Abstract
Recent studies have reported that regions of homozygosity (ROH) in the genome are detectable in outbred populations and can be associated with an increased risk of malignancy. To examine whether homozygosity is associated with an increased risk of developing childhood B-cell precursor acute lymphoblastic leukemia (BCP-ALL), we analyzed 824 ALL cases and 2398 controls genotyped for 292 200 tagging SNPs. Across the genome, cumulative distribution of ROH was not significantly different between cases and controls. Four common ROH at 10p11.2-10q11.21, 1p31.1, 19p13.2-3, and 20q11.1-23 were, however, associated with ALL risk at P less than .01 (including 1 ROH to which the erythropoietin receptor [EPOR] gene maps, P = .005) but were nonsignificant after adjusting for multiple testing. Our findings make it unlikely that levels of measured homozygosity, caused by autozygosity, uniparental isodisomy, or hemizygosity, play a major role in defining BCP-ALL risk in predominantly outbred populations.
Introduction
Although acute lymphoblastic leukemia (ALL) is the commonest childhood malignancy, accounting for approximately 80% of leukemia in the pediatric age group, its etiology is largely unknown.1 B-cell precursor (BCP)–ALL is the major form of the disease, accounting for approximately 85% of all pediatric ALL
Two recent genome-wide association (GWA) studies of ALL identified several common single nucleotide polymorphisms (SNPs) at 7p12.2 (IKZF1), 10q21.2 (ARID5B), and 14q11.2 (CEBPE) that influence the risk of BCP-ALL.2,3 The variants so far identified by these GWA studies are common in the general population (minor allele frequency, > 5%), but have, individually, small effects on disease risk,2,3 with odds ratios typically less than 1.6. Despite the relatively small predisposing effects conferred, the variants identified provide important and novel insights into the disease biology. Specifically, these risk variants map to genes involved in transcriptional regulation and differentiation of B-cell progenitors, suggesting dysfunctional B-cell pathway gene expression as an etiologic basis for BCP-ALL development.
The majority of cancer predisposition genes that have been identified to date through GWA studies act in a codominant fashion, and studies have found no good evidence for recessively acting disease loci. Although this may be reflective of the biology, it may also be a consequence of GWA studies having suboptimal ability to detect recessively acting disease alleles. Clues that tumor susceptibility may have a recessive basis come from reports of an increased incidence associated with consanguinity and in populations characterized by a high degree of inbreeding.4-9 Further evidence for the role of homozygosity in cancer predisposition is provided by experimental animal inbreeding (eg, backcrossing mice) increasing tumor incidence.10 Specific situations of homozygosity have also been directly associated with cancer, such as uniparental disomy through altered imprinting.11
Common regions of homozygosity (ROH), the result of autozygosity (ie, the occurrence of 2 alleles at the same locus originating from a common ancestor by way of nonrandom mating), have recently been shown to occur at a high frequency in outbred populations as a result of selection.12 Searching for ROH on a genome-wide basis therefore provides a means of exposing recessively acting disease genes. Recently, Assié et al studied patients with breast, prostate, or head and neck cancer of Northern/Western European ancestry by whole-genome loss of heterozygosity analysis using microsatellite markers.13 A significant increase in the frequency of homozygosity in combined cases compared with controls was reported. In a separate study of colorectal cancer using Affymetrix SNP arrays, Bacolod et al showed that cases harbored significantly more homozygous regions than healthy persons.14 Findings from these studies support the hypothesis that there exist multiple, recessive, cancer-predisposing loci, which are not readily detected using a conventional GWA approach based on analysis of individual SNPs. A possible explanation for this is that relative risks per locus are too low and/or that the disease-associated variants are not in strong linkage disequilibrium (LD) with tagSNPs, perhaps because of low allele frequencies.
