Abstract
Posttransplantation non-Hodgkin lymphoma is a life-threatening complication after transplantation. Although pharmacologically suppressed adaptive immunity plays a major role in its development, the role of innate immunity in posttransplantation lymphoma is unknown. We assessed the 158 V/F polymorphism in the Fc-γ receptor 3A gene (FCGR3A), killer cell immunoglobulin-like receptor (KIR) genotype, KIR ligand status, and a single nucleotide polymorphism affecting the production of interferon-γ (IFN-γ; +874 A/T) in 236 patients with posttransplantation lymphoma reported to the Collaborative Transplant Study. In addition, polymorphisms in the interleukin-10 (IL-10) and transforming growth factor-β (TGF-β) genes previously associated with lymphoma development were also typed. Using a split-cohort approach, gene/allele frequency was related to the 5-year patient survival after the diagnosis of lymphoma and compared with 100 control solid organ transplant recipients. FCGR3A and KIR genotype significantly influenced survival after diagnosis of posttransplantation lymphoma: the hazard of dying was reduced in homozygous carriers of the high-affinity V allele (hazard ratio 0.49, 95% confidence interval 0.29-0.82, P = .006), whereas carrying a genotype including KIR2DL2/KIR2DS2 increased the risk of dying (hazard ratio 1.49, 95% confidence interval 1.07-2.05, P = .02). KIR ligands and cytokine polymorphisms had no effect on survival. None of the genetic loci analyzed emerged as risk factors for lymphoma development.
Introduction
Posttransplantation non-Hodgkin lymphoma (NHL) represents a life-threatening complication in organ transplant recipients. Compared with a nontransplanted population, the incidence of lymphoma is increased more than 10-fold in the first 10 years after solid organ transplantation, and has remained remarkably stable over the last 2 decades, despite major changes in immunosuppressive regimens used.1 Insufficient Epstein-Barr virus (EBV)–specific adaptive immunity, resulting from iatrogenic immunosuppression, has been firmly established as a key factor in the pathogenesis of posttransplantation lymphoma.2 Consequently, research has largely focused on uncovering mechanisms leading to defective adaptive immune control of EBV.3 In contrast, little is known about how innate immunity impacts on the development of posttransplantation lymphoma.
Antibody-dependent cellular cytotoxicity (ADCC) is an important effector mechanism linking adaptive (humoral) immunity to the innate immune response. Natural killer (NK) cells expressing the high-affinity variant of the Fc-γ receptor 3A (FCGR3A, CD16) have been shown to mediate ADCC more effectively,4 and patients homozygous for this receptor respond better to treatment of certain lymphomas with anti-CD20 monoclonal antibodies (mAbs).5,6 In addition to Fc-receptor–mediated cellular reactivity, NK-cell function is centrally regulated by activating and inhibitory killer cell immunoglobulin-like receptors (KIR). Lack of activating KIR has been linked to an increased risk of viral infection in solid organ transplant recipients.7 No clinical data have so far associated KIR with EBV infection. However, the activating KIR2DS1 has been shown to bind EBV peptide/major histocompatibility antigen complexes in vitro.8 Inhibitory KIR/human histocompatibility leukocyte antigen (HLA) interactions may also play a role in the pathogenesis of posttransplantation lymphoma; HLA-A3, the ligand for the inhibitory KIR3DL2, has been proposed to be associated with lymphoma development.9
Cytokines are important mediators of both innate and adaptive immunity. Polymorphisms in genes encoding interferon-γ (IFN-γ),10,11 transforming growth factor-β (TGF-β),12,13 and interleukin-10 (IL-10)12 have been suggested to play a role in the development of posttransplantation lymphoma.
Establishing associations between genes encoding for functionally defined molecules and posttransplantation lymphoma development and survival may point toward pathogenetic mechanisms and thereby support the development of novel therapeutic strategies. In a candidate gene approach, we assessed FCGR3A gene polymorphisms, established the KIR genotype and KIR ligand status, and investigated cytokine polymorphisms in a large cohort of lymphoma patients and a control cohort of transplant recipients without lymphoma.
