-AML1-positive leukemia and the polymorphisms studied, with the exception of MTHFR 1298 where the presence of the C-allele appeared to be related to a decreased risk of TEL-AML1-positive leukemia (OR = 0.52; 95% CI, 0.33-0.81; P = .01).
Number (%) of cases and controls, Ors, and 95% CIs by leukemia subgroup for MTHFR 677C>T, MTHFR 1298A>C, MTR 2756A>G, SHMT1 1420C>T, TS 1494del6, and TS 28-bp repeat
| . | Controls, n (%) . | MLL . | TEL-AML1 . | Hyperdiploidy . | |||
|---|---|---|---|---|---|---|---|
| n (%) . | OR (95% CI) . | n (%) . | OR (95% CI) . | n (%) . | OR (95% CI) . | ||
| Total* | 824 | 34 | 103 | 316 | |||
| MTHFR (677 C>T) | |||||||
| CC | 359 (47.2) | 13 (44.8) | 1.00 | 38 (42.2) | 1.00 | 124 (45.9) | 1.00 |
| CT | 317 (41.7) | 12 (41.4) | 1.05 (0.47-2.32) | 36 (40.0) | 1.07 (0.66-1.73) | 117 (43.3) | 1.07 (0.80-1.43) |
| TT | 84 (11.1) | 4 (13.8) | 1.32 (0.42-4.14) | 16 (17.8) | 1.80 (0.96-3.38) | 29 (10.8) | 1.00 (0.63-1.60) |
| CC versus CT/TT | 401 (52.8) | 16 (55.2) | 1.10 (0.52-2.32) | 52 (57.8) | 1.23 (0.79-1.91) | 146 (54.1) | 1.05 (0.80-1.39) |
| MTHFR (1298A>C) | |||||||
| AA | 350 (46.1) | 16 (55.2) | 1.00 | 56 (62.2) | 1.00 | 137 (50.9) | 1.00 |
| AC | 332 (43.7) | 8 (27.6) | 0.53 (0.22-1.25) | 28 (31.1) | 0.53 (0.33-0.85)a | 109 (40.5) | 0.84 (0.63-1.12) |
| CC | 77 (10.2) | 5 (17.2) | 1.42 (0.51-1.40) | 6 (6.7) | 0.49 (0.20-1.17) | 23 (8.6) | 0.76 (0.46-1.27) |
| AA versus AC/CC | 409 (53.9) | 13 (44.8) | 0.70 (0.33-1.47) | 34 (37.8) | 0.52 (0.33-0.81)b | 132 (49.1) | 0.82 (0.62-1.09) |
| SHMT1 (1420C>T) | |||||||
| CC | 351 (46.1) | 15 (48.4) | 1.00 | 35 (38.5) | 1.00 | 148 (52.7) | 1.00 |
| CT | 318 (41.8) | 13 (41.9) | 0.96 (0.45-2.04) | 44 (48.3) | 1.39 (0.87-2.22) | 98 (34.9) | 0.73 (0.54-0.98) |
| TT | 92 (12.1) | 3 (9.7) | 0.76 (0.22-2.69) | 12 (13.2) | 1.31 (0.65-2.62) | 35 (12.4) | 0.90 (0.58-1.39) |
| CC versus CT/TT | 410 (53.9) | 16 (51.6) | 0.91 (0.45-1.87) | 56 (61.5) | 1.37 (0.88-2.14) | 133 (47.3) | 0.77 (0.58-1.01) |
| TS 28-bp repeat† | |||||||
| 2R/2R | 181 (23.9) | 3 (10.3) | 1.00 | 20 (24.7) | 1.00 | 70 (26.4) | 1.00 |
| 2R/3R | 368 (48.7) | 14 (48.3) | 2.30 (0.65-8.09) | 40 (49.4) | 0.98 (0.56-1.73) | 116 (43.8) | 0.82 (0.58-1.15) |
| 3R/3R | 205 (27.1) | 12 (41.4) | 3.53 (0.98-12.71) | 21 (25.9) | 0.93 (0.49-1.77) | 79 (29.8) | 1.00 (0.68-1.46) |
| 2R/2R versus all others | 575 (76.1) | 26 (89.7) | 2.73 (0.82-9.12) | 61 (75.3) | 0.96 (0.56-1.63) | 195 (73.6) | 0.88 (0.64-1.21) |
| TS (1494 del6) | |||||||
| 6-bp+/6-bp+ | 373 (49.0) | 18 (54.5) | 1.00 | 55 (56.7) | 1.00 | 137 (47.7) | 1.00 |
| 6-bp+/6-bp− | 331 (43.4) | 10 (30.3) | 0.63 (0.28-1.38) | 34 (35.1) | 0.70 (0.44-1.10) | 114 (39.7) | 0.94 (0.70-1.25) |
| 6-bp−/6-bp− | 58 (7.6) | 5 (15.2) | 1.79 (0.64-5.00) | 8 (8.2) | 0.94 (0.42-2.06) | 36 (12.6) | 1.69 (1.07-2.68)c |
| 6-bp+/6-bp+ versus 6-bp+/6-bp−/6-bp−/6-bp− | 389 (51.0) | 15 (45.5) | 0.80 (0.40-1.61) | 42 (43.3) | 0.73 (0.48-1.12) | 150 (52.3) | 1.05 (0.80-1.38) |
| MTR 2756 (A>G) | |||||||
| AA | 510 (67.2) | 12 (41.4) | 1.00 | 58 (59.8) | 1.00 | 181 (60.5) | 1.00 |
| AG | 223 (29.4) | 14 (48.3) | 2.67 (1.21-5.86)d | 34 (35.1) | 1.34 (0.85-2.11) | 101 (33.8) | 1.28 (0.96-1.71) |
| GG | 26 (3.4) | 3 (10.3) | 4.90 (1.30-18.45)e | 5 (5.1) | 1.69 (0.63-4.57) | 17 (5.7) | 1.84 (0.98-3.47) |
| AA versus AG/GG | 249 (32.8) | 17 (58.6) | 2.90 (1.36-6.17)f | 39 (40.1) | 1.37 (0.88-2.14) | 118 (39.1) | 1.34 (1.01-1.76)g |
| . | Controls, n (%) . | MLL . | TEL-AML1 . | Hyperdiploidy . | |||
|---|---|---|---|---|---|---|---|
| n (%) . | OR (95% CI) . | n (%) . | OR (95% CI) . | n (%) . | OR (95% CI) . | ||
| Total* | 824 | 34 | 103 | 316 | |||
| MTHFR (677 C>T) | |||||||
| CC | 359 (47.2) | 13 (44.8) | 1.00 | 38 (42.2) | 1.00 | 124 (45.9) | 1.00 |
| CT | 317 (41.7) | 12 (41.4) | 1.05 (0.47-2.32) | 36 (40.0) | 1.07 (0.66-1.73) | 117 (43.3) | 1.07 (0.80-1.43) |
| TT | 84 (11.1) | 4 (13.8) | 1.32 (0.42-4.14) | 16 (17.8) | 1.80 (0.96-3.38) | 29 (10.8) | 1.00 (0.63-1.60) |
| CC versus CT/TT | 401 (52.8) | 16 (55.2) | 1.10 (0.52-2.32) | 52 (57.8) | 1.23 (0.79-1.91) | 146 (54.1) | 1.05 (0.80-1.39) |
| MTHFR (1298A>C) | |||||||
| AA | 350 (46.1) | 16 (55.2) | 1.00 | 56 (62.2) | 1.00 | 137 (50.9) | 1.00 |
| AC | 332 (43.7) | 8 (27.6) | 0.53 (0.22-1.25) | 28 (31.1) | 0.53 (0.33-0.85)a | 109 (40.5) | 0.84 (0.63-1.12) |
| CC | 77 (10.2) | 5 (17.2) | 1.42 (0.51-1.40) | 6 (6.7) | 0.49 (0.20-1.17) | 23 (8.6) | 0.76 (0.46-1.27) |
| AA versus AC/CC | 409 (53.9) | 13 (44.8) | 0.70 (0.33-1.47) | 34 (37.8) | 0.52 (0.33-0.81)b | 132 (49.1) | 0.82 (0.62-1.09) |
| SHMT1 (1420C>T) | |||||||
| CC | 351 (46.1) | 15 (48.4) | 1.00 | 35 (38.5) | 1.00 | 148 (52.7) | 1.00 |
| CT | 318 (41.8) | 13 (41.9) | 0.96 (0.45-2.04) | 44 (48.3) | 1.39 (0.87-2.22) | 98 (34.9) | 0.73 (0.54-0.98) |
| TT | 92 (12.1) | 3 (9.7) | 0.76 (0.22-2.69) | 12 (13.2) | 1.31 (0.65-2.62) | 35 (12.4) | 0.90 (0.58-1.39) |
| CC versus CT/TT | 410 (53.9) | 16 (51.6) | 0.91 (0.45-1.87) | 56 (61.5) | 1.37 (0.88-2.14) | 133 (47.3) | 0.77 (0.58-1.01) |
| TS 28-bp repeat† | |||||||
| 2R/2R | 181 (23.9) | 3 (10.3) | 1.00 | 20 (24.7) | 1.00 | 70 (26.4) | 1.00 |
| 2R/3R | 368 (48.7) | 14 (48.3) | 2.30 (0.65-8.09) | 40 (49.4) | 0.98 (0.56-1.73) | 116 (43.8) | 0.82 (0.58-1.15) |
| 3R/3R | 205 (27.1) | 12 (41.4) | 3.53 (0.98-12.71) | 21 (25.9) | 0.93 (0.49-1.77) | 79 (29.8) | 1.00 (0.68-1.46) |
| 2R/2R versus all others | 575 (76.1) | 26 (89.7) | 2.73 (0.82-9.12) | 61 (75.3) | 0.96 (0.56-1.63) | 195 (73.6) | 0.88 (0.64-1.21) |
| TS (1494 del6) | |||||||
| 6-bp+/6-bp+ | 373 (49.0) | 18 (54.5) | 1.00 | 55 (56.7) | 1.00 | 137 (47.7) | 1.00 |
| 6-bp+/6-bp− | 331 (43.4) | 10 (30.3) | 0.63 (0.28-1.38) | 34 (35.1) | 0.70 (0.44-1.10) | 114 (39.7) | 0.94 (0.70-1.25) |
| 6-bp−/6-bp− | 58 (7.6) | 5 (15.2) | 1.79 (0.64-5.00) | 8 (8.2) | 0.94 (0.42-2.06) | 36 (12.6) | 1.69 (1.07-2.68)c |
| 6-bp+/6-bp+ versus 6-bp+/6-bp−/6-bp−/6-bp− | 389 (51.0) | 15 (45.5) | 0.80 (0.40-1.61) | 42 (43.3) | 0.73 (0.48-1.12) | 150 (52.3) | 1.05 (0.80-1.38) |
| MTR 2756 (A>G) | |||||||
| AA | 510 (67.2) | 12 (41.4) | 1.00 | 58 (59.8) | 1.00 | 181 (60.5) | 1.00 |
| AG | 223 (29.4) | 14 (48.3) | 2.67 (1.21-5.86)d | 34 (35.1) | 1.34 (0.85-2.11) | 101 (33.8) | 1.28 (0.96-1.71) |
| GG | 26 (3.4) | 3 (10.3) | 4.90 (1.30-18.45)e | 5 (5.1) | 1.69 (0.63-4.57) | 17 (5.7) | 1.84 (0.98-3.47) |
| AA versus AG/GG | 249 (32.8) | 17 (58.6) | 2.90 (1.36-6.17)f | 39 (40.1) | 1.37 (0.88-2.14) | 118 (39.1) | 1.34 (1.01-1.76)g |
OR indicates odds ratio.
Total includes cases for whom a result was not available, and varied between single nucleotide polymorphisms.
Includes 3 controls with 3R/4R genotype.
aP = .01, bP = .001, cP = .02, dP = .01, eP = .02, fP = .006, gP = .04.
When we investigated associations between the 6 polymorphisms, we observed, as expected, associations between MTHFR677 and MTHFR1298 and between TS 6-bp deletion and TS 28-bp repeat polymorphisms. In addition, we also detected an interaction between the MTR 2756 and TS 6-bp deletion polymorphisms among ALL cases (P = .05), such that heterozygotes for the MTR 2756 polymorphism were more likely to have at least 1 copy of the allele with the 6-bp deletion present. When we examined the effects of gene-gene interactions on leukemia risk, we observed some evidence of an interaction between MTHFR 1298 and SHMT1 1420 polymorphisms and between MTHFR 1298 and TS 28-bp repeat polymorphisms for ALL (P = .09 and P = .11, respectively) and also B-lineage ALL (P = .16 and P = .07, respectively). There were too few persons in the other subsets of cases to support this analysis.
The relationship between maternal genotype and risk of childhood leukemia was also explored. No association between MTHFR677, MTHFR1298, or MTR 2756 polymorphisms and any leukemia subtype was observed in either univariate (Table 3) or multiple regression models (data not shown). Furthermore, no differences were seen when data were stratified by sex of the child or cytogenetic subtype (data not shown). When maternal and child genotype data were included in a single regression model, results were generally similar to when the child's genotype alone was considered, although for some polymorphisms the findings were more striking. For example, homozygosity for MTR 2756 (GG) in both mother and child was more strongly associated with MLL-positive leukemia compared with other leukemias (OR = 8.78; 95% CI, 1.92-40.13; P = .005) and leukemias with a normal cytogenetic profile (OR = 18.75; 95% CI, 1.60-220.00; P = .02), than when the child's genotype alone was considered (all leukemias combined: MTR 2756 GG, OR = 2.60; 95% CI, 0.71-9.49; normal cytogenetic profile: MTR 2756 GG, OR = 3.94; 95% CI, 0.78-19.88).
The distribution of MTHFR 677C>T, MTHFR 1298A>C, and MTR 2756A>G polymorphisms among mothers of children with acute leukemia
| . | Controls n . | Case mothers . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ALL . | AML . | ||||||||
| Total ALL . | B-cell ALL . | T-cell ALL . | |||||||
| n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | ||
| Total* | 378 | 685 | 573 | 54 | 58 | ||||
| MTHFR (677 C>T) | |||||||||
| CC | 157 (44.7) | 315 (47.1) | 1.00 | 261 (46.7) | 1.00 | 26 (49.1) | 1.00 | 24 (45.3) | 1.00 |
| CT | 159 (45.2) | 277 (41.4) | 0.87 (0.66-1.14) | 229 (41.0) | 0.87 (0.65-1.15) | 21 (39.6) | 0.80 (0.43-1.48) | 24 (45.3) | 0.99 (0.54-1.81) |
| TT | 35 (10.0) | 77 (11.5) | 1.10 (0.70-1.71) | 69 (12.3) | 1.19 (0.75-1.86) | 6 (11.3) | 1.04 (0.40-2.70) | 5 (9.4) | 0.93 (0.33-2.62) |
| CC versus CT/TT | 194 (55.2) | 354 (52.9) | 0.91 (0.70-1.18) | 298 (53.3) | 0.92 (0.71-1.21) | 27 (50.9) | 0.84 (0.47-1.50) | 29 (54.6) | 0.98 (0.55-1.75) |
| MTHFR (1298A>C) | |||||||||
| AA | 157 (44.9) | 334 (50.1) | 1.00 | 285 (51.2) | 1.00 | 27 (50.9) | 1.00 | 24 (46.2) | 1.00 |
| AC | 151 (43.1) | 254 (38.1) | 0.79 (0.60-1.04) | 205 (36.9) | 0.75 (0.56-1.00) | 24 (45.3) | 0.92 (0.51-1.67) | 18 (34.6) | 0.78 (0.41-1.49) |
| CC | 42 (12.0) | 78 (11.8) | 0.87 (0.57-1.33) | 66 (11.9) | 0.87 (0.56-1.34) | 2 (3.8) | 0.28 (0.06-1.21) | 10 (19.2) | 1.56 (0.69-3.51) |
| AA versus AC/CC | 193 (55.1) | 332 (49.9) | 0.81 (0.62-1.05) | 231 (48.8) | 0.77 (0.59-1.01) | 26 (49.1) | 0.78 (0.44-1.40) | 28 (53.8) | 0.95 (0.53-1.70) |
| MTR 2756 (A>G) | |||||||||
| AA | 239 (68.3) | 429 (65.7) | 1.00 | 358 (64.4) | 1.00 | 35 (66.0) | 1.00 | 31 (57.4) | 1.00 |
| AG | 97 (27.7) | 201 (31.2) | 1.15 (0.86-2.50) | 168 (30.2) | 1.16 (0.86-1.56) | 16 (30.2) | 1.13 (0.60-2.13) | 21 (38.9) | 1.67 (0.91-3.05) |
| GG | 14 (4.0) | 33 (5.1) | 1.31 (0.69-2.50) | 30 (5.4) | 1.43 (0.74-2.75) | 2 (3.8) | 0.98 (0.21-4.48) | 2 (3.7) | 1.10 (0.24-5.08) |
| AA versus AG/GG | 111 (31.7) | 234 (36.3) | 1.17 (0.89-1.55) | 198 (35.6) | 1.19 (0.90-1.58) | 18 (34.0) | 1.11 (0.60-2.04) | 23 (42.6) | 1.60 (0.89-2.87) |
| . | Controls n . | Case mothers . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ALL . | AML . | ||||||||
| Total ALL . | B-cell ALL . | T-cell ALL . | |||||||
| n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | n . | OR (95% CI) . | ||
| Total* | 378 | 685 | 573 | 54 | 58 | ||||
| MTHFR (677 C>T) | |||||||||
| CC | 157 (44.7) | 315 (47.1) | 1.00 | 261 (46.7) | 1.00 | 26 (49.1) | 1.00 | 24 (45.3) | 1.00 |
| CT | 159 (45.2) | 277 (41.4) | 0.87 (0.66-1.14) | 229 (41.0) | 0.87 (0.65-1.15) | 21 (39.6) | 0.80 (0.43-1.48) | 24 (45.3) | 0.99 (0.54-1.81) |
| TT | 35 (10.0) | 77 (11.5) | 1.10 (0.70-1.71) | 69 (12.3) | 1.19 (0.75-1.86) | 6 (11.3) | 1.04 (0.40-2.70) | 5 (9.4) | 0.93 (0.33-2.62) |
| CC versus CT/TT | 194 (55.2) | 354 (52.9) | 0.91 (0.70-1.18) | 298 (53.3) | 0.92 (0.71-1.21) | 27 (50.9) | 0.84 (0.47-1.50) | 29 (54.6) | 0.98 (0.55-1.75) |
| MTHFR (1298A>C) | |||||||||
| AA | 157 (44.9) | 334 (50.1) | 1.00 | 285 (51.2) | 1.00 | 27 (50.9) | 1.00 | 24 (46.2) | 1.00 |
| AC | 151 (43.1) | 254 (38.1) | 0.79 (0.60-1.04) | 205 (36.9) | 0.75 (0.56-1.00) | 24 (45.3) | 0.92 (0.51-1.67) | 18 (34.6) | 0.78 (0.41-1.49) |
| CC | 42 (12.0) | 78 (11.8) | 0.87 (0.57-1.33) | 66 (11.9) | 0.87 (0.56-1.34) | 2 (3.8) | 0.28 (0.06-1.21) | 10 (19.2) | 1.56 (0.69-3.51) |
| AA versus AC/CC | 193 (55.1) | 332 (49.9) | 0.81 (0.62-1.05) | 231 (48.8) | 0.77 (0.59-1.01) | 26 (49.1) | 0.78 (0.44-1.40) | 28 (53.8) | 0.95 (0.53-1.70) |
| MTR 2756 (A>G) | |||||||||
| AA | 239 (68.3) | 429 (65.7) | 1.00 | 358 (64.4) | 1.00 | 35 (66.0) | 1.00 | 31 (57.4) | 1.00 |
| AG | 97 (27.7) | 201 (31.2) | 1.15 (0.86-2.50) | 168 (30.2) | 1.16 (0.86-1.56) | 16 (30.2) | 1.13 (0.60-2.13) | 21 (38.9) | 1.67 (0.91-3.05) |
| GG | 14 (4.0) | 33 (5.1) | 1.31 (0.69-2.50) | 30 (5.4) | 1.43 (0.74-2.75) | 2 (3.8) | 0.98 (0.21-4.48) | 2 (3.7) | 1.10 (0.24-5.08) |
| AA versus AG/GG | 111 (31.7) | 234 (36.3) | 1.17 (0.89-1.55) | 198 (35.6) | 1.19 (0.90-1.58) | 18 (34.0) | 1.11 (0.60-2.04) | 23 (42.6) | 1.60 (0.89-2.87) |
ALL indicates acute lymphoblastic leukemia; AML, acute myeloid leukemia; and OR, odds ratio.
Totals include persons for whom a result was not available, and varied between single nucleotide polymorphisms.
Discussion
We have demonstrated that the MTR 2756 A>G genetic polymorphism is associated with increased risk of both childhood ALL and AML and that this risk is further increased in the subset of cases with an MLL chromosomal abnormality. However, in contrast to others,9-11,13,18,38 our data do not support the hypothesis that the MTHFR 677 C>T polymorphism modifies the risk of ALL or AML in the United Kingdom. Furthermore, no associations were observed for either SHMT 1420 C>T or TS 28-bp polymorphisms. In addition, our findings provide no evidence for a role for maternal genetic variation in folate metabolism in the etiology of childhood leukemia.
MTR encodes a vitamin B12–dependent enzyme, which catalyzes the remethylation of homocysteine to methionine, the precursor to SAM, the universal methyl group donor.27 The activity of MTR is dependent on vitamin B12 being available, as well as methionine synthase reductase, which maintains the methionine synthase-bound B12 in its fully reduced active state as methyl-cob(III)alamin.39 The MTR reaction also releases tetrahydrofolate, which is remethylated to 5,10-MeTHF for nucleotide synthesis. It has been suggested that the MTR 2756 polymorphism may alter enzyme activity27 and that the G-variant could enhance the flux of one-carbon moieties available for DNA methylation processes.40 This may provide a possible mechanism by which this polymorphism could mediate risk as hypermethylation is important in acute leukemia.41 In addition, in a prospective cohort study investigating pregnancy complications, the presence of the fetal MTR 2756 G-allele was associated with uteroplacental insufficiency,42 suggesting that it plays a part in normal fetal development, which combined with the knowledge that childhood ALL and AML originate in utero provides further support for its role in disease etiology. The only other study to investigate the association between MTR 2756 and childhood leukemia is that by Gast et al43 ; and although no evidence was found to support a role for MTR, a protective effect with polymorphisms in methionine synthase reductase was observed.
MTHFR 677 C>T and 1298 A>C polymorphisms have been the focus of many investigations of genetic variation in the folate metabolic pathway. However, results are conflicting, with some studies reporting protective effects for MTHFR 677 TT9-11,13,18,38 and 1298 CC,9-11 whereas others, including our own, have yielded little or no evidence of effect at least for MTHFR 677.12,15,17,44,45 There are several possible reasons for these inconsistencies, one of which relates to the small case population of most previous studies. Here, however, we present data on almost 1000 childhood ALLs, which is the largest single study to date. In addition, it is probable that the complexity of the folate metabolic pathway may be important as MTHFR is only 1 of more than 30 enzymes involved in the pathway.
An alternative explanation, suggested in relation to colorectal cancer,46 relates to differences in circulating folate levels between populations. In the mid to late 1990s fortification of foods with folic acid became mandatory in several countries, including the United States, but not the United Kingdom, resulting in marked increases in folate intake. At the same time, recommendations were made for folate supplementation during pregnancy. Interestingly, when data from a Canadian study of 270 cases of ALL were stratified by year of birth to take account of these recommendations (before and after 1996), protective effects of the MTHFR 677 T-allele and MTHFR 1298 C-allele were only observed in children born before 1996.11 Analogous findings have been observed for colorectal cancer where associations between polymorphisms in genes involved in the folate pathway and colorectal cancer risk appear to be modified by folate levels.47,48
MTHFR genotypes with lower enzyme activity (677 TT and 1298 CC) favor increased availability of the nonmethylated form of folate (5,10-MeTHF) for DNA synthesis and decreased levels of 5-MeTHF for DNA methylation; that is, decreased MTHFR activity alters the normal intracellular distribution of folate substrates in favor of precursors for nucleotide synthesis. Thus, if adequate levels of folate are available, even if MTHFR activity is low, there is sufficient conversion of 5-MeTHF for DNA methylation while still shunting 5,10-MeTHF toward the synthesis of deoxyuridine monophosphate to deoxythymidine monophosphate and therefore preventing uracil incorporation and chromosomal damage. This suggests that differences in folate availability may influence functional effects of MTHFR polymorphisms, which could possibly account for different findings between studies. In the absence of folate intake data, it is not possible to investigate this further within the UKCCS.
Folate plays an important role in embryogenesis and early fetal development via its effects on DNA methylation and synthesis.49 As such, the well-documented in utero origin of ALL has led to hypotheses that folate intake may be important in disease etiology. However, unlike for Down syndrome and neural tube defects, few studies of leukemia have investigated maternal genotype and folate intake,8,16,19 focusing instead on the role of the child's genotype.9-13,15,17,18,38,44,45 Our findings for mothers are, however, consistent with those reported by the only other studies to have investigated this topic, albeit on smaller populations and fewer polymorphisms.11,16
This is the largest study to date to investigate the association between genetic variation in the folate metabolic pathway and the risk of childhood leukemia. The pathway is, however, complex, and our analyses were restricted to several key enzymes and excluded other possible candidates, including methionine synthase reductase as well as reduced folate carrier, which has previously been linked to risk of childhood ALL.18 In conclusion, although our data do not support a role for MTHFR 677 C>T, they suggest that methionine synthase may be important for both ALL and AML, especially in cases with MLL translocations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The United Kingdom Childhood Cancer Study (UKCCS) is sponsored and administered by the Leukaemia Research Fund (LRF). This study was conducted by 12 teams of investigators (10 clinical and epidemiological, and 2 biological). The work was coordinated by a Management Committee chaired by Sir Richard Doll. It was supported by the UK Children's Cancer Study Group of paediatric oncologists and by the National Radiological Protection Board. We thank the members of the UK Childhood Cancer Study Group for their support and staff of local hospitals, general practitioners, general practice staff, and UKCCS interviewers and technicians. We especially thank the families of the children included in the study, without whose participation this investigation would not have been possible. We thank all consultants, hospital staff, general practitioners, and interviewees who participated in the study. Our thanks also go to all the staff involved in sample collection and processing.
Financial support was provided by Cancer Research UK (previously Imperial Cancer Research Fund and Cancer Research Campaign), the Leukemia Research Fund and the Medical Research Council through grants to their units; by the Leukaemia Research Fund, the Department of Health, the Electricity Association, the Irish Electricity Supply Board (ESB), the National Grid Company plc and Westlakes Research (Trading) Ltd through grants for the general expenses of the study; by the Kay Kendall Leukaemia Fund for the associated laboratory studies. The investigation in Scotland was funded by The Scottish Office, Scottish Power plc, Scottish Hydro-Electric plc, and Scottish Nuclear Ltd.
Data and sample collection for the lymphoma case control study, as well as DNA amplification of the UKCCS samples, were supported by Leukaemia Research. Genotyping of all UKCCS samples was funded by Cancer Research UK and Leukaemia Research. Genotyping for the lymphoma case control was funded through National Institutes of Health grant CA104862 from the US National Cancer Institute and by the National Foundation for Cancer Research.
National Institutes of Health
Authorship
Contribution: T.J.L. designed and secured funding for the investigation, contributed to UKCCS genotyping, and was responsible for conception of the article, interpretation of data, and producing the first draft and revisions; W.T.J. devised and carried out statistical analyses; D.P. carried out laboratory analyses of UKCCS samples; J.S. was responsible for data coordination and management; E.R. was responsible for UKCCS data integrity, was involved in all aspects of UKCCS design and conduct, and oversaw data acquisition and interpretation; C.F.S. designed the original assays and carried out laboratory analysis of control samples; M.T.S. was involved in design and conduct of the analysis of the control samples; J.M.A. contributed to the study design, UKCCS genotyping, and pre- and post-WGA quality control; G.M.T. was sample custodian for the UKCCS, secured funding for the WGA, and contributed to the study design; and all authors reviewed and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tracy J. Lightfoot, Epidemiology & Genetics Unit, Area 3, Seebohm Rowntree Bldg, Department of Health Sciences, University of York, York, YO10 5DD, United Kingdom; e-mail: tracy.lightfoot@egu.york.ac.uk.