Although GWA studies have limited ability to identify recessive disease alleles through single SNP analyses, these datasets can potentially be exploited to search for recessively acting disease loci through whole genome homozygosity analysis. Hence to examine whether homozygosity is associated with an increased risk of developing childhood BCP-ALL and to search for novel recessively acting disease loci, we conducted a whole genome homozygosity analysis of 824 BCP-ALL cases and 2398 controls genotyped for 292 200 tagging SNPs.2
Methods
Patients and DNA samples
Cases analyzed had been diagnosed with BCP-ALL and constitute 90% of the patients analyzed in the GWA study of childhood ALL we have recently reported.2 Full details of the study are provided in previously published material.2 Briefly, we analyzed the constitutional DNA of 824 pediatric patients with BCP-ALL ascertained from the United Kingdom (464 male, 360 female; mean age at diagnosis, 5.4 years; SD = 3.6 years). These composed 459 cases derived from the United Kingdom Childhood Cancer study (UKCCS),15 an epidemiologic study of childhood malignancies conducted between 1991 and 1998, 342 cases derived from the United Kingdom Medical Research Council (MRC) ALL 97-99 trial, and 23 cases from the Northern Institute of Cancer Research. Immunophenotyping and genotyping of patient samples were undertaken using standard diagnostic methodologies. To minimize population stratification, cases with self-reported non–Western European ancestry were excluded. Cytogenetic data were available on 632 persons with BCP-ALL: hyperdiploid ALL (≥ 50 chromosomes-B-hyperdiploid, n = 293); B-cell lineage with the ETV6/RUNX1 fusion (alias TEL/AML1; n = 127), and B-cell other (n = 217).
Control series
We used data from 2 publicly accessible data series for population SNP genotype frequencies: persons from the 1958 Birth Cohort (58C, also known as the National Child development study)16 and persons from a United Kingdom GWA study of colorectal cancer.17 Because the prevalence of childhood ALL survivors in adults is less than 1 in 2000 in the United Kingdom, both control series can be considered representative of the non-ALL United Kingdom population.
Ethics
Collection of blood samples and clinicopathologic information from subjects was undertaken with informed consent and approval from the ethical review board of all participating institutions in accordance with the tenets of the Declaration of Helsinki.
Genotyping
As previously described,2 DNA was extracted and quantified from ethylenediaminetetraacetic acid-venous blood samples using conventional methodologies and a genome-wide scan of tagging SNPs conducted using Illumina Infinium HD Human370 Duo BeadChips according to the manufacturer's protocols (Illumina). We restricted our analysis to the autosomal SNPs. We considered that a DNA sample had failed if it did not generate a genotype for more than 95% of loci. Similarly, an SNP was considered a failure if less than 95% of DNA samples generated a genotype at the locus. To ensure quality of genotyping, a series of duplicate samples were genotyped on the same arrays, with concordance rates of 99.9%. The overall genotyping call rate was 99.84%.
Quality control
To identify samples showing relatedness, identity-by-state values were calculated for pairs of persons and for any pair with more than 80% identical SNP genotypes, we removed the sample with the lower call rate from the analysis. We excluded SNPs on the basis of deviation from Hardy-Weinberg equilibrium using a threshold of P < 1 × 10−5 in either the cases or controls. We also removed SNPs with minor allele frequency less than 0.05. To identify and exclude persons with non-Western European ancestry, case and control data were merged with persons of different ethnicities from the International HapMap Project, genome-wide identity-by-state value distances for markers shared between HapMap and our SNP panel determined, and dissimilarity measures used to perform principal component analysis. After imposing these stringent quality control measures, 292 200 SNP genotypes were available on 824 BCP-ALL cases and 2356 controls, which formed the basis of our analysis.
Statistical and bioinformatics analysis
We detected ROH using PLINK,18 Version 1.06. The ROH tool moves a sliding window of SNPs across the entire genome. To allow for genotyping error or other sources of artificial heterozygosity, such as paralogous sequences, within a stretch of truly homozygous SNPs and, hence, to prevent underestimating the number and size of ROH, 2% heterozygous SNPs were allowed in each window. We left the remaining options set to the default values (including allowing 5 missing calls per window), except that we varied the parameter homozyg-snp according to our heuristic preferences for defining the ROH as detailed in the next section. Subsequent statistical analyses were performed using packages available in R (Version 2.7.0) and specifically written Perl code. Comparison of the distribution of categorical variables was performed using the χ2 test. To compare the difference in average number of ROH between cases and controls, we used the Student t test. Naive adjustment for multiple testing was based on the Bonferroni correction.