Methods
Patient selection
Patients were included in this study if they had developed posttransplantation NHL after kidney, heart, or liver transplantation and if patient DNA was available for genotyping. DNA samples and clinical data were provided by centers participating in the Collaborative Transplant Study (CTS).14 Transplantations were performed between 1988 and 2004. Data on lymphoma occurrence in each patient were collected at 3, 6, and 12 months during the first year after transplantation and yearly thereafter. To minimize underreporting of tumor data, each center was asked to provide yearly written confirmation of the accuracy and completeness of reported data. Only patients with documented NHL were included in the analysis; benign proliferation and hyperplasia were excluded. Lymphomas occurring up to the time of graft loss were analyzed.
Allelic frequency data for the polymorphisms analyzed are comparable within white ethnic populations, which constitute the majority of patients analyzed, but differ greatly between racial groups.15-17 As controls, we therefore used an ethnically matched cohort of transplant recipients without diagnosis of posttransplantation lymphoma or posttransplantation lymphoproliferative disease, all treated at a single center that contributes to the CTS (Basel University Hospital). All data and sample collection and analysis were approved by the Basel University institutional review board.
Genotyping
Polymorphisms of the FCGR3A (158 V/F) and of the cytokines IFN-γ (+874 A/T), IL-10 (−1082 A/G), and TGF-β (codon 25 C/G) were assessed by sequence-specific primer polymerase chain reaction (SSP PCR) using published primers.18 PCR products were visualized by electrophoresis on agarose gel.
KIR genotypes were determined in the first 100 lymphoma patients and the control cohort by multiplex PCR, followed by a reverse sequence-specific oligonucleotide method according to the manufacturer's instructions (One Lambda). KIR genes analyzed included 2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1. The framework KIR genes 2DL4, 3DL2, and 3DL3 and the pseudogenes 2DP1 and 3DP1 were not analyzed. The value of P for associations with KIR genes in the discovery cohort were adjusted for multiple comparisons. In the validation cohort (n = 136), presence of KIR2DL2, KIR2DL3, and KIR2DS2 was assessed by SSP PCR according to a published protocol.19 HLA-A and HLA-B antigens were typed at transplant centers contributing to the CTS. The polymorphism at position 77 and 80 in HLA-C antigens, which determines binding to KIR2D, was typed retrospectively by SSP PCR for all patients using a previously published method.20
Statistical analysis
To assess the impact on lymphoma incidence of genes and alleles analyzed in this study, hazard ratios (HR) were calculated by Cox modeling that adjusted for the type of immunosuppression. All genotypes were entered into Cox models as categorized covariates (eg, FCGR3A VV vs VF vs FF), with homozygous carriers of the more prevalent allele (eg, FCGR3A FF patients) defined as having an HR of 1.00.
We used a split-sample cohort approach to internally validate the data to minimize the risk of false-positive findings.21 Polymorphisms and KIR genotypes were analyzed in a discovery cohort of 100 lymphoma patients and compared with frequencies found in 100 control patients without posttransplantation lymphoma. In addition, all polymorphisms and genes were analyzed regarding their association with survival after diagnosis. Genes and polymorphisms that were distributed significantly differently in the discovery and the control cohort were tested in an independent second cohort of 136 patients with lymphoma. For survival analysis, Kaplan-Meier estimates of patient survival from the day of lymphoma diagnosis were calculated. In multivariable survival analysis, HR were calculated by Cox modeling that included as covariates disease localization/extension, type of immunosuppression, transplant function as a time-dependent covariate, and the year of lymphoma diagnosis to account for advances in lymphoma diagnosis and treatment.
Results
Study population
Patient and transplant recipient characteristics are summarized in Table 1. In the discovery cohort (n = 100) only recipients of kidney grafts were contained. All liver (n = 12) or heart (n = 24) transplant recipients were included in the validation cohort (overall n = 136). The lymphoma-negative control cohort contained a higher proportion of living donor transplant recipients, was transplanted later (median year 2005 vs 1993 for patients with lymphoma), and accordingly differed in immunosuppressive regimens (based mainly on tacrolimus/mycophenolate mofetil/anti-IL-2 mAbs, whereas in lymphoma patients immunosuppressive regimens based on cyclosporine A/azathioprine/steroids prevailed). The ethnic background was comparable in all 3 cohorts.
Patient characteristics
| . | Discovery cohort . | Validation cohort . | Control cohort . |
|---|---|---|---|
| Number | 100 | 136 | 100 |
| Patient sex, male/female | 60/40 | 37/99 | 62/38 |
| Ethnic background, white/other/na | 92/6/2 | 128/5/3 | 97/3/0 |
| Transplant type, living/deceased donor | 8/92 | 13/123 | 36/64 |
| Median year of transplantation (range) | 1993 (1988-2004) | 1993 (1988-2004) | 2005 (2004-2007) |
| Median interval transplant to diagnosis of lymphoma, mo (range) | 63 (1-197) | 59(1-201) | |
| Lymphoma localization | |||
| Kidney | 9 (9%) | 9 (6.6%) | |
| Gastrointestinal | 12 (12%) | 16 (12%) | |
| Central nervous system | 4 (4%) | 7 (5%) | |
| Other single site | 53 (53%) | 69 (51%) | |
| Disseminated | 14 (14%) | 15 (11%) | |
| Unknown | 8 (8%) | 20 (15%) | |
| Median age at lymphoma diagnosis, y (range) | 43 (4-72) | 46 (4-73) | |
| Transplanted organ | |||
| Kidney | 100 (100%) | 100 (74%) | 100 (100%) |
| Heart | 24 (24%) | ||
| Liver | 12 (12%) | ||
| Immunosuppression | |||
| Cyclosporine A | 77 (77%) | 114 (84%) | 4 (4%) |
| Tacrolimus | 11 (11%) | 12 (9%) | 94 (94%) |
| Azathioprin | 62 (62%) | 107 (79%) | 13 (13%) |
| Mycophenolate | 14 (14%) | 10 (7%) | 86 (86%) |
| Steroids | 90 (90%) | 126 (92%) | 62 (62%) |
| Rapamycin | 4 (4%) | 2 (2%) | 43 (43%) |
| Antithymocyte globulin | 30 (30%) | 31 (22%) | 11 (11%) |
| Anti-IL2 Ab | 3 (3%) | 9 (7%) | 66 (66%) |
| No data | 4 (4%) | 1 (1%) | 0 (0%) |
| CMV serology | |||
| D−/R− | 27 (27%) | 11 (8%) | 22 (22%) |
| D−/R+ | 18 (18%) | 29 (21%) | 14 (14%) |
| D+/R− | 11 (11%) | 31 (23%) | 24 (24%) |
| D+/R+ | 27 (27%) | 47 (35%) | 39 (39%) |
| na | 17 (17%) | 18 (13%) | 1 (1%) |
| . | Discovery cohort . | Validation cohort . | Control cohort . |
|---|---|---|---|
| Number | 100 | 136 | 100 |
| Patient sex, male/female | 60/40 | 37/99 | 62/38 |
| Ethnic background, white/other/na | 92/6/2 | 128/5/3 | 97/3/0 |
| Transplant type, living/deceased donor | 8/92 | 13/123 | 36/64 |
| Median year of transplantation (range) | 1993 (1988-2004) | 1993 (1988-2004) | 2005 (2004-2007) |
| Median interval transplant to diagnosis of lymphoma, mo (range) | 63 (1-197) | 59(1-201) | |
| Lymphoma localization | |||
| Kidney | 9 (9%) | 9 (6.6%) | |
| Gastrointestinal | 12 (12%) | 16 (12%) | |
| Central nervous system | 4 (4%) | 7 (5%) | |
| Other single site | 53 (53%) | 69 (51%) | |
| Disseminated | 14 (14%) | 15 (11%) | |
| Unknown | 8 (8%) | 20 (15%) | |
| Median age at lymphoma diagnosis, y (range) | 43 (4-72) | 46 (4-73) | |
| Transplanted organ | |||
| Kidney | 100 (100%) | 100 (74%) | 100 (100%) |
| Heart | 24 (24%) | ||
| Liver | 12 (12%) | ||
| Immunosuppression | |||
| Cyclosporine A | 77 (77%) | 114 (84%) | 4 (4%) |
| Tacrolimus | 11 (11%) | 12 (9%) | 94 (94%) |
| Azathioprin | 62 (62%) | 107 (79%) | 13 (13%) |
| Mycophenolate | 14 (14%) | 10 (7%) | 86 (86%) |
| Steroids | 90 (90%) | 126 (92%) | 62 (62%) |
| Rapamycin | 4 (4%) | 2 (2%) | 43 (43%) |
| Antithymocyte globulin | 30 (30%) | 31 (22%) | 11 (11%) |
| Anti-IL2 Ab | 3 (3%) | 9 (7%) | 66 (66%) |
| No data | 4 (4%) | 1 (1%) | 0 (0%) |
| CMV serology | |||
| D−/R− | 27 (27%) | 11 (8%) | 22 (22%) |
| D−/R+ | 18 (18%) | 29 (21%) | 14 (14%) |
| D+/R− | 11 (11%) | 31 (23%) | 24 (24%) |
| D+/R+ | 27 (27%) | 47 (35%) | 39 (39%) |
| na | 17 (17%) | 18 (13%) | 1 (1%) |
D indicates donor; R, recipient; and na, not available.
FCGR3A
Compared with patient groups homozygous for the F allele (FF, HR 1.00), carrying an FV and VV genotype was not associated with a significantly altered risk of developing posttransplantation lymphoma (HR in discovery cohort 0.77 [0.48-1.22, P = .26] and 1.02 [0.56-1.84, P = .95]; HR in validation cohort 0.87 [0.54-1.40, P = .55] and 0.80 [0.53-1.18, P = .26]).
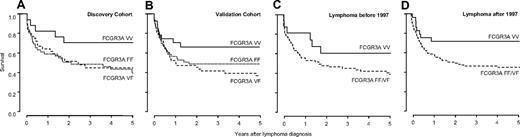
In contrast, survival analysis showed increased patient survival after diagnosis in carriers homozygous for FCGR3A, coding for the high-affinity receptor for IgG1 and IgG3 (V allele). The survival advantage was evident in both discovery and validation cohorts (HR 0.59, 95% confidence interval [CI] 0.25-1.39; Figure 1A and HR 0.38, 95% CI 0.18-0.78; Figure 1B), respectively. This resulted in 5-year survival rates after the diagnosis of lymphoma of 66% (± 8%) in homozygous carriers of the V allele, 44% (± 5%) for homozygous carriers of the low-affinity F allele, and 35% (± 5%) for heterozygous patients (HR 0.49 for VV vs FF/VF, 95% CI 0.29-0.82, P = .006).
Survival of transplant recipients after lymphoma diagnosis is influenced by FCGR3A genotype. Homozygous carriers of the high-affinity V allele show improved survival in discovery (VV 69% ± 11%; VF 36% ± 8%; FF 42% ± 8%; A) and validation (VV 64% ± 10%; VF 33% ± 7%; FF 46% ± 7%; B) cohort. Improved survival of VV carrying patients is not exclusively related to increased efficacy of rituximab, as a similar effect on survival is evident before (VV 58% ± 13%; VF/FF 35% ± 6%; C) and after (VV 70% ± 9%; VF/FF 42% ± 5%; D) the introduction of rituximab in 1997.
Survival of transplant recipients after lymphoma diagnosis is influenced by FCGR3A genotype. Homozygous carriers of the high-affinity V allele show improved survival in discovery (VV 69% ± 11%; VF 36% ± 8%; FF 42% ± 8%; A) and validation (VV 64% ± 10%; VF 33% ± 7%; FF 46% ± 7%; B) cohort. Improved survival of VV carrying patients is not exclusively related to increased efficacy of rituximab, as a similar effect on survival is evident before (VV 58% ± 13%; VF/FF 35% ± 6%; C) and after (VV 70% ± 9%; VF/FF 42% ± 5%; D) the introduction of rituximab in 1997.
No data on pharmacologic treatment of posttransplantation lymphoma patients are reported to the CTS. Hence a possible role of rituximab in the survival advantage of VV carriers could not be directly analyzed. However, rituximab was approved by the Food and Drug Administration in 1997, and according to information from the manufacturer of rituximab, no patient with posttransplantation lymphoma received rituximab before 1997 (A. J. Grillo-Lopez, retired Chief Medical Officer, IDEC Pharmaceuticals, personal e-mail communication, January 17, 2010). Indeed, the first report in the literature on treatment of posttransplantation lymphoma with rituximab dates back to 1999.22 After stratification of patients according to date of diagnosis, the survival advantage was slightly greater in patients diagnosed before 1997 (HR 0.44, 95% CI 0.16-1.23; Figure 1C) than in patients diagnosed after 1997 (HR 0.49, 95% CI 0.26-0.92; Figure 1D), suggesting that improved survival of VV carriers was not (exclusively) based on increased efficacy of rituximab treatment in VV allele carriers.
KIR genes
The 2 KIR genes significantly altered the risk of developing posttransplantation lymphoma in the exploratory cohort: carrying KIR2DL2 and KIR2DS2 (concordant in all patients) was protective for lymphoma development (HR 0.54, 95% CI 0.36-0.83, P = .005). For all other KIR genes, no significant effects were detected. The effect of KIR2DLA/KIR2DS could, however, not be replicated in the validation cohort (HR 1.26, 95% CI 0.84-1.89, P = .26).
HLA-A3, which in conjunction with EBV-derived peptides is one of the ligands of inhibitory KIR3DL2, has recently been suggested to be present at an increased frequency in patients developing posttransplantation lymphoma.9 In our population, carrying HLA-A3 did not confer an increased risk for posttransplantation lymphoma development (HR in discovery cohort 0.92 [0.58-1.46, P = .72]; and HR in validation cohort 1.15 [0.78-1.68, P = .49]).
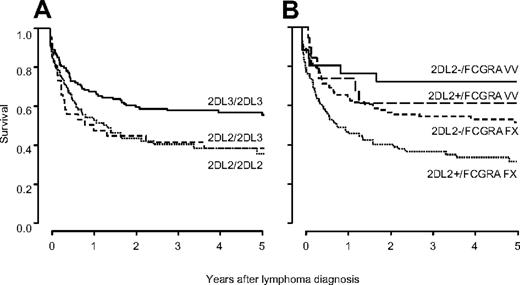
Survival analysis of patients stratified by KIR genotype revealed a significantly inferior survival for patients carrying KIR2DL2/KIR2DS2 (HR 1.49, 95% CI 1.07-2.05, P = .02). As KIR2DL2 and KIR2DL3 segregate as alleles, the validation cohort was typed for KIR2DL3. This analysis indicated inferior survival for homozygous carriers of KIR2DL2 (HR vs KIR2DL3 homozygotes 1.60, 95% CI 0.99-2.60) and for heterozygous KIR2DL2/KIR2DL3 patients (HR 1.44, 95% CI 0.99-2.09; Figure 2A). Stratifying patients by HLA-C KIR ligands, the detrimental association with survival of KIR2DL2/KIR2DS2 was smaller in C1C1 patients (n = 92, HR 1.07, 95% CI 0.62-1.88), than in C1C2 (n = 115, HR 1.70, 95% CI 1.01-2.85) and C2C2 patients (n = 29, HR 1.89, 95% CI 0.53-6.82). However, in a Cox model the interaction between KIR and HLA-C was not statistically significant (P = .09), arguing against a strong impact of HLA-C on the effect of KIR on survival.
Survival of transplant recipients after lymphoma diagnosis is influenced by KIR genotype. Survival is best in KIR2DL3 homozygous patients, who do not carry KIR2DL2 (2DL3/2DL3 53% ± 5%; 2DL2/2DL3 35% ± 8%; 2DL2/2DL2 32% ± 6%; A). KIR2DL2 and FCGR3A are independent and additive risk factors for death after lymphoma diagnosis. Stratifying patients by both KIR2DL2 and FCGR3A yields 4 prognostic groups, in which patients carrying KIR2DL2 and the FCGR3A alleles FF/VF show lowest survival (28% ± 5%), patients not carrying KIR2DL2 with FCGR3A VV genotype show best survival (71% ± 9%), and the remaining patients show intermediate survival (59% ± 12% for KIR2DL2+/FCGR3A VV patients and 49% ± 5% for KIR2DL2−/FCGR3A FF/VF patients, respectively; B).
Survival of transplant recipients after lymphoma diagnosis is influenced by KIR genotype. Survival is best in KIR2DL3 homozygous patients, who do not carry KIR2DL2 (2DL3/2DL3 53% ± 5%; 2DL2/2DL3 35% ± 8%; 2DL2/2DL2 32% ± 6%; A). KIR2DL2 and FCGR3A are independent and additive risk factors for death after lymphoma diagnosis. Stratifying patients by both KIR2DL2 and FCGR3A yields 4 prognostic groups, in which patients carrying KIR2DL2 and the FCGR3A alleles FF/VF show lowest survival (28% ± 5%), patients not carrying KIR2DL2 with FCGR3A VV genotype show best survival (71% ± 9%), and the remaining patients show intermediate survival (59% ± 12% for KIR2DL2+/FCGR3A VV patients and 49% ± 5% for KIR2DL2−/FCGR3A FF/VF patients, respectively; B).
When KIR genotype and FCGR3A polymorphism were combined into a multivariable survival model, the 2 factors independently predicted survival (HR 1.47, 95% CI 1.05-2.07, P = .02 for KIR; and HR 0.50, 95% CI 0.30-0.82, P = .007 for FCGR3A). In accordance with an additive effect, stratifying patients by the 2 factors yielded 4 prognostic groups in which the risk of dying increased cumulatively (Figure 2B).
IFN-γ +874A/T
In contrast to published data,10,11 no significantly increased risk for posttransplantation lymphoma development was apparent in heterozygous or homozygous carriers of the low IFN-producing A allele (HR 1.04, 95% CI 0.59-1.81-2.07, P = .90 for heterozygous AT carriers; and HR 0.81, 95% CI 0.44-1.47, P = .48 for homozygous AA carriers). No significant association of the IFN-γ polymorphism on survival after lymphoma diagnosis was observed (5-year survival: 44%, 44%, and 49% for AA, AT, and TT allele carriers, respectively; P = .83).
IL-10 −1082A/G
Carrying the IL-10 high-producing G allele has previously been associated with increased risk for posttransplantation lymphoma.12 We could not replicate this finding in our population (HR vs AA patients 1.07 [0.78-1.46, P = .68] for AG patients; and HR 0.84 [0.59-1.20, P = .33] for GG patients). The 5-year survival after diagnosis was also not significantly influenced by the IL-10 polymorphism (survival rates: 43%, 38%, and 52% for carriers of AA, AG, and GG alleles, respectively; P = .90).
TGF-β-1 codon 25 C/G
For carriers of the C allele in the codon 25 single nucleotide polymorphism of TGF-β that has previously been associated with an increased risk of posttransplantation lymphoma,12,13 we found only a nonsignificant risk increase (HR vs GG patients 1.10 [0.59-2.03, P = .77] for GC patients; and HR 1.86 [0.80-4.33, P = .15] for CC patients). Although homozygocity for CC at this locus came closest to being a significant risk factor for lymphoma development, the frequency of patients with this genotype was low both in groups with and those without lymphoma (3% and 6%, respectively). The polymorphism at TGF-β codon 25 did not influence survival after diagnosis of posttransplantation lymphoma (5-year survival rates: 45%, 42%, and 33% for GG, GC, and CC genotypes, respectively; P = .42).
Discussion
Immunogenetic association studies have provided substantial insight into the function of the immune system, both in the setting of infection and malignancy.23,24 Importantly however, an impressive number of irreproducible associations have been published, highlighting the need for sufficiently powered and well-designed studies. Here we present data derived from a cohort of posttransplantation lymphoma patients that is greater by one order of magnitude than previously published series. As distribution of the polymorphisms analyzed varies between racial groups and may theoretically also vary between healthy donor and transplant recipients, we used a cohort of solid organ recipients with a similar ethnic background as controls.
Using this scheme we failed to detect a genetic association between receptors expressed by NK cells and the development of posttransplantation lymphoma. Furthermore, previous studies suggesting an impact of allelic polymorphisms in cytokine genes and HLA on the development of posttransplantation lymphoma could not be confirmed in our study.10-13 We made, however, the intriguing observation that polymorphisms in receptors expressed by NK cells were significantly associated with survival after the diagnosis of lymphoma. Homozygous carriers of the high-affinity 158V allele of the FCGR3A (VV patients) showed a significantly improved 5-year survival rate compared with VF and FF patients (Figure 2A-B). One might speculate that increased efficiency of ADCC triggered via endogenous antibodies may be capable of mediating clinically relevant antitumor activity. The hypothesis that endogenous rather than therapeutic antibodies may trigger such activity is supported by the fact that the association was present, even somewhat more pronounced, in posttransplantation lymphoma patients diagnosed before rituximab became available. The 158V allelic variant of FCGR3A has previously been shown to bind immunoglobulin-G antibodies in vitro with higher affinity than the 158F variant.25 This translates into improved response rates after therapeutic administration of mAbs in homozygous 158V patients.5,6 Increased rates of antibody-mediated autoimmune diseases in 158V carriers suggest that the polymorphism also plays a relevant role in the binding of endogenous antibodies.26 An association of the 158 V/F polymorphism with herpes family virus infections and oncogenesis has recently been documented, as the polymorphism influences the susceptibility to human herpesvirus-8 infection and HIV-associated Kaposi sarcoma.27 No previous studies have investigated a possible influence of FCGR3A alleles on the immune response to EBV infection, nor on the survival after NHL in the absence of mAb treatment. Strikingly, despite a strong association of FCGR3A genotype with survival after lymphoma diagnosis, no association could be detected between this genetic locus and either EBV reactivation in control patients (data not shown) or incidence of lymphoma.
The KIR genotype was also significantly associated with survival after diagnosis, as patients carrying KIR2DL2/KIR2DS2 showed increased mortality after diagnosis of posttransplantation lymphoma. This effect was not significantly influenced by the patients' HLA phenotype. Previous studies have shown that carrying KIR2DL2 (as opposed to KIR2DL3) compromises the capacity to eliminate hepatitis C virus,24 and that KIR2DL2/KIR2DS2 negatively affects control of herpes virus type 1 infection.28 A convincing pathogenetic model for the association of KIR with viral infection is still lacking. It remains open whether KIR2DL2 or KIR2DS2 are involved in the recognition of EBV, the virus associated with the development of posttransplantation lymphoma. No association was detected between KIR2DL/S2 carrier status and frequency of EBV reactivation in the control cohort (data not shown).
Although the large number of cases analyzed in this study is its major strength, the approach chosen also has limitations. Lack of detailed clinical and histologic information limits in-depth interpretation of the survival analysis, as does the lack of information on possible confounding variables such as comorbidities and infectious outcomes. Analysis of cause of death in cancer patients is notoriously difficult, as patients may die indirectly from cancer, eg from therapy-related side effects. We therefore did not analyze mortality directly attributable to cancer, even though this information was available in the database. “Cancer” was given as direct cause of death in 75% of patients. This result and the plateau reached in survival at 2 years after lymphoma diagnosis suggest that the large majority of deaths are directly attributable to lymphoma.
In conclusion, in the largest cohort of posttransplantation NHL patients analyzed to date, a polymorphism in FCGR3A as well as the KIR genotype were found to be associated with survival after the diagnosis of lymphoma, but not linked to lymphoma development itself. KIR ligands and allelic polymorphisms in the genes encoding for IFN-γ, IL-10, and TGF-β were not distributed differently in posttransplantation lymphoma patients and control patients, nor were they associated with survival. Our study has the limitation of being retrospective, but efforts were made to exclude the possibility of false-positive findings. If confirmed in an independent cohort, the findings presented provide insight into the control of posttransplantation lymphoma and may have therapeutic implications.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to the laboratories and transplant centers that provided the data to the Collaborative Transplant Study on which this analysis was based. We thank Dr A. J. Grillo-Lopez for providing detailed information on clinical studies carried out with rituximab before 1997.
This work was supported by Oncosuisse (OCS-02266-08-2008) (M.S.). C.H. is supported by the Swiss National Science Foundation (PP00B-114850).
Authorship
Contribution: All authors analyzed data and contributed to writing of the manuscript; M.S. performed genotyping; G.O. and C.H. designed the study; and G.O. and B.D. provided samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Stern, Division of Hematology, University Hospital Basel, Petersgraben 4, 4031 Basel, Switzerland; e-mail: sternm@uhbs.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal