Abstract
Gene transfer vectors based on retroviruses are commonly used in gene therapy applications because of their unique ability to integrate efficiently into host genomes. This ability also forms the basis of a transformation event that can be induced in transduced cells by transactivation of proto-oncogenes near the vector integration sites. Here, we report on the development of lymphoma in mice generated from embryonic stem cells transduced with an enhanced green fluorescent protein. The cells expressed B220, CD5, Mac1, and IgM on their surfaces and expressed transcription factors characteristic of B-cell lymphoma. Importantly, each mouse had a single copy of the provirus in its genome; the copy was integrated into the second intron of the dopamine receptor 3 (D3) gene, and high-level expression of D3 was detected only in the lymphoma cells. Ectopic expression of D3 in murine marrow cells resulted in preferential proliferation of cells at the pre–B-cell stage in response to a D3-specific agonist, but this proliferation was not observed in vivo. Cells cotransduced with D3 and Bcl-xL genes had a phenotype similar to that of lymphoma in vivo, suggesting that the leukemogenesis induced by retroviral integration required “second hit” mutations of additional genes.
Introduction
Gene therapy has the potential as an alternative form of therapy for diseases that are not amenable to conventional medical approaches. Various types of gene transfer methods, including the use of viral vectors, have been devised and tested in animal models and in gene therapy clinical trials. These clinical trials have proven that retroviral (including lentiviral) vectors are among the most effective gene transfer vehicles, especially for inherent genetic disorders.1-4 This success is attributed mainly to the unique ability of retroviral vectors to integrate into the host DNA, which allows the stable presence of the transferred gene in the genome of the transduced cell and therefore promises to allow continued expression of the therapeutic genes. On the other hand, unwanted instances of retroviral gene transfer into hematopoietic stem cells, which is known as “genotoxicity of retroviral integration,” have possibly occurred.5,6 The most serious case is leukemogenesis by transactivation of genes neighboring the integration sites of the retroviral vectors in the genomes.3,7,8 Indeed, in France and the United Kingdom, a considerable fraction of patients with X-linked severe combined immunodeficiency (X-SCID) diseases who received autologous CD34+ cells genetically modified by the retroviral vectors to express the common γ-chain complementary DNA (cDNA) have developed T-cell leukemia, although almost all the patients recovered their immunologic function, and their clinical signs of the diseases were ameliorated by the gene therapy.8,9
The accepted explanation for mechanisms of leukemogenesis is that the retroviral vectors were integrated into the sites of host genomes near the proto-oncogenes such as LMO2, CCND2, or Bmi1, and this resulted in aberrant expression of the proto-oncogenes through T-cell differentiation, which caused malignant transformation.7-9 Because some of the transformed cells harbored the multiple copies number of proviruses, however, little is known about the involvement of an integration event in leukemogenesis. In addition to abnormal expression of proto-oncogenes, some patients showed other chromosomal aberrances, which seemed to be the “second hits” in the course of tumorigenesis.8,9
Using an improved retroviral gene transfer system in which the viral promoter/enhancer regions are less susceptible to methylation in immature cells, including hematopoietic stem cells10 and neural stem cells,11 we have shown that stable expression of enhanced green fluorescent protein (EGFP) can be obtained in mice generated from embryonic stem (ES) cells transduced with EGFP cDNA.12 Such chimeric mice had multiple provirus copies in their chromosomes. However, because each provirus was transmitted to their gametes independently, sequential mating with wild-type mice resulted in offspring with a single proviral integration.12 Interestingly, one strain of mice with a single provirus integrated into the second intron of the dopamine receptor 3 (D3) gene developed B-cell lymphoma at approximately 1 year after birth. In the present study, we have attempted to elucidate the mechanism of leukemogenesis caused by retroviral integration in these mice.
Methods
Mice
C57BL/6N (B6) mice and Ly5.1 B6 mice were purchased from Nihon Clea, and nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice were purchased from Sankyo Lab Service. All experiments were approved by the Institutional Review Committee and performed in accordance with the guidelines of the University of Tsukuba.
Retroviral vector construction and preparation
Full-length mouse D3 cDNA was synthesized by polymerase chain reaction (PCR) of the 5′ part (472 bp) and the 3′ part (940 bp) using the total RNA extracted from Ly5.1 B6 brain, followed by ligation of the amplification fragments. Primer sets and PCR conditions used are shown in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). An XhoI fragment containing the full-length D3 cDNA was cloned into GCDsap retroviral vector,12 and an XhoI-ClaI fragment containing the internal ribosomal entry site and humanized Kusabira Orange (huKO)10 was inserted into the vector (GCD/D3/huKO). We also constructed a vector to express the genes encoding Bcl-xL13 and truncated human nerve growth factor receptor (NGFR)14 genes (GCD/Bcl-xL/NGFR). All vectors were converted into the corresponding retroviruses by transduction into the packaging cell line 293gpg, as described.10 The titers of retroviruses were 9.0 × 106 IU/mL on Jurkat cells.
Cell cultures
Lymphomatous B cells developed in transgenic mice were cultured in RPMI 1640 with 10% fetal calf serum (FCS; HyClone), 2mM l-glutamine, 100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate, and 50 μg/mL 2-mercaptoethanol. 293gpg cells and the stromal cell line PA6 were maintained as previously described.15,16
For coculture experiments, 3 × 105 c-KIT+/lineage− (KL) cells isolated from the bone marrow (BM) of Ly5.1 B6 mice were cultured on day 0 in StemPro34 (Invitrogen) in the presence of 50 ng/mL mouse stem cell factor, 100 ng/mL human thrombopoietin, and 10 ng/mL human Flt3-ligand (R&D Systems) in 24-well plates coated with human fibronectin fragment CH296 (RetroNectin; Takara Bio). On days 1 and 2, cells were infected with the recombinant retroviruses by adding 50 μL of the concentrated virus supernatants. At 2 hours after transduction, the supernatants were replaced with fresh media supplemented with cytokines. On day 3, the transduced KL cells (1 × 106) were cocultured in 6-well plates with PA6 cells in RPMI 1640 with 10% FCS in the presence of 20 ng/mL stem cell factor, 10 ng/mL mouse interleukin-7 (IL-7; R&D Systems), and 1nM D3-specific agonist 7-hydroxy-2-dipropylaminotetralin (DPAT; Alexis). Both floating and adherent cells were collected from the culture every 4 days and analyzed individually by cell counting and flow cytometry with a FACSCalibur (BD Biosciences).
For BM transplantation, 3 × 105 transduced KL cells were intravenously inoculated into Ly5.2 B6 mice that had been irradiated with 550 cGy twice with a 4-hour interval. Peripheral blood samples of recipients were analyzed every 4 weeks by cell counting and surface marker analysis.
In transplantation experiments with B lymphoma cells, 5 × 105 B lymphoma cells were inoculated into sublethally irradiated (250 cGy) NOD/SCID mice via the tail vein. The numbers of leukocytes in the peripheral blood of recipient mice were monitored every 4 weeks after transplantation. When the cell counts rose above 12 000 leukocytes/mL, splenic leukocytes obtained from the mice were intravenously inoculated into NOD/SCID mice that had been irradiated with 100 cGy.
All other culture reagents were purchased from Sigma-Aldrich.
Proliferation and apoptosis assay
Transduced KL cells were cultured on PA6 cells for 4 days. Pre-B cells (B220+/Mac1−), myeloid cells (B220−/Mac1+), or hematopoietic progenitor cells (CD43−/c-KIT+) were purified from floating cells, and pro-B cells (CD43+/c-KIT−) were purified from cultured adhered cells by using a FACSVantage (BD Biosciences) cell sorter. A total of 10 000 sorted cells were then cultured in RPMI 1640 with 10% FCS (HyClone) supplemented with 2-mercaptoethanol, 20 ng/mL stem cell factor, and 10 ng/mL mouse IL-7 in the presence of 1nM DPAT (Alexis) in 96-well plates. After 24 hours of incubation, 0.037 Mbq (1 μCi) of [methyl-3H]-thymidine (GE Healthcare) was added to each well, and the mixture was incubated for a further 16 hours. Cultures were harvested with a Macro96 Cell Harvest (Molecular Devices), and the radioactivity in each well was measured with a liquid scintillation counter (LS6500; Beckman Coulter).
To assess the DNA fragmentation induced by apoptosis, genomic DNA isolated from B lymphoma cells and their derivatives transduced with the Bcl-xL was treated with RNase and electrophoresed in a 1.5% agarose gel. For the viability assay, cells were stained with 1 mg/mL propidium iodide (Sigma-Aldrich) and analyzed with a FACSCalibur (BD Biosciences).
Morphologic analysis and karyotyping
Cytospin cell preparations were made on slide glasses and stained with a May-Gruenwald-Giemsa reagent (Merck). Samples were analyzed and images captured with an Axioplan 2 (Carl Zeiss). For karyotyping, B lymphoma cells were incubated in the presence of 0.02 mg/mL Colcemid (Sigma-Aldrich) for 30 minutes. After hypotonic treatment, cells were fixed with a solution of methanol–acetic acid (at a ratio of 3:1) solution. Giemsa-stained chromosome samples were analyzed and imaged under a DM2000 (Leica Microsystems), and the images were imported into the manufacturer's software as a series of .tif files.
Cell-surface analysis
The antibodies used were as follows: fluorescein isothiocyanate–conjugated anti–mouse B220 (RA3-6B2), c-KIT (2B8), CD45.1 (A20), and Mac1 (M1/70); phycoerythrin (PE)–conjugated anti–mouse CD4 (RM4-5), CD5 (53-7.3), CD8 (53-6.7), CD25 (PC61), and Mac1 (M1/70; BD Biosciences); c-KIT (2B8), immunoglobulin M (1B4B1), IL-7Ra (A7R34), and Ter-119 (eBioscience); biotinylated anti–mouse CD5 (53-7.3), CD24 (30-F1), CD43 (S7), and streptavidin-PE (BD Biosciences), CD40 (1C10, HM40-3), CD80 (16-10A1), CD86 (GL1), c-KIT (2B8), and MHC class II (M5/114.15.2; eBioscience); PE-Cy5–conjugated anti–mouse CD3 (145-2C11), B220 (RA3-6B2), Mac1 (M1/70), streptavidin–PE-Cy5, and allophycocyanin-conjugated anti–mouse B220 (BioLegend); and anti–human NGFR (Miltenyi Biotec). Cells were stained with antibodies after lysis of red blood cells and staining with anti–mouse CD16/32 (93; Beckman Coulter) for Fc receptor blocking. This was followed by analysis with a FACSCalibur (BD Biosciences).
PCR
Reverse transcription–PCR (RT-PCR) was used to analyze expression of transcription factors related to B-cell proliferation/differentiation in splenic B cells purified from B6 mice by negative selection using biotinylated anti-mouse CD4, CD8, Gr-1, Mac1, and Ter-119 antibodies and streptavidin-magnetic beads (MyOne Streptavidin C1; Invitrogen) and in B lymphoma cells. For quantitative RT-PCR, 1 μg of the first-strand cDNA prepared from splenic B cells or B lymphoma cells was mixed with mouse D3 or β-actin TaqMan MGB probe, each primer set, and TaqMan Universal PCR Master Mix and assayed in an ABI PRISM 7900HT (Applied Biosystems). For clonality analysis, genomic DNA was isolated from splenic B cells or B lymphoma cells using SepaGene (Sanko Junyaku). After incubation of the genomic DNA with RNase (Sigma-Aldrich), 100 ng of the DNA was used for analysis of V(D)J rearrangement. To assess the integration sites, linear amplification–mediated PCR (LAM-PCR) was performed as previously described,17 with some modifications. PCR products cloned by LAM-PCR were sequenced, and the genomic coordinates of the integration sites were determined by interrogation of the mouse genome database of the National Center for Biotechnology Information (NCBI).18 Primer sets and PCR conditions used in these experiments are shown in supplemental Methods.
Southern and Northern blot analyses
A total of 20 μg of genomic DNA obtained from the tails of mice was digested with EcoRI or BamHI (Takara Bio). The membrane to which the DNA was transferred (Biodyne Nylon Membrane; Pall) was hybridized to deoxycytidine [α-32P] (GE Healthcare)–labeled EGFP and D3 probes. The BamHI cuts once within the vector sequence: therefore, the number offragments hybridized with the EGFP probe corresponds to the number of proviruses integrated into the host genome. Hybridization and the following steps were performed in Perfect Hyb buffer (TOYOBO) in accordance with the manufacturer's instructions. For Northern blotting, 30 μg of total RNA obtained from the brains of B6 mice or B-lymphoma cells was pretreated with 3-[N-morpholino] propanesulfonic acid buffer, formaldehyde, and formamide (Sigma-Aldrich), then electrophoresed in a 1.8% agarose gel. The transferred membrane (Hybond-N+; GE Healthcare) was hybridized to deoxycytidine [α-32P]–labeled D3 probes. Hybridization and the following steps were performed in ULTRAhyb Ultrasensitive Buffer (Applied Biosystems) in accordance with the manufacturer's instructions.
Microarray-based gene expression profiling
An AllPrep Mini kit (QIAGEN) was used to extract total RNA from 2 cell lines that were independently established from mice developing lymphoma (L1 and L2) and from splenocytes from 2 B6 mice as controls (S1 and S2). A detailed protocol of the microarray analysis is given in the supplemental Methods. Briefly, labeled complementary RNA was prepared from the total RNA by using the Agilent labeling protocol (Agilent Technologies). Genes/transcripts were considered to be up- or down-regulated in the lymphoma compared with normal splenocytes if the normalized signal values of the genes/transcripts were more than 100 in each sample, and if the fold-change values were consistently higher than 2.0 or lower than 0.5. Gene Ontology analysis for the differentially expressed genes was conducted by using the DAVID Web site.19,20 All microarray data may be found on the Gene Expression Omnibus (GEO) public database under accession number GSE20661 (National Center for Biotechnology Information [NCBI], http://www.ncbi.nlm.nih.gov, submitted March 5, 2010).
Statistical analysis
Kaplan-Meier estimation with the SAS-type log-rank test was used for survival analysis. All other statistical analyses were performed with the Mann-Whitney U test.
Results
Development of B-cell lymphoma in transgenic mice
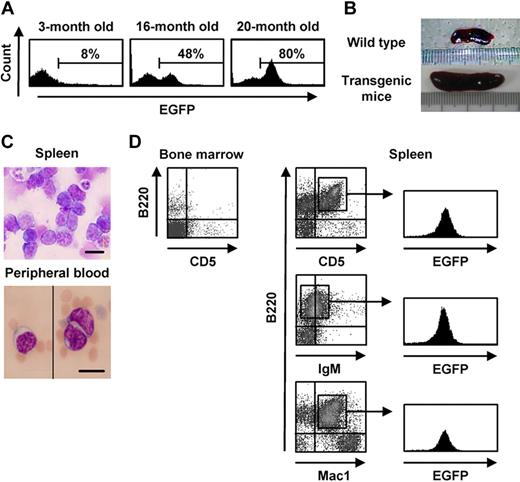
We used the retroviral vector GCDsap to generate transgenic mice from ES cells that were retrovirally transduced with EGFP. These mice demonstrated stable expression of the gene and the ability to transfer this feature to their progeny, although the proportion of EGFP-expressing cells and the mean intensity of expression varied, even in siblings that harbored proviruses integrated into the same chromosome sites, suggesting that gene silencing of the vector occurred in a discontinuous and stochastic manner during cell division.21 Interestingly, approximately half of the F1 mice with EGFP expression that were born from a chimeric mouse (9 of 19 mice) showed some sort of hematologic abnormalities, such as splenomegaly and the appearance of CD5+B220+ cells in their peripheral blood, and they showed a gradual increase of the proportion of EGFP+ cells in their peripheral blood with age. Typically, the percentage of EGFP+ cells rose from 8% at the age of 3 months to up to 80% at 20 months after birth (Figure 1A). Concomitant with the increase in the percentage of EGFP+ cells, mice lost body weight and showed hematopoietic abnormalities, including marked leukocytosis and severe anemia, with ascites and massive splenomegaly (Figure 1B). Morphologic analyses revealed that atypical lymphocytes with coarse and pachychromatic nuclei were the prevalent population in the peripheral blood and spleen (Figure 1C). Few cells with these morphologic features were observed in the BM. When these cells were isolated from the peripheral blood, spleen, and lymph nodes, they expressed B220, CD5, and IgM, as well as EGFP (Figure 1D). Development of the same abnormal cell phenotype was observed in one-half of the progeny generated by mating the F1 mice (6 of 12 mice in the F2) and in a third of the progeny generated by mating the F2 mice (7 of 21 mice in the F3) with wild-type B6 mice, suggesting that some proviruses integrated and transmitted to the progeny were involved in leukemogenesis to B-cell lymphoma.
Hematologic analysis of the transgenic mice affected with B lymphoma. (A) EGFP expression in peripheral blood of a transgenic mouse affected with typical hematologic abnormalities at the indicated ages. (B) Spleens of 20-month-old wild-type and transgenic mice. (C-D) Morphologic appearance of spleen and peripheral blood cells (May-Gruenwald-Giemsa staining 63×/1.4 NA oil objective; C) and flow cytometric analysis of bone marrow and spleen cells (D) of transgenic mice exhibiting remarkable expansion of EGFP+ cells. Bars represent 10 μm in panel C.
Hematologic analysis of the transgenic mice affected with B lymphoma. (A) EGFP expression in peripheral blood of a transgenic mouse affected with typical hematologic abnormalities at the indicated ages. (B) Spleens of 20-month-old wild-type and transgenic mice. (C-D) Morphologic appearance of spleen and peripheral blood cells (May-Gruenwald-Giemsa staining 63×/1.4 NA oil objective; C) and flow cytometric analysis of bone marrow and spleen cells (D) of transgenic mice exhibiting remarkable expansion of EGFP+ cells. Bars represent 10 μm in panel C.
Characteristics of B-cell lymphoma
Because the emerging cells expressed B220 and IgM on their surfaces, we performed further analyses of markers and transcription factors characteristic of B lymphocyte differentiation. Expression of CD24, CD80, CD86, and MHC class II on B-lymphoma cells from EGFP-transgenic mice was similar to that on splenic B cells from wild-type B6 mice, whereas CD43 and CD5 expression was more prevalent in the B-lymphoma cells than in the wild-type splenic B cells (Table 1). Analysis of transcription factors, such as Pax5 and the NF-κB p50 and p65 components, also indicated that the lymphoma was derived from immature B cells (Table 2). Study of the immunoglobulin gene rearrangement pattern showed a monoclonal configuration (Figure 2A). Interestingly, although the cells proliferated vigorously in vitro (Figure 2B), they were prone to apoptosis, as shown by DNA fragmentation and propidium iodide staining. These changes were rescued by overexpression of the antiapoptotic Bcl-xL molecule (Figure 2C-D). To assess the growth activity of the lymphoma cells in vivo, we transplanted them into NOD/SCID mice. When the NOD/SCID mice were intravenously inoculated with 1 × 105 cells, they showed aggressive leukocytosis and severe anemia with weight loss, massive splenomegaly, and ascites by 3 weeks after transplantation. Lymphoma cells obtained from spleen, lymph nodes, and BM of the NOD/SCID mice that underwent transplantation still expressed B220, CD5, and EGFP, as observed before transplantation (Figure 2E). Because the karyotype analysis revealed that most of the cells were diploid (2n = 40), it was not likely that large chromosomal aberrations such as deletion, inversion, and translocation had caused the transformation events (Figure 2F-G). Lymphoma cells were able to establish disease in mice that received transplants after 12 passages in vivo, suggesting that their malignant potential was of a high grade.
Expression of surface antigens on B-lymphoma cells
| . | CD5 . | CD24 . | CD25 . | CD40 . | CD43 . | CD80 . | CD86 . | c-KIT . | IgM . | MHC II . |
|---|---|---|---|---|---|---|---|---|---|---|
| Splenic B cell, % | 21 | 78 | 0.7 | 32 | 13 | 59 | 57 | 4.1 | 92 | 99 |
| B-lymphoma cell, % | 99 | 74 | 1.4 | 19 | 88 | 85 | 95 | 1.2 | 76 | 94 |
| . | CD5 . | CD24 . | CD25 . | CD40 . | CD43 . | CD80 . | CD86 . | c-KIT . | IgM . | MHC II . |
|---|---|---|---|---|---|---|---|---|---|---|
| Splenic B cell, % | 21 | 78 | 0.7 | 32 | 13 | 59 | 57 | 4.1 | 92 | 99 |
| B-lymphoma cell, % | 99 | 74 | 1.4 | 19 | 88 | 85 | 95 | 1.2 | 76 | 94 |
Mean percentages of surface antigens on splenic B cells obtained from wild-type mice or B-lymphoma cells generated from EGFP-transgenic mice are shown.
Expression of transcription factors in B-lymphoma cells
| . | Blimp-1 . | CIIA . | c-Rel . | Oct-2 . | p50 . | p65 . | Pax5 . |
|---|---|---|---|---|---|---|---|
| Splenic B cell | − | + | + | + | + | + | + |
| B-lymphoma cell | − | + | − | − | + | + | + |
| . | Blimp-1 . | CIIA . | c-Rel . | Oct-2 . | p50 . | p65 . | Pax5 . |
|---|---|---|---|---|---|---|---|
| Splenic B cell | − | + | + | + | + | + | + |
| B-lymphoma cell | − | + | − | − | + | + | + |
The results of RT-PCR analysis for expression of transcription factors are shown. RT-PCR was performed using total RNA samples obtained from splenic B cells of wild-type mice or B-lymphoma cells generated from EGFP-transgenic mice according to the protocol in “Methods.” The results are described as to whether PCR products are confirmed or not in electrophoresis (+ or −, respectively).
Characteristics of B-lymphoma cells generated from the transgenic mice. (A) PCR analysis of immunoglobulin gene rearrangement in splenic B cells of wild-type mice and B-lymphoma cells of transgenic mice. Lane 1 shows J558-JH4; lane 2, 7183-JH4; lane 3, Q52-JH4; lane 4, D-JH4; and lane 5, distilled water. (B) Morphologic appearance of B-lymphoma cells in vitro (left panel shows bright field in culture; right panel, May-Gruenwald-Giemsa staining of a cytospun sample; 63×/1.4 NA oil objective). Bars represent 10 μm. (C-D) Electrophoresis of genomic DNA (C) and propidium iodide staining (D) of untransduced or Bcl-xL–transduced B-lymphoma cells. (E) Flow cytometric analysis of marrow, spleen, and lymph node cells in NOD/SCID mice that underwent transplantation with B-lymphoma cells at 4 weeks after transplantation. (F) Karyotype analysis of the B-lymphoma cells by Giemsa staining (100×/1.4 NA oil objective). (G) The number of chromosomes of B-lymphoma cells. MW indicates molecular weight marker in panels A and C.
Characteristics of B-lymphoma cells generated from the transgenic mice. (A) PCR analysis of immunoglobulin gene rearrangement in splenic B cells of wild-type mice and B-lymphoma cells of transgenic mice. Lane 1 shows J558-JH4; lane 2, 7183-JH4; lane 3, Q52-JH4; lane 4, D-JH4; and lane 5, distilled water. (B) Morphologic appearance of B-lymphoma cells in vitro (left panel shows bright field in culture; right panel, May-Gruenwald-Giemsa staining of a cytospun sample; 63×/1.4 NA oil objective). Bars represent 10 μm. (C-D) Electrophoresis of genomic DNA (C) and propidium iodide staining (D) of untransduced or Bcl-xL–transduced B-lymphoma cells. (E) Flow cytometric analysis of marrow, spleen, and lymph node cells in NOD/SCID mice that underwent transplantation with B-lymphoma cells at 4 weeks after transplantation. (F) Karyotype analysis of the B-lymphoma cells by Giemsa staining (100×/1.4 NA oil objective). (G) The number of chromosomes of B-lymphoma cells. MW indicates molecular weight marker in panels A and C.
Identification of the integration site relevant to the leukemogenesis
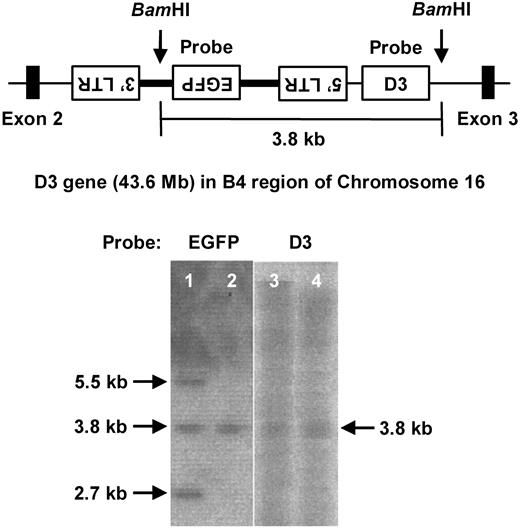
The number of provirus copies integrated into the chromosomes of the original chimeric mice decreased in their progeny, thus resulting in the generation of mice with a single provirus copy.12 To identify an integration site relevant to the leukemogenesis, chimeric mice developing B-cell lymphoma were mated with B6 mice to obtain a single provirus progeny (Figure 3). The progeny mice developed the same hematologic signs, including leukocytosis, anemia, ascites, and splenomegaly, by the age of 1 year and were found to be affected by B-cell lymphoma with the identical phenotypes of surface antigens (EGFP, B220, and CD5) as the malignancy that developed in the original chimeric mice. To identify the integration site of the provirus, high-molecular-weight DNA was obtained and analyzed by LAM-PCR. BLAST analysis of the NCBI mouse DNA database resulted in identification of the provirus integration site between exons 2 and 3 of the D3 gene in the reverse orientation (Figure 3 top panel). This result was also confirmed by Southern blot analysis in which the middle of 3 bands that hybridized with the EGFP probe also hybridized with the D3 probe in chimeric mice (lanes 1 and 3 in Figure 3 bottom panel). A single band hybridized with the EGFP probe in the F2 progeny was also hybridized with the D3 probe (lanes 2 and 4 in Figure 3 bottom panel, respectively).
Identification of retroviral integration sites. (Top) A scheme of an integration site of the provirus into D3 locus determined on the basis of the results of LAM-PCR analysis. BamHI sites and probes used in Southern blotting are also shown. (Bottom) Southern blotting of BamHI-digested genomic DNA obtained from F1 (lanes 1 and 3) and F2 (lanes 2 and 4) of transgenic mice using an EGFP (lanes 1-2) or D3 (lanes 3-4) probe.
Identification of retroviral integration sites. (Top) A scheme of an integration site of the provirus into D3 locus determined on the basis of the results of LAM-PCR analysis. BamHI sites and probes used in Southern blotting are also shown. (Bottom) Southern blotting of BamHI-digested genomic DNA obtained from F1 (lanes 1 and 3) and F2 (lanes 2 and 4) of transgenic mice using an EGFP (lanes 1-2) or D3 (lanes 3-4) probe.
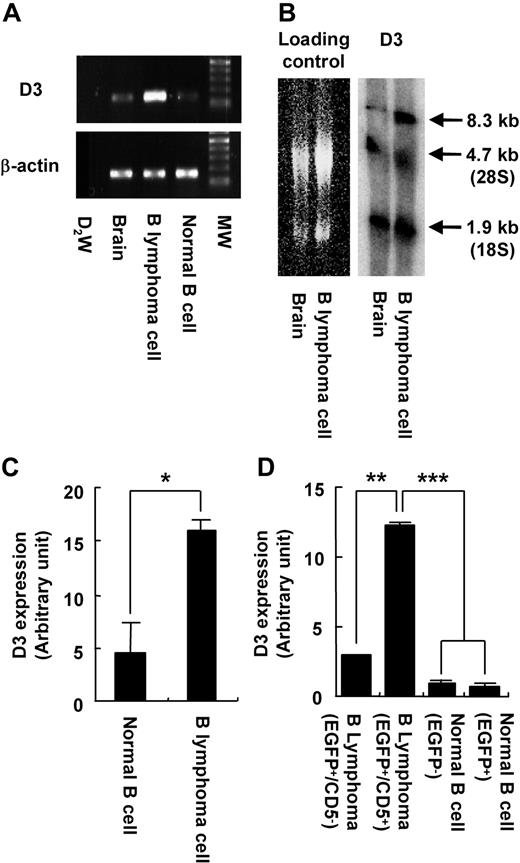
As expected, the B-cell lymphoma cells expressed D3 at a higher level than normal splenic B cells (Figure 4A). The transcript size of the D3 gene in the lymphoma cells was identical to that in the brain (Figure 4B), suggesting that there was no aberrance of the splicing mechanism. Quantitative RT-PCR revealed that the expression level was approximately 4 times that in normal B cells (P < .05; Figure 4C). Importantly, overexpression of the D3 gene was observed only in B-cell lymphoma cells (EGFP+/CD5+; Figure 4D). Although EGFP+/CD5− B cells also had the provirus integrated into the D3 locus, the level of D3 expression in these cells was significantly lower than that in the lymphoma cells and nearly as low as that in the splenic B cells of wild-type mice (Figure 4D), suggesting that integration of the provirus into the D3 locus was necessary, but not sufficient for leukemogenesis.
Activation of the D3 gene in the transgenic mice. (A) RT-PCR analysis of D3 expression in brain and splenic B cells of wild-type mice, and B-lymphoma cells of transgenic mice. β-actin is used as an internal control. MW indicates molecular weight marker. (B) Northern blotting of total RNA obtained from brain or B-lymphoma cells using a D3 probe. Left panel is shown as a loading control. D3 transcripts are found at the size of 8.3 kb in the right panel. 28S and 18S indicate ribosomal RNA. (C) Quantitative RT-PCR analysis of D3 expression in normal splenic B cells and B-lymphoma cells. (D) Quantitative RT-PCR analysis of D3 expression in B-lymphoma cells (CD5− or CD5+ in EGFP+ population) and normal B cells (EGFP− or EGFP+) in transgenic mice. Error bars are ± SD. *P < .05; **P < .01; ***P < .005 in panels C and D.
Activation of the D3 gene in the transgenic mice. (A) RT-PCR analysis of D3 expression in brain and splenic B cells of wild-type mice, and B-lymphoma cells of transgenic mice. β-actin is used as an internal control. MW indicates molecular weight marker. (B) Northern blotting of total RNA obtained from brain or B-lymphoma cells using a D3 probe. Left panel is shown as a loading control. D3 transcripts are found at the size of 8.3 kb in the right panel. 28S and 18S indicate ribosomal RNA. (C) Quantitative RT-PCR analysis of D3 expression in normal splenic B cells and B-lymphoma cells. (D) Quantitative RT-PCR analysis of D3 expression in B-lymphoma cells (CD5− or CD5+ in EGFP+ population) and normal B cells (EGFP− or EGFP+) in transgenic mice. Error bars are ± SD. *P < .05; **P < .01; ***P < .005 in panels C and D.
Involvement of D3 expression in B-cell differentiation
Mice with provirus integrations in the D3 locus selectively developed B-cell lymphoma, suggesting that overexpression of D3 could affect proliferation and/or differentiation of the B-cell lineage. D3, one of the members of the dopamine receptor family, which are G protein–coupled and 7-transmembrane receptors, inhibits neural activity by suppressing both the activity of the potassium ion channel and the production of cyclic adenosine monophosphate via inactivation of adenylate cyclase.22 Although D3 is expressed in the brain (especially in the olfactory bulb), recent studies have reported that human peripheral blood lymphocytes such as naive CD8+ T cells also express D3.23 Simultaneously, a constant amount of dopamine is secreted and accumulated in the lymph nodes and plasma.24-26 However, thus far there have been no reports of D3 expression in B cells and B-cell lymphoma. To assess the effect of D3 expression on the differentiation of hematopoietic progenitor cells (HPCs) into B cells, KL cells were transduced with the D3 cDNA by using the retroviral vector GCDsap and then cultured on the stromal cell line PA6 supplemented with IL-7 in the presence of the D3-specific agonist DPAT. D3-transduced cells proliferated and preferentially differentiated into pre-B cells over time; cells transduced with the huKO gene showed less proliferation and differentiation (Figure 5A). Importantly, D3-transduced pre-B cells in the presence of DPAT proliferated significantly (P < .05) more than huKO-transduced cells (Figure 5B). These results demonstrated that D3 signaling positively regulated not only the differentiation of HPCs into the B-cell lineage, but also the proliferation of pre-B cells.
A role of D3 expression in B-cell development. (A) Coculture experiments of D3-transduced KL cells on PA6 cells in the presence of DPAT. The number of HPCs, pro-B, pre-B, and myeloid cells yielded from huKO- or D3-transduced KL cells (□ or ■, respectively) were shown. Representative data are shown in 2 independent experiments. (B) HPCs, pro-B, and pre-B cells derived from the transduced huKO-transduced (□) or D3-tranduced KL cells (■) cultured on PA6 cells were isolated and their proliferations were determined by the proliferation assay using [3H]-thymidine in the presence or absence of DPAT (Ago + or −, respectively). Data are shown as normalized values to DPAT-free, huKO-transduced cultures in each cell population. Representative data are shown in 2 independent experiments. Error bars are ± SD. *P < .05.
A role of D3 expression in B-cell development. (A) Coculture experiments of D3-transduced KL cells on PA6 cells in the presence of DPAT. The number of HPCs, pro-B, pre-B, and myeloid cells yielded from huKO- or D3-transduced KL cells (□ or ■, respectively) were shown. Representative data are shown in 2 independent experiments. (B) HPCs, pro-B, and pre-B cells derived from the transduced huKO-transduced (□) or D3-tranduced KL cells (■) cultured on PA6 cells were isolated and their proliferations were determined by the proliferation assay using [3H]-thymidine in the presence or absence of DPAT (Ago + or −, respectively). Data are shown as normalized values to DPAT-free, huKO-transduced cultures in each cell population. Representative data are shown in 2 independent experiments. Error bars are ± SD. *P < .05.
Involvement of D3 expression in development of B-cell lymphoma
Having found that D3 expression preferentially induced the differentiation of HPCs into the B-cell lineage, we assessed whether D3-transduced cells transformed into B-cell lymphoma in vivo. Contrary to expectation, mice that received transplants of KL cells transduced with the D3 gene showed neither preferential proliferation of B cells nor development of B-cell lymphoma (data not shown). Because the lymphoma observed in transgenic mice was highly sensitive to apoptosis, which was capable of rescue by the overexpression of the Bcl-xL, KL cells were double-transduced with D3 and Bcl-xL together with huKO and NGFR, respectively, and transplanted into lethally irradiated mice. Expression of both D3 and Bcl-xL was confirmed by RT-PCR using peripheral blood cells obtained from mice 20 weeks after transplantation (Figure 6A). Whereas mice that underwent transplantation with D3-transduced or Bcl-xL–transduced cells showed no abnormal hematopoiesis during a period of 40 weeks after transplantation, 3 mice represented as open symbols in Figure 6B, which had undergone transplantation with cells transduced with both D3 and Bcl-xL together with other 2 mice without any hematologic abnormality (Figure 6B filled symbols), showed gradual expansion of leukocytes coexpressing Mac1 and B220 cells from 24 to 32 weeks after transplantation (Figure 6B left panel). In particular, one mouse represented as open diamonds in the right panel of Figure 6B showed a strong leukocytosis from 36 weeks after transplantation.
Hematologic abnormality in mice that received transplants of KL cells genetically modified to express D3 and Bcl-xL. (A) RT-PCR analysis of D3 and Bcl-xL expression in peripheral blood cells from mice that underwent transplantation with D3-transduced or D3- and Bcl-xL–transduced KL cells. Left and right panels show D3 and Bcl-xL expression, respectively. β-actin is used as an internal control. D3 mice indicates mice that received transplants of D3-transduced KL cells; D3/Bcl-xL mice, mice that received transplants of D3- and Bcl-xL–transduced KL cells; and MW, molecular weight marker. A vertical line has been inserted to indicate repositioned gel lanes of β-actin and Bcl-XL. (B-C) A total of 5 mice received transplants of KL cells transduced with both D3 and Bcl-xL. The percentages of leukocytes expressing Mac1 (B) and the total number of leukocytes (C) in the peripheral blood are shown. Each open or filled symbol represents a mouse with or without hematologic abnormality, respectively.
Hematologic abnormality in mice that received transplants of KL cells genetically modified to express D3 and Bcl-xL. (A) RT-PCR analysis of D3 and Bcl-xL expression in peripheral blood cells from mice that underwent transplantation with D3-transduced or D3- and Bcl-xL–transduced KL cells. Left and right panels show D3 and Bcl-xL expression, respectively. β-actin is used as an internal control. D3 mice indicates mice that received transplants of D3-transduced KL cells; D3/Bcl-xL mice, mice that received transplants of D3- and Bcl-xL–transduced KL cells; and MW, molecular weight marker. A vertical line has been inserted to indicate repositioned gel lanes of β-actin and Bcl-XL. (B-C) A total of 5 mice received transplants of KL cells transduced with both D3 and Bcl-xL. The percentages of leukocytes expressing Mac1 (B) and the total number of leukocytes (C) in the peripheral blood are shown. Each open or filled symbol represents a mouse with or without hematologic abnormality, respectively.
Characterization of cells transduced with D3 and Bcl-xL
In 1 of the 3 mentioned mice that displayed a proliferation of Mac1+/B220+ cells, the population of cells expressing both D3 and Bcl-xL on their surfaces dominated that of cells expressing Bcl-xL alone at 24 weeks after transplantation (Figure 6C). Whereas 17% of cells expressing Bcl-xL alone were positive for B220, almost half of the cells expressing both D3 and Bcl-xL showed high-level expression of B220 (Figure 6C). Furthermore, another of the mice that underwent transplantation with D3- and Bcl-xL–transduced KL cells exhibited body weight loss, low activity levels, hunched posture, splenomegaly, and lymph node swelling (Figure 7A) as well as abnormal hematologic values, including leukocytosis (white blood cells, 9.5 × 104/mL), severe anemia (red blood cells, 5.4 × 104/mL), and thrombocytopenia (platelets, 6.9 × 104/mL) with atypical lymphocytes, at 40 weeks after transplantation (Figures 6B,7B and data not shown). Flow cytometric analysis revealed that cells expressing both B220 and Mac1 proliferated in the peripheral blood, spleen, and lymph nodes, but not in the BM; the proliferated cells expressing both B220 and Mac1 had higher levels of expression of D3 on their surfaces than cells expressing only B220 (Figure 7C). Mice that underwent transplantation with both D3- and Bcl-xL–transduced KL cells (filled squares in Figure 7D) had shorter survival curves than mice that underwent transplantation with cells transduced with huKO, D3, or Bcl-xL (Figure 7D; P < .01, D3 and Bcl-xL compared with the other groups).
Analysis of mice affected with B lymphoma after transplantation with D3- and Bcl-xL–transduced KL cells. (A-B) Morphologic appearance of spleen, lymph nodes (A), and peripheral blood cells (May-Gruenwald-Giemsa staining; B) in mice that received transplants of D3- and Bcl-xL–transduced KL cells at 40 weeks after transplantation (63×/1.4 NA oil objective). Bar in panel B represents 10 mm. (C) Flow cytometric analysis of marrow or peripheral blood cells (left and middle panels, respectively). D3 expression in Mac1− or Mac1+ cells in B220+ population was further analyzed (right panel). (D) Survival analysis of mice that underwent transplantation with huKO-transduced (○), D3-transduced (●), Bcl-xL–transduced (□), or D3- and Bcl-xL–transduced KL cells (■). BMT indicates BM transplantation. *P < .01 compared with the other groups.
Analysis of mice affected with B lymphoma after transplantation with D3- and Bcl-xL–transduced KL cells. (A-B) Morphologic appearance of spleen, lymph nodes (A), and peripheral blood cells (May-Gruenwald-Giemsa staining; B) in mice that received transplants of D3- and Bcl-xL–transduced KL cells at 40 weeks after transplantation (63×/1.4 NA oil objective). Bar in panel B represents 10 mm. (C) Flow cytometric analysis of marrow or peripheral blood cells (left and middle panels, respectively). D3 expression in Mac1− or Mac1+ cells in B220+ population was further analyzed (right panel). (D) Survival analysis of mice that underwent transplantation with huKO-transduced (○), D3-transduced (●), Bcl-xL–transduced (□), or D3- and Bcl-xL–transduced KL cells (■). BMT indicates BM transplantation. *P < .01 compared with the other groups.
Gene expression profiling of the lymphoma
The correlation of coefficients of the expression profiles between each of the 2 lymphoma cell lines and the controls were 0.990 (L1) and 0.964 (L2), indicating that the lymphoma cell lines harbored similar global gene expression patterns, although they were established independently. Among 41 278 probes, 19 282 (L1, 12 532 nonredundant [nr] genes) or 18 320 (L2, 11 827 nr genes) exhibited signal values more than 100 in the lymphoma or controls. The number of genes that were up- or down-regulated in the lymphoma was 3354 (25.9% of the 12 532 nr genes; L1) or 3243 (27.4% of 11 827 nr genes; L2). Gene Ontology analysis of the results of the top 10% of the up- or down-regulated genes (1182 and 1253, respectively) showed that genes related to “cell cycle,” “mitosis,” and “amino acid/amine metabolic processes” were abundant among the up-regulated genes, whereas mainly those related to “immune response” were abundant among the “down-regulated genes” (supplemental Tables 1-2). Transcription of the D3 gene in lymphoma was approximately twice that in normal splenocytes.
Discussion
We demonstrated one of the possible mechanisms by which hematologic malignancies can develop in vivo by characterizing a B-cell lymphoma that developed spontaneously in mice derived from retrovirally transduced ES cells. The mice and their progeny with a single copy of the provirus integrated in the D3 locus showed preferential proliferation of the B-cell lineage and finally died of malignant B-cell lymphoma. We showed that although dopamine signaling through D3 induced HPCs to differentiate preferentially into pre-B cells, this stimulation was insufficient for spontaneous development of lymphoma, which needed the additional expression of the Bcl-xL gene. Taken together, these results suggest that the lymphoma caused by deregulation of D3 expression is an example of cancer developing in accordance with the “2-hit” theory.
One of the key questions in gene therapy is whether retroviral integration per se is sufficient for leukemogenesis. As shown in clinical trials up to the present, retroviruses are integrated near actively transcribed regions and sometimes cause leukemogenesis by the activation of proto-oncogenes near the integration sites. However, recent studies have shown that leukemia cells in SCID-X1 patients carry additional genetic changes, such as a gain-of-function mutation in NOTCH, deletion of the tumor suppressor gene locus cyclin-dependent kinase 2A (CDKN2A), STIL-TAL1 rearrangement, and 6q interstitial losses, suggesting that the leukemia observed in SCID-X1 cases develops in accordance with multistep tumorigenesis theory.7,8
Gilliland27 proposed that, as is often the case with solid tumors, hematologic malignancies are also subject to the “2-hit” theory.28 They suggested that hematologic malignancy results from sequential mutations of class I genes (genes for proliferation and/or survival advantage) and class II genes (differentiation-related genes). If this is the case, then the presence of the provirus at the D3 locus represented the mutation of a class II gene (overexpression of the D3 gene), and other genetic changes that function as class I genes should be required for lymphoma transformation. Although these class I genes remain unidentified, it is highly likely that they are related to cell cycle–associated molecules such as cyclin D1,29 or the cyclin-dependent kinase inhibitors p14ARF and p16INK4a,30 or that they are oncogenes such as c-Myc and Ras family genes,31-33 or antiapoptotic genes such as Bcl-xL.33,34 Indeed, our microarray-based gene expression profiling revealed that several genes were up- or down-regulated in the lymphoma cell lines. In particular, it should be noted that several genes related to “cell cycle,” such as CDKN2A, CDKN2B, and Trp63 (p63), were highly up-regulated in the lymphoma.
Although the results were reminiscent of the mechanism by which leukemia develops in human SCID-X1 cases, why was the substantial amount of time needed for the development of lymphoma in vivo? Baum et al said that malignant transformation is not a necessary consequence of insertional proto-oncogene up-regulation but results from a complex series of multiple factors (eg, the genes conferring the selective advantage on gene-corrected cells, the culture conditions favoring expansion of promalignant clones, and the engraftment conditions generating stress hematopoiesis, associated with an antiapoptotic cytokine milieu that might favor the selection of pretransformed mutants).35 Given that EGFP did not give rise to such selective advantage, gene-modified ES cells were cultured without any strong selective pressure, and the very limited numbers of transduced ES cells (< 10 cells) were used to generate transgenic mice; however, the probability of lymphomagenesis in the present study has been considered to be extremely low. Therefore, the substantial amount time might account for a pause needed for the occurrence of other genetic mutations in pre–B-cell lineage that proliferated slowly by the aberrant expression of D3.
To our knowledge, there have been no reports of B-cell lymphoma related to the aberrant expression of D3. D3 is one of 5 7-transmembrane G protein–coupled receptors, referred to as D1 to D5, all of which influence cell biology by modulating adenylate cyclase; typically, the D1 subfamily, containing D1 and D5, stimulates adenylate cyclase and forms cyclic AMP, whereas the D2 subfamily, containing D2, D3, and D4, inhibits adenylate cyclase and modulates Ca2+ signaling by inhibiting Ca2+ entry through voltage-sensitive Ca2+ channels.22 Numerous studies have elucidated the constant communication between the nervous and immune systems, and the existence of dopamine receptors on lymphocytes has been analyzed by RT-PCR,36 by the binding assay using dopamine ligands,37 and by immunostaining using specific antibodies.38 According to the results, which are still inconclusive, murine and human B cells, unlike T lymphocytes, hardly express D3. Although one paper has described the cytostatic effect of dopamine on cycling B cells such as lymphoma cell lines, the effect was independent of dopamine receptors, and oxidative stress constituted the primary mechanisms.39 Considering the fact that neurotransmitters, including dopamine, elicit various functions in T cells via their receptors (including proliferation, adhesion, and cytokine secretion), B cells, if forced to express excessive D3, would be subjected to the multiple effects of dopamine, some of which would induce the proliferation of pre-B cells.
Our study also contributes information on the issue of the safety of vectors used in gene therapy clinical trials. Recently, vectors carrying weaker promoter/enhancer units than in the wild-type of retroviral LTRs have been suggested as safer gene transfer tools for use in clinical trials.40 In keeping with this, Modlich et al, using a very sophisticated assay, reported that lentiviral vectors were much safer than gammaretroviral vectors in transformation of primary hematopoietic cells.41 They also suggested altering the vector's enhancer-promoter elements (eg, by using the target gene's own promoter element for the new vector design). As shown in our study, however, such vectors also need to be integrated into the host genome to express the therapeutic gene—an action that can be seen as a possible “first hit” toward leukemogenesis. Therefore, we should be aware that even self-inactivating lentiviral vectors,42 which are constructed by deletion of the U3 region in the 3′ LTR, with genetic insulator elements,43 can be liable to cause hematologic malignancies. The need remains to develop other types of vectors for the correction of mutated genes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Richard C. Mulligan (Harvard Medical School) for providing 293gpg, Shin-Ichi Nishikawa (RIKEN, the Center for Developmental Biology) for providing PA6, K. Hata and K. Nakabayashi for analyzing the microarray data, and Dr F. Candotti for providing a critical review of the manuscript. We also thank Ms Naoko Okano and Junko Zenkoh for their excellent secretarial assistance.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Ministry of Health, Labor and Welfare of Japan (M.O.).
Authorship
Contribution: Y.H. and S.H. performed the experiments; and Y.H. and M.O. designed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masafumi Onodera, Department of Genetics, National Research Institute for Child Health and Development, 2-10-1 Okura, Setagaya-ku, Tokyo 157-8575, Japan; e-mail: monodera@nch.go.jp.

![Figure 5. A role of D3 expression in B-cell development. (A) Coculture experiments of D3-transduced KL cells on PA6 cells in the presence of DPAT. The number of HPCs, pro-B, pre-B, and myeloid cells yielded from huKO- or D3-transduced KL cells (□ or ■, respectively) were shown. Representative data are shown in 2 independent experiments. (B) HPCs, pro-B, and pre-B cells derived from the transduced huKO-transduced (□) or D3-tranduced KL cells (■) cultured on PA6 cells were isolated and their proliferations were determined by the proliferation assay using [3H]-thymidine in the presence or absence of DPAT (Ago + or −, respectively). Data are shown as normalized values to DPAT-free, huKO-transduced cultures in each cell population. Representative data are shown in 2 independent experiments. Error bars are ± SD. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/19/10.1182_blood-2009-08-240077/4/m_zh89991052440005.jpeg?Expires=1767709471&Signature=wKPJyA0PoqUKnSnb-Ja-GABovAQT-OacKYrRAaaQcpOWxcQQO9z9emSwquP0jnU38zQ1J~CehNBsBbfy9-oKz-jOc3b~gRjrQ5RaA~joqg22gX0E3HteM4rNLq8XGwtip5AcvqqzKTONVvWKAGLKISsd6wrTZtT0O~8jT5yBkKKvt4nB38aCEHoVCYuD3GgbsTS4IRZbp01T32kSGOiIWe4ab7yQ~41KIE9YpuwJ72j0wbsr4I0cmh5P56OLQQT2RHEsFBfWU9kUgdGqFiy2zVdsjGm4CEL-QLPdy1sp85Fe3C4dQw4FozRMNgla~YHjj1a-KaINbp2ZiUGSIHVlUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