We used 3 metrics to investigate the selection pressure on each ROH. Integrated Haplotype Score (iHS) is based on LD surrounding a positively selected allele compared with background, providing evidence of recent positive selection at a locus.19 An iHS score more than 2.0 reflects that haplotypes on the ancestral background are longer compared with the derived allelic background. Episodes of selection tend to skew SNP frequencies in different directions, and Tajima's D is based on the frequencies of SNPs segregating in the region of interest.20 Fixation index (Fst) measures the degree of population differentiation at a locus, taking values from 0 to 1.0.21 iHS, Tajima's D, and Fst metrics were obtained from Haplotter Software.19
Identification of ROH
To focus on commonly occurring ROH and to empower our analysis to identify meaningful associations, only ROH in which 10 or more persons share the same ROH were retained for analysis (ie, minimum frequency of ROH in each series ∼ 0.1%). The initial search for ROH was performed using PLINK18 with a specified length of 75 consecutive SNPs (homozyg-snp parameter). This ROH length was chosen to be more than an order of magnitude larger than the mean haploblock size in the human genome without being too large as to be very rare. The likelihood of observing 75 consecutive chance events can be calculated as follows.12 Mean heterozygosity in the controls was calculated to be 35%. Thus, given 292 200 SNPs and 3180 persons, a minimum length of 55 would be required to produce less than 5% randomly generated ROH across all subjects ([1 − 0.35]55 × 292 200 × 3180 = 0.048). A consequence of LD is that the SNP genotypes are not always independent, thereby inflating the probability of chance occurrences of biologically meaningless ROH. Analysis based on PLINK's pairwise LD SNP pruning function showed 228 714 separable tag groups, representing a 21.7% reduction of information compared with the original number of SNPs. Thus, ROH of length 75 were used to approximate the degrees of freedom of 55 independent SNP calls.
Once all ROH of at least 75 SNPs in length were identified, these were pruned to only those ROH, which occurred in more than 10 persons. To ensure that a minimum length and minimum number of SNPs in each ROH were maintained, each person's SNP data were recoded as 1 if the SNP was in an ROH for that person, and 0 otherwise. Then, for each SNP, those SNPs with less than 10 persons coded as 1 were recoded to 0 before removing any ROH that because of this recoding were now less than the required number of SNPs in length. This process therefore resulted in a list of “common” ROH having a minimum of 75 consecutive ROH calls across 10 or more samples and with each ROH having the same start and end locations across all persons where that ROH is observed.
Results
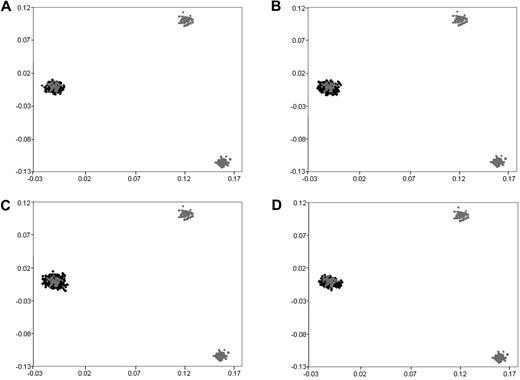
We have previously subjected cases and controls to rigorous quality control in terms of excluding samples and SNPs with poor call rates. Furthermore, we excluded SNPs showing significant departure from Hardy-Weinberg equilibrium. Before pooling data from the 2 GWA studies, we critically evaluated datasets for ancestral differences by principal component analysis and removed all outliers. Figure 1 shows that the final sample series used were ancestrally comparable and hence could be pooled without introducing systematic bias.
Comparison of ethnicity in each of the sample series. The first 2 principal components of the analysis were plotted based on genotypes from 6000 randomly selected SNPs. HapMap persons are plotted in gray. CEU indicates Utah residents with ancestry from northern and western Europe (far left); CHB + JPT, Han Chinese in Beijing, China; Japanese in Tokyo, Japan (bottom right); YRI, Yoruba in Ibadan, Nigeria (top right): (A) UKCCS BCP-ALL cases, (B) Northern Institute of Cancer Research and United Kingdom MRC BCP-ALL cases, (C) 1958 birth cohort controls, and (D) colorectal cancer controls.
Comparison of ethnicity in each of the sample series. The first 2 principal components of the analysis were plotted based on genotypes from 6000 randomly selected SNPs. HapMap persons are plotted in gray. CEU indicates Utah residents with ancestry from northern and western Europe (far left); CHB + JPT, Han Chinese in Beijing, China; Japanese in Tokyo, Japan (bottom right); YRI, Yoruba in Ibadan, Nigeria (top right): (A) UKCCS BCP-ALL cases, (B) Northern Institute of Cancer Research and United Kingdom MRC BCP-ALL cases, (C) 1958 birth cohort controls, and (D) colorectal cancer controls.
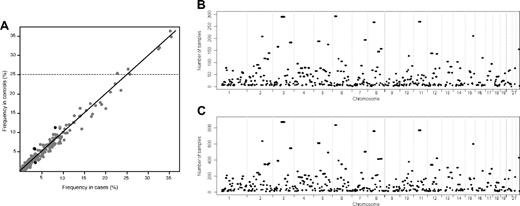
A total of 396 common ROH were identified in samples (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), encompassing approximately 40% of the genome as measured by both the total chromosomal length and the number of included SNPs. Figure 2 shows the similarity between the genome-wide plots of the location of each ROH among the genomes of BCP-ALL cases and controls and the correlation between the frequency of individual ROH in the cases and the controls.
Plots showing the similarity between the ROH identified in cases and controls. (A) Comparison between the frequency of the 396 ROH identified in the cases and controls. The 4 ROH colored black are those that are significantly associated with BCP-ALL risk (P < .01), and the horizontal line indicates the 7 ROH with frequency more than 25% in controls. The location of the ROH among the genomes of the cases (B) and the controls (C).
Plots showing the similarity between the ROH identified in cases and controls. (A) Comparison between the frequency of the 396 ROH identified in the cases and controls. The 4 ROH colored black are those that are significantly associated with BCP-ALL risk (P < .01), and the horizontal line indicates the 7 ROH with frequency more than 25% in controls. The location of the ROH among the genomes of the cases (B) and the controls (C).
The 18 longest ROH exceeded 12 Mb in length and included ROH encompassing the centromeric regions of chromosomes 2, 3, 4, 5, 6, 8, 11, 12, 16, and 19. The lengths of these ROH are partly a consequence of long regions for which there are no annotating SNPs. This is however unlikely to be the sole explanation, as in each case these centromeric regions were flanked by large homozygous regions containing numerous SNPs. One of these centromeric regions (chromosome 8) has been previously highlighted in several genome-wide studies of selective sweeps, thus providing validation of our methodology.19,22-24 Eight noncentromeric regions harboring ROH greater than 12 Mb in length were identified in our study at 2q12.2-14.2, 2q24.1-3, 3q25.31-26.2, 4q13.1-3, 5p14.3-13.3, 6p22.2-21.31, 7q31.1-32.1, and 8q21.1-22.1 (supplemental Table 1).
The ROH covering the largest genomic region (28 Mb) was found to be ROH92 spanning the centromere of chromosome 3, a region previously shown to be characterized by a high frequency of ROH in the European population.23 The ROH containing the largest number of SNPs was ROH162 spanning a 12-Mb section of chromosome 6, encompassing the region to which the human leukocyte antigen (HLA) immune regulation genes localize.
There are 7 ROH that were very common (> 25% frequency) in the control series (Table 1). Three of these are included in the 9 most common ROH found in Lencz et al12 and harbor several gene categories identified in various studies, which appear to be influenced by a high degree of selective pressure.19,22-24 Publicly available data from HapMap do not indicate that these regions have excessive copy number variation or segmental duplication, nor do they have very low recombination rates.22 However, the high iHS, D, and Fst metrics for each region are compatible with positive selection in white samples (Table 1).
List of 7 ROH with frequency of more than 25% in the controls
| ROH . | Chromosome . | Start, bp . | End, bp . | Length, bp . | No. of SNPs . | No. (%) of controls . | iHSmax* . | Tajima Dmax* . | Fstmax* . | No. of deletions/duplications/hotspots . |
|---|---|---|---|---|---|---|---|---|---|---|
| ROH92 | 3p12.3-3q13.11 | 77 828 854 | 106 110 833 | 28 281 979 | 1623 | 872 (37.0) | 2.24 | 2.90 | 0.79 | 4/4/100 |
| ROH162 | 6p22.2-6p21.31 | 24 213 309 | 36 712 176 | 12 498 867 | 1971 | 835 (35.4) | 2.84 | 2.10 | 0.86 | 3/8/61 |
| ROH283 | 11p11.2-11q12.2 | 45 131 106 | 60 672 543 | 15 541 437 | 801 | 768 (32.6) | 2.31 | 2.59 | 0.80 | 5/5/32 |
| ROH218 | 8p11.21-8q11.23 | 41 842 707 | 55 590 142 | 13 747 435 | 651 | 760 (32.3) | 3.80 | 2.20 | 0.84 | 5/6/53 |
| ROH62 | 2q21.2-2q22.1 | 134 030 848 | 142 232 977 | 8 202 129 | 822 | 635 (26.9) | 4.25 | 2.33 | 0.68 | 0/2/65 |
| ROH136 | 5p13.1-5q11.2 | 39 299 418 | 53 546 258 | 14 246 840 | 919 | 608 (25.8) | 1.75 | 2.30 | 0.70 | 5/2/44 |
| ROH351 | 15q23-15q25.1 | 69 391 304 | 76 490 686 | 7 099 382 | 571 | 599 (25.4) | 2.26 | 3.15 | 0.72 | 5/1/37 |
| ROH . | Chromosome . | Start, bp . | End, bp . | Length, bp . | No. of SNPs . | No. (%) of controls . | iHSmax* . | Tajima Dmax* . | Fstmax* . | No. of deletions/duplications/hotspots . |
|---|---|---|---|---|---|---|---|---|---|---|
| ROH92 | 3p12.3-3q13.11 | 77 828 854 | 106 110 833 | 28 281 979 | 1623 | 872 (37.0) | 2.24 | 2.90 | 0.79 | 4/4/100 |
| ROH162 | 6p22.2-6p21.31 | 24 213 309 | 36 712 176 | 12 498 867 | 1971 | 835 (35.4) | 2.84 | 2.10 | 0.86 | 3/8/61 |
| ROH283 | 11p11.2-11q12.2 | 45 131 106 | 60 672 543 | 15 541 437 | 801 | 768 (32.6) | 2.31 | 2.59 | 0.80 | 5/5/32 |
| ROH218 | 8p11.21-8q11.23 | 41 842 707 | 55 590 142 | 13 747 435 | 651 | 760 (32.3) | 3.80 | 2.20 | 0.84 | 5/6/53 |
| ROH62 | 2q21.2-2q22.1 | 134 030 848 | 142 232 977 | 8 202 129 | 822 | 635 (26.9) | 4.25 | 2.33 | 0.68 | 0/2/65 |
| ROH136 | 5p13.1-5q11.2 | 39 299 418 | 53 546 258 | 14 246 840 | 919 | 608 (25.8) | 1.75 | 2.30 | 0.70 | 5/2/44 |
| ROH351 | 15q23-15q25.1 | 69 391 304 | 76 490 686 | 7 099 382 | 571 | 599 (25.4) | 2.26 | 3.15 | 0.72 | 5/1/37 |
Chromosomal coordinates were derived from the National Center for Biotechnology Information, build 36.
ROH indicates regions of homozygosity; and SNP, single nucleotide polymorphism.
Maximal values for several metrics of positive selection, derived from Haplotter (http://hg-wen.uchicago.edu/selection/haplotter.htm). The number of deletions, duplications, and recombination hotspots are derived from HapMap release 27 (http://hapmap.org).
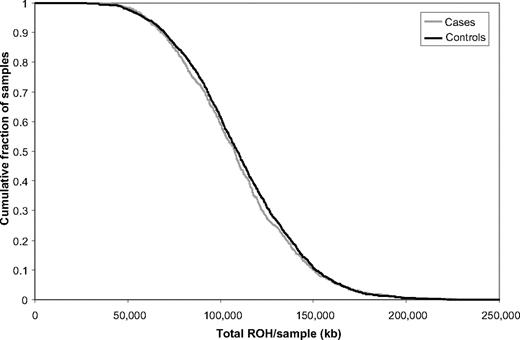
The total number of common ROH observed in each person was calculated to permit genome-wide comparison between the case and control groups. Each person therefore was assigned a value between 0 and 396. Overall, patients with BCP-ALL (mean = 14.84, SD = 4.33) and controls (mean = 15.11, SD = 4.0) showed no significant difference in the average number of ROH (t3178 = 1.6217, P = .11). To also examine whether there were differences in the distributions of ROH in the genomes of cases and controls, we computed the cumulative distributions of both series (Figure 3). This analysis also provides no support for a difference in autozygosity profiles between cases and controls on a genome-wide basis.
Cumulative distributions of ROH in BCP-ALL cases and controls. The graph is presented in such a way that each data point represents the cumulative fraction (y-axis) of the samples with the corresponding minimum cumulative run of homozygosity (x-axis).
Cumulative distributions of ROH in BCP-ALL cases and controls. The graph is presented in such a way that each data point represents the cumulative fraction (y-axis) of the samples with the corresponding minimum cumulative run of homozygosity (x-axis).
At an individual level 4 ROH, none of which includes an excessive number of copy number variants, differed significantly (P < .01) between cases and controls (Table 2). Although these associations were not individually statistically significant, after adjusting for multiple testing using the Bonferroni correction, imposing such an adjustment is highly conservative and can lead to type 2 error. Three of these 4, marginally significant, ROH were more common in the controls than in the cases. The fourth, ROH380, was identified in 2.2% of cases (n = 18) compared with 0.9% of controls (n = 22; P = .005). More than 40 genes or predicted transcripts map the region encompassed by this ROH, including the gene encoding erythropoietin receptor (EPOR; MIM 133171) protein. Although speculative, it is intriguing to note that overexpression of EPOR has been documented in ETV6/RUNX1-positive ALL.25 Although there was no overrepresentation of ROH380 in our ETV6/RUNX1-positive ALL cases, we explored the possibility of a relationship between EPOR genotype and ALL risk through single point analysis based on SNPs which mapping within 25 kb of the gene (Table 3). Of the 5 SNPs tested, evidence for an association between EPOR genotype and ETV6/RUNX1-positive ALL was provided by rs4804164 and rs317913, which map 7 kb and 15 kb centromeric to EPOR, respectively (Table 3). The strongest association was provided by rs4804164, with odds ratio of 0.58 and P value from Cochran-Armitage trend test of .008.
Four ROH significantly associated with BCP-ALL risk (P < .01)
| ROH . | Chromosome . | Start, bp . | End, bp . | Length, bp . | No. of SNPs . | No. (%) of cases . | No. (%) of controls . | χ2 . | P . |
|---|---|---|---|---|---|---|---|---|---|
| ROH261 | 10p11.21-10q11.21 | 36 774 789 | 44 190 246 | 7 415 457 | 317 | 26 (3.4) | 140 (5.9) | 9.58 | < .002 |
| ROH15 | 1p31.1-1p31.1 | 70 726 887 | 76 563 751 | 5 836 864 | 463 | 27 (3.2) | 137 (5.8) | 8.04 | < .005 |
| ROH380 | 19p13.2-19p13.13 | 11 145 302 | 12 633 165 | 1 487 863 | 115 | 18 (2.2) | 22 (0.9) | 7.69 | < .006 |
| ROH388 | 20q11.1-20q11.23 | 28 121 437 | 35 496 979 | 7 375 542 | 401 | 67 (8.1) | 270 (11.4) | 7.14 | > .007 |
| ROH . | Chromosome . | Start, bp . | End, bp . | Length, bp . | No. of SNPs . | No. (%) of cases . | No. (%) of controls . | χ2 . | P . |
|---|---|---|---|---|---|---|---|---|---|
| ROH261 | 10p11.21-10q11.21 | 36 774 789 | 44 190 246 | 7 415 457 | 317 | 26 (3.4) | 140 (5.9) | 9.58 | < .002 |
| ROH15 | 1p31.1-1p31.1 | 70 726 887 | 76 563 751 | 5 836 864 | 463 | 27 (3.2) | 137 (5.8) | 8.04 | < .005 |
| ROH380 | 19p13.2-19p13.13 | 11 145 302 | 12 633 165 | 1 487 863 | 115 | 18 (2.2) | 22 (0.9) | 7.69 | < .006 |
| ROH388 | 20q11.1-20q11.23 | 28 121 437 | 35 496 979 | 7 375 542 | 401 | 67 (8.1) | 270 (11.4) | 7.14 | > .007 |
ROH indicates regions of homozygosity; BCP-ALL, B-cell precursor acute lymphoblastic leukemia; and SNP, single nucleotide polymorphism.
Relationship between SNP mapping within 25 kb of EPOR and BCP-ALL risk
| SNP . | Location, bp . | Ancestral allele . | ETV6-RUNX1-positive cases . | Hyperdiploid cases . | Other cases . | |||
|---|---|---|---|---|---|---|---|---|
| Odds ratio . | P . | Odds ratio . | P . | Odds ratio . | P . | |||
| rs7251786 | 11 325 879 | T | 1.16 | .264 | 1.04 | .675 | 0.94 | .416 |
| rs318699 | 11 362 240 | G | 1.11 | .423 | 1.12 | .207 | 1.02 | .818 |
| rs4804164 | 11 363 291 | G | 0.58 | .008 | 0.92 | .588 | 0.97 | .838 |
| rs2291516 | 11 369 177 | G | 0.83 | .409 | 1.10 | .526 | 1.03 | .827 |
| rs317913 | 11 373 314 | C | 0.60 | .019 | 0.97 | .850 | 0.94 | .680 |
| SNP . | Location, bp . | Ancestral allele . | ETV6-RUNX1-positive cases . | Hyperdiploid cases . | Other cases . | |||
|---|---|---|---|---|---|---|---|---|
| Odds ratio . | P . | Odds ratio . | P . | Odds ratio . | P . | |||
| rs7251786 | 11 325 879 | T | 1.16 | .264 | 1.04 | .675 | 0.94 | .416 |
| rs318699 | 11 362 240 | G | 1.11 | .423 | 1.12 | .207 | 1.02 | .818 |
| rs4804164 | 11 363 291 | G | 0.58 | .008 | 0.92 | .588 | 0.97 | .838 |
| rs2291516 | 11 369 177 | G | 0.83 | .409 | 1.10 | .526 | 1.03 | .827 |
| rs317913 | 11 373 314 | C | 0.60 | .019 | 0.97 | .850 | 0.94 | .680 |
Odds ratios were calculated with the ancestral allele as the reference allele, and P values are from the Cochran-Armitage trend test.
SNP indicates single nucleotide polymorphism; and BCP-ALL, B-cell precursor acute lymphoblastic leukemia.
Discussion
Recent studies have provided evidence that signatures of autozygosity correlate to cancer incidence and that these regions showing identity by descent may be the locations of genes contributing to tumor heritability.13,14 These data have been interpreted as providing an explanation for the increased cancer rates often reported in inbred populations.
Here we have used a high-density genomic scan to compare the structure of genetic variation in patients with BCP-ALL with healthy controls. This same sample series has recently been used to robustly identify 3 predisposition loci for BCP-ALL. By imposing stringent quality control, we have ensured that persons in our study were from an apparently panmictic population (ie, population where all persons are potential partners) with no evidence of stratification. Our data provide further evidence that ROH, ranging in size from 1 to 28 Mb, are common in persons from an outbred population.26-29 As documented in Table 1, the common ROH we identified are representative of autozygosity because of distant consanguinity and not chromosomal abnormalities or common copy number variants. Moreover, these homozygous regions are too common and small to be a consequence of recent consanguinity and are consistent with the possibility that they mark regions under selective pressure.30 Based on our analysis, there was no evidence for an association between homozygosity and BCP-ALL risk on the basis of total ROH size per person. Although not formally statistically significant, after adjustment for multiple testing, the associations between ALL risk and a number of specific ROH, as demonstrated by EPOR, may reflect regions that warrant further investigation.
The assertion that increased autozygosity correlates with cancer incidence provides an attractive explanation for reported increased cancer risk in inbred populations. However, as recently articulated, several criticisms can be leveled at such an idea.31 The observation of an increased cancer risk associated with consanguinity has often been based on studies of a small number of persons in an isolated community or a single large family with a high level of inbreeding. Thus, the relevance of inbreeding to the population risk of cancer is unclear as inbreeding and founder effects may be confounded. Sample sizes in the molecular studies,13,14 which have sought to establish a relationship between ROH and cancer risk, have generally been small and, crucially, cases and controls groups ethnically heterogeneous or unmatched. Here we have addressed these possible shortcomings in our study of ALL by analyzing a large set of cases and controls that have been genotyped for several hundred thousand SNPs and imposed a high level of quality control both in terms of genotyping and sample ancestry.
In conclusion, our findings make it unlikely that levels of measured common homozygosity, from autozygosity, uniparental isodisomy, or hemizygosity, play a significant role in defining the risk of developing childhood BCP-ALL in a predominantly outbred population. Moreover, it is unlikely that there exist large numbers of recessive alleles that predispose to ALL and are unmasked by autozygosity in most European populations. This analysis does not, however, exclude the possibility that recessively acting disease alleles exist for ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sue Richards and Julie Burrett (Clinical Trials Service Unit, Oxford); Christine Harrison, Lucy Chilton, and Anthony Moorman (Leukemia Research Cytogenetics Group, Northern Institute for Cancer Research, Newcastle University); Jill Simpson (University of York); Pamela Thomson and Adiba Hussain (Cancer Immunogenetics, School of Cancer Sciences, University of Manchester) for assistance with data harmonization; Irene Roberts and the Children's Cancer and Leukemia Group Biological Studies Steering Group for access to MRC ALL Trial samples; all the patients and persons for their participation; and the clinicians, other hospital staff, and study staff who contributed to the blood sample and data collection for this study.
This work was supported by Leukemia Research and the Kay Kendall Leukemia Fund, which provided principal funding, and Cancer Research UK (C1298/A8362, supported by the Bobby Moore Fund).
This study made use of genotyping data on the 1958 Birth Cohort. Genotyping data on 1958 controls were generated and generously supplied to us by Panagiotis Deloukas of the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the 1958 data is available at www.wtccc.org.uk.
Authorship
Contribution: R.S.H. and F.J.H. designed the study and drafted the manuscript; F.J.H. performed statistical analyses; E.P. oversaw laboratory analyses; E.S. and S.E.K. performed curation and sample preparation of MRC ALL 97 trial samples; T.L. and E.R. managed and maintained UKCCS sample data; M.T. performed curation and sample preparation of UKCCS samples; J.M.A. and J.A.E.I. performed ascertainment, curation, and sample preparation of Northern Institute for Cancer Research case series; R.S.H. and M.G. obtained funding and designed parent project; and I.P.T. performed generation and management of United Kingdom colorectal cancer control genotypes.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, Section of Cancer Genetics, Institute of Cancer Research, 15 Cotswold Rd, Sutton, Surrey, SM2 5NG, United Kingdom; e-mail: richard.houlston@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal