Abstract
Previous studies have shown that single-stranded RNA (ssRNA) mixed with protamine forms particles and activates immune cells through Toll-like receptors (TLRs). We have found that the size of protamine-RNA particles generated depends on the electrolyte content when mixing the 2 components. Moreover, we have evidenced that (1) nanometric particles induce production of interferon-α, whereas (2) micrometric particles mainly induce production of tumor necrosis factor-α (TNF-α) in human immune cells. We found that the mechanisms underlying these observations are (1) nanoparticles but not microparticles are selectively phagocytosed by plasmacytoid dendritic cells (pDCs), which produce interferon-α and (2) monocytes that produce TNF-α have a higher activation threshold than that of pDCs. Thus, at the same time as sensing pathogen-associated molecular patterns such as ssRNA, the immune system distinguishes the size of the associated structure in such a way as to trigger the adapted antivirus (nanometric) or antibacterial/antifungal (micrometric) immune response. Our results introduce a new dimension in danger signaling—how size qualitatively affects innate response.
Introduction
The innate immune system is capable of sensing invading pathogens and abnormal cells by the detection of “danger signals.” Danger signals are molecules originating specifically from invaders and disturbed cell metabolism. Studies show 3 forms of nucleic acid are recognized by the mammalian immune system as danger signals: DNA containing unmethylated CG sequences (CpG), unmodified single-stranded RNA (ssRNA), and double-stranded RNA (dsRNA). Such foreign nucleic acids mislocalized in the intracellular compartments are recognized by endosome-resident Toll-like receptor 9 (TLR9), TLR-7, TLR-8, and TLR-3, respectively (reviewed in Takeda et al1 ). When engaged, those receptors deliver an activation signal to the cell. Subsequently, cytokines and costimulation molecules are produced and certain homing and chemokine receptors are up-regulated.
The first identified nucleic acid danger signal, dsRNA, was validated as an antitumor immunotherapeutic compound in mice in the 1960s and further tested as an immunostimulating compound in cancer patients in the 1970s.2,3 However, low clinical efficacy as well as toxicity issues precluded the further evaluation of dsRNA for antitumor immunotherapy. Nevertheless, dsRNA is nowadays available in the form of a less toxic, interrupted polyinosinic-polycytidylic acid duplex (Ampligen; Hemispherx Biopharma) and is used for treating chronic fatigue.
Although unmethylated CpG-containing DNA is the most recently identified nucleic acid danger signal,4 it is the most thoroughly studied and clinically evaluated up to now. One relevant but unexplained feature of CpG oligodeoxynucleotides (ODNs) is that they trigger a sequence-dependent immunostimulation profile5 : ODN of the “A” class mostly activate plasmacytoid dendritic cells (pDCs) and induce the production of interferon-α, whereas those of the “B” class mostly activate B cells and induce the production of TNF-α. The “C” class CpG ODNs activate both pDCs and B cells, stimulating the production of both of these cytokines.
The ssRNA molecules were found to be mildly immunostimulating in the 1960s.6 Using rabbit kidney cultures, Billiau et al showed in 1969 that ssRNA mixed with polybasic substances such as protamine activates the interferon pathway and thereby induces protection against vesicular stomatitis virus or Semliki Forest virus.7 More recently we and others have shown that stabilized ssRNA (phosphorothioate oligoribonucleotide or mRNA premixed with protamine) strongly activates human and mouse immune cells through TLRs.8-10 The identification of TLR-7 and TLR-8 as the receptors for ssRNA has boosted the study and development of this danger signal.11-13 No precise sequence motif is needed in the RNA to stimulate TLR-7 and TLR-8; however, U-residues, and to a lesser extent G-residues, are most potent.11,14,15 Up to now, the specific immunostimulation profile triggered by ssRNA has not been investigated. Here, we present the finding that when using protamine to formulate RNA, we can generate nanoparticles (diameter ∼ 220 nm) or microparticles (∼ 1200 nm) depending on the conditions of formulation. Most importantly, we show that nano- and microparticles induce qualitatively different innate immune responses. Our data introduce the new concept that not only the nature of the danger signal but also its form, in either nano- or microstructures, determines the type of innate immunity that is triggered.
Methods
Oligonucleotides
RNA oligonucleotides were all nonmodified and nonphosphorylated. Sequences (5′ to 3′) are as follows: U-rich (21m) AGUGUUAUUCUUGUAUGGUUG; RNA18 AGUGUUAUUCUUGUAUGG; U-rich (16m) GUGUUAUUCUUGUAUG; U-rich (14m) GUUAUUCUUGUAUG; U-rich (12m) GUUAUUCUUGUA; U-rich (10m) GUUAUUCUUG; A-rich (12m) GAAAAACAAGAA; C-rich (12m) GCCACCCCCGCA; G-rich (12m) GGGAGGCGGAGA; U-rich (12m) GUUAUUCUUGUA; Purine (12m) AGAGAGAGAGAG; and Pyrimidine (12m) CUCUCUCUCUCU. CpG 1826 ODN is fully phosphorothioate. All oligonucleotides were produced by Thermo or by Microsynth.
Preparation and physical analysis of protamine-RNA particles
Five micrograms of RNA were mixed with the indicated amount of protamine (Valeant), at the indicated concentration in the indicated solution. One milliliter of phosphate-buffered saline (PBS, 10mM phosphate, 138mM NaCl, [pH 7.4]) was added, and the size distribution of the protamine-RNA complexes was characterized by dynamic light scattering with a Zetasizer 3000HSA (Malvern). After the measurement, the preparation was further diluted with 1 mL of PBS and subjected to electrophoretic light scattering measurement, also using a Zetasizer, to determine the zetapotential of the complexes. Data were analyzed using the built-in DTS software. For electron microscopy the above-described components were mixed, and a 10-μL aliquot was allowed to settle for 1 minute onto Formvar and carbon-coated copper grids, which had been freshly glow discharged. Excess fluid was blotted. For transmission electron microscopy, grids were stained with 2% Uranylacetat in distilled water for 3 × 15 seconds. Grids were then blotted and allowed to air dry before imaging at 80 kV in a Phillips CM 208 transmission electron microscope. For scanning electron microscopy the grids were washed twice in distilled water. After blotting and air drying, grids were mounted, sputter-coated with 4 nm platinum, and imaged in a Zeiss Supra 50 VP Scanning Electron Microscope at 4 kV in the secondary electron mode.
Preparation, isolation, and culture of human PBMCs
Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of different donors by Ficoll density gradient centrifugation (PAA Laboratories). After washing in PBS, cells were resuspended at 1 million/200 μL of complete medium (RPMI 1640, 10% fetal calf serum, 2mM l-Glutamine, 10 μg/mL streptomycin, 10 U/mL penicillin). Cells were distributed in 96-well plates (200 μL/well) containing stimulating RNA (2 μg of RNA in 2 μL) complexed with protamine (2 μg of protamine in 2 μL) or CpG DNA (2 μg) or polyinosinic-polycytidylic acid (2 μg; Invitrogen) or R848 (1 μg; Alexis Biochemicals).
Cytokine measurement
After 24 hours of incubation at 37°C, the amount of cytokine in 20 μL of supernatant was quantified using enzyme-linked immunosorbent assay (ELISA) kits for interferon-α (Bender) or TNF-α (Abazyme) or using the Multiplex Kit (Bio-Rad) following the instructions of the manufacturer.
Cell staining
Cells were incubated with fluorescent nanospheres (Molecular Probes) for 4 hours then stained for 30 minutes in the dark at 4°C with allophycocyanin (APC)–labeled CD11c (BD PharMingen) or APC-labeled anti–blood dendritic cell antigen-2 (BDCA-2; Miltenyi Biotec) monoclonal antibodies. For intracellular staining of interferon-α, fresh human PBMCs were incubated for 6 hours with protamine-RNA particles. Cells were permeabilized and fixed (Cytofix/Cytoperm kit; BD PharMingen), according to the manufacturer's instructions, before being stained with fluorescein isothiocyanate-labeled anti–interferon-α antibody (BD PharMingen) and APC-labeled anti–BDCA-2 antibody.
Generation of mouse dendritic cells
All animal experiments were approved by the University Hospital of Zürich. Bone marrow–derived dendritic cells were obtained by flushing the bones of the hind leg of 6- to 12-week-old C57BL/6 (Charles River Laboratories) and TLR7−/− mice (TLR-7 knockout [KO] mice), kindly provided by Dr Karl Lang (Institute of Experimental Immunology, University Hospital of Zürich) with Iscove modified Dulbecco medium (PAN Biotech). Cells were then cultured for 6 days in Iscove modified Dulbecco medium with 10% heat-inactivated fetal calf serum (both from PAN Biotech), 2mM l-Glutamine (BioWhittaker), 10 μg/mL streptomycin, 10 U/mL penicillin (PEN-STREP; BioWhittaker) and granulocyte-macrophage colony-stimulating factor (4000 ng/mL final concentration) in Petri dishes at a concentration of 0.5 × 106 cells/mL. Dendritic cells were replated on day 6 and used for experiments on day 7.
Results
The size of protamine-RNA particles depends on formulation conditions
Being cationic, protamine spontaneously associates with anionic RNA generating a turbid suspension immediately upon mixing. We have previously shown10 that particles in such a solution are large heterogenous complexes of several micrometers. These poorly defined structures are difficult to qualify as a pharmaceutical product; they precipitate quickly and spontaneously and are difficult to take up and deliver through a syringe. We now observed that the electrolyte content in RNA and protamine is critical in generating precisely sized, homogenous particles. By dissolving RNA pellets at 1 mg/mL in pure water and diluting Protamine 5000 (pharmaceutical protamine used as an anti–heparin drug) from 14 to 1 mg/mL in pure water, we generated homogenous nanoparticles upon mixing, as detected by dynamic light scattering (Figure 1A) or transmission electron microscopy (air drying of nanoparticles on microscopy grids leads to clustering; however, the particles are dispersed in solution; Figure 1D). Thus, the electrolytes present in the solutions of RNA and protamine influence the structure of the resulting protamine-RNA complexes. The size of the RNA does not have much impact on this phenomenon: from 18 mer oligonucleotides (Figure 1A,D) up to long RNA of a few hundred bases (Figure 1B) can be used to form nanoparticles averaging 220 nm. Longer RNA, however, tends to generate slightly more heterogenous particles (from 100 to 600 nm) than oligonucleotides (compare Figure 1A to B). Particles of less than 500 nm remain polydispersed in solution and do not precipitate spontaneously. However, they can be pelleted by centrifugation. Introducing electrolytes in the 1-mg/mL single-component solutions, for example by diluting the RNA and Protamine 5000 in Ringer lactate solution, results in the production of larger protamine-RNA particles (Figure 1C top, E). These particles form a pellet within minutes when the reaction tube is on the bench. Increasing the protamine to RNA ratio produces a slight reduction of the size of the microcomplexes (Figure 1C) but does not affect the size of the nanocomplexes (Figure 1A).
The size of protamine-RNA complexes depends on the ionic conditions of formulation. An 18-residue RNA (RNA18) oligonucleotide (A,C-F) or mRNA coding enhanced green fluorescent protein (B) were diluted to 1 mg/mL using water (A-B,D) or Ringer lactate solution (C,E) and mixed at indicated mass ratios (A,C) or at 1:1 mass ratios (B,D-F) with Protamine 5000 diluted to 1 mg/mL using water (A-B,D) or Ringer lactate solution (C,E). The size of the particles (A, left, B-C) was assessed by dynamic light scattering (the number shown in each graphic indicates the average hydrodynamic diameter) and electron microscopy (transmission in panel D and scanning in panel E). Data shown are representative of 10 independent experiments. The zetapotentials of the nanoparticles (A right) are measured using a Zetasizer (Malvern). (F) A summary of the results from dynamic light scattering measurements of particles produced with protamine and RNA diluted to 1 mg/mL using different solutions of varying ionic strength. Each formulation was repeated 3 times and measured. Error bars indicate the standard variation within those triplicate experiments.
The size of protamine-RNA complexes depends on the ionic conditions of formulation. An 18-residue RNA (RNA18) oligonucleotide (A,C-F) or mRNA coding enhanced green fluorescent protein (B) were diluted to 1 mg/mL using water (A-B,D) or Ringer lactate solution (C,E) and mixed at indicated mass ratios (A,C) or at 1:1 mass ratios (B,D-F) with Protamine 5000 diluted to 1 mg/mL using water (A-B,D) or Ringer lactate solution (C,E). The size of the particles (A, left, B-C) was assessed by dynamic light scattering (the number shown in each graphic indicates the average hydrodynamic diameter) and electron microscopy (transmission in panel D and scanning in panel E). Data shown are representative of 10 independent experiments. The zetapotentials of the nanoparticles (A right) are measured using a Zetasizer (Malvern). (F) A summary of the results from dynamic light scattering measurements of particles produced with protamine and RNA diluted to 1 mg/mL using different solutions of varying ionic strength. Each formulation was repeated 3 times and measured. Error bars indicate the standard variation within those triplicate experiments.
By diluting RNA and Protamine 5000 to a final concentration of 1 mg/mL using solutions containing various salt concentrations, we can generate protamine-RNA nanoparticles of different sizes as shown in Figure 1F. The higher the NaCl concentration in RNA and protamine, the larger the generated protamine-RNA particles. The concentration of protamine and RNA before mixing also affects the size of the particles as shown in Table 1. The minimum possible size of the protamine-RNA particles was approximately 220 nm. This was obtained using Protamine 5000 diluted 14 times in water (final concentration ∼ 1 mg/mL) and RNA at 1 mg/mL in water. Further dilution of the 2 compounds with water did not result in any reduction in the size of the particles (Table 1). However, when protamine and RNA were in Ringer lactate solution, lowering their concentrations reduced the size of the particles: approximately 1250 nm when both components were at 1 mg/mL versus approximately 550 nm when they were at 0.25 mg/mL.
The concentration of protamine and RNA affects particle size
| . | Particle size in nm . | |||
|---|---|---|---|---|
| 2 mg/mL . | 1 mg/mL . | 0.5 mg/mL . | 0.25 mg/mL . | |
| Water | 575 | 267 | 274 | 263 |
| 25mM NaCl | 492 | 277 | ||
| 75mM NaCl | 1045 | 512 | ||
| 225mM NaCl | 1288 | 869 | ||
| Ringer lactate | 1363 | 1432 | 926 | 571 |
| . | Particle size in nm . | |||
|---|---|---|---|---|
| 2 mg/mL . | 1 mg/mL . | 0.5 mg/mL . | 0.25 mg/mL . | |
| Water | 575 | 267 | 274 | 263 |
| 25mM NaCl | 492 | 277 | ||
| 75mM NaCl | 1045 | 512 | ||
| 225mM NaCl | 1288 | 869 | ||
| Ringer lactate | 1363 | 1432 | 926 | 571 |
Protamine 5000 was diluted 7 times (∼ 2 mg/mL final), 14 times (∼ 1 mg/mL final), 28 times (∼ 0.5 mg/mL final), or 56 times (∼0.25 mg/mL final) with water, 25mM NaCl, 75mM NaCl, 225mM NaCl, or ringer lactate. Similarly, the RNA oligonucleotide RNA18 was diluted to the same final concentrations as the above-mentioned 5 solutions. Protamine and RNA were mixed at a 1:1 mass ratio. Subsequently, the size of the particle was measured by dynamic light scattering.
Mixing protamine and RNA at a 1:1 mass ratio generated particles with a slightly positive surface charge (∼ + 7 mV zetapotential; Figure 1A). Higher amounts of protamine versus RNA increased the surface charge, whereas lower amounts of protamine versus RNA (1:2 protamine to RNA in mass) resulted in negatively charged particles. The same results were observed for nano- and microparticles. A surface charge of near neutrality is known to be optimal for particle stability, inocuity, and bioavailability.16 Thus we performed all subsequent immunostimulating experiments using a 1:1 protamine-RNA ratio.
The size of protamine-RNA particles influences their immunostimulating profile
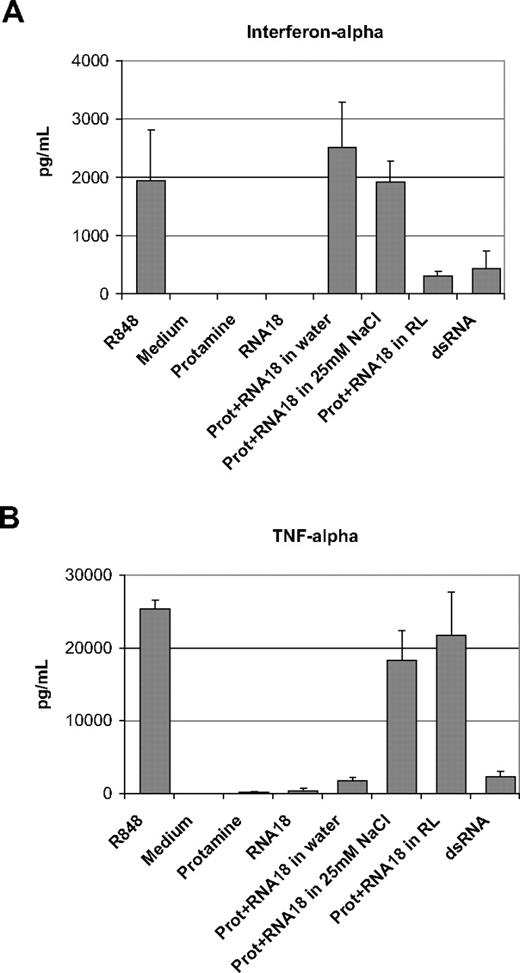
Using total human PBMCs prepared from healthy donors, we compared the immunostimulating capacity of protamine-RNA particles of 220 nm, approximately 500 nm, and 1200 nm. An 18-residue RNA oligonucleotide containing all 4 bases (RNA18) was used. The final concentration of protamine-RNA in cell culture was 20 μg/mL (10 μg/mL protamine and 10 μg/mL RNA). As shown in Figure 2, the smaller the particles, the more they triggered the production of interferon-α (Figure 2A). In contrast, the larger the particles were the more they triggered TNF-α production (Figure 2B). The resiquimod R848 induced production of high amounts of interferon-α and TNF-α, whereas dsRNA was a poor inducer of these 2 cytokines. Similar results were obtained with all tested PBMC donors. RNA alone at 10 μg/mL or protamine alone at 10 μg/mL did not induce detectable production of TNF-α or interferon-α in human PBMCs. Thus, the size of the particle as defined by the formulation conditions (mainly the electrolyte concentration) has a direct impact on the profile of the immune response in human cells. To analyze in more detail the differential immunostimulation of nano- versus microparticle protamine-RNA, we studied the production of several cytokines by human PBMCs in culture with nano- or microparticles using Multiplex (Bio-Rad) as a readout. As shown in supplemental Figure 2C (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), interleukin-1β (IL-1β), IL-1α, interleukin-6, monocyte chemoattractant protein-1, macrophage inhibitory protein-1α, and macrophage inhibitory protein-1β were similarly induced by all particles and by R848. Only interleukin-10, and granulocyte colony-stimulating factor were differentially induced; nanoparticles stimulated less production of these cytokines than did either microparticles or R848. The production of interleukin-12 (p70) was poorly induced by nanoparticles and R848, whereas production was best triggered by protamine-RNA microparticles.
Protamine-RNA particles of different sizes induce different immunostimulation profiles. PBMCs prepared from 3 different healthy donors were incubated overnight with protamine-RNA particles (Prot+RNA) produced by mixing 1:1 (mass ratios) protamine and RNA (RNA18 oligonucleotide) at 1 mg/mL. Protamine 5000 and RNA were diluted with water (220 nm), 25mM NaCl solution (500 nm), or Ringer lactate solution (1200 nm). Cells stimulated by R848 and dsRNA were used as positive controls. The content of interferon-α (A) or TNF-α (B) in the supernatants was then assessed by ELISA. The data show the mean content of cytokines for the 3 donors. Error bars indicate the observed deviation between donors. The data shown are representative of 12 independent experiments.
Protamine-RNA particles of different sizes induce different immunostimulation profiles. PBMCs prepared from 3 different healthy donors were incubated overnight with protamine-RNA particles (Prot+RNA) produced by mixing 1:1 (mass ratios) protamine and RNA (RNA18 oligonucleotide) at 1 mg/mL. Protamine 5000 and RNA were diluted with water (220 nm), 25mM NaCl solution (500 nm), or Ringer lactate solution (1200 nm). Cells stimulated by R848 and dsRNA were used as positive controls. The content of interferon-α (A) or TNF-α (B) in the supernatants was then assessed by ELISA. The data show the mean content of cytokines for the 3 donors. Error bars indicate the observed deviation between donors. The data shown are representative of 12 independent experiments.
Protamine-RNA particles are recognized by mouse TLR-7
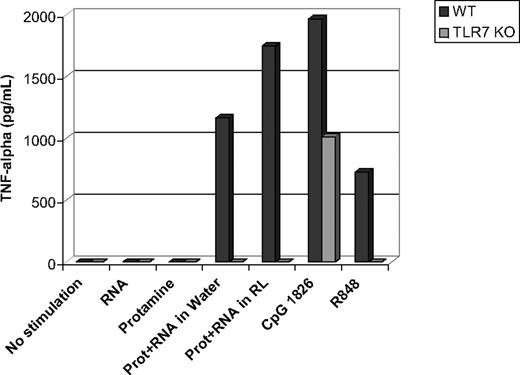
TLR-7 and TLR-8 have previously been shown to recognize ssRNA molecules. In mice, TLR-8 is mostly nonfunctional. Using bone marrow–derived dendritic cells from TLR-7 KO mice, we showed (Figure 3) that protamine-RNA nano- and microparticles can stimulate through TLR-7: TNF-α was produced by wild-type cells stimulated with nano- and microparticle protamine-RNA, R848, and CpG DNA, whereas TLR-7 KO cells were stimulated only with CpG DNA but not with protamine-RNA particles or R848. Using commercially available kits with a limit of detection of approximately 20 pg/mL, no mouse interferon-α was detected in culture supernatants (data not shown).
TLR-7 is necessary for activation of mouse dendritic cells. Bone marrow–derived dendritic cells from wild-type (WT) and TLR-7 KO mice were incubated overnight with RNA18, protamine, protamine-RNA particles (Prot+RNA) of 220 nm or 1200 nm, CpG DNA, or R848. Thereafter cell culture supernatants were tested by ELISA for their TNF-α content. Representative results from 5 independent experiments are shown.
TLR-7 is necessary for activation of mouse dendritic cells. Bone marrow–derived dendritic cells from wild-type (WT) and TLR-7 KO mice were incubated overnight with RNA18, protamine, protamine-RNA particles (Prot+RNA) of 220 nm or 1200 nm, CpG DNA, or R848. Thereafter cell culture supernatants were tested by ELISA for their TNF-α content. Representative results from 5 independent experiments are shown.
Stability of protamine-RNA particles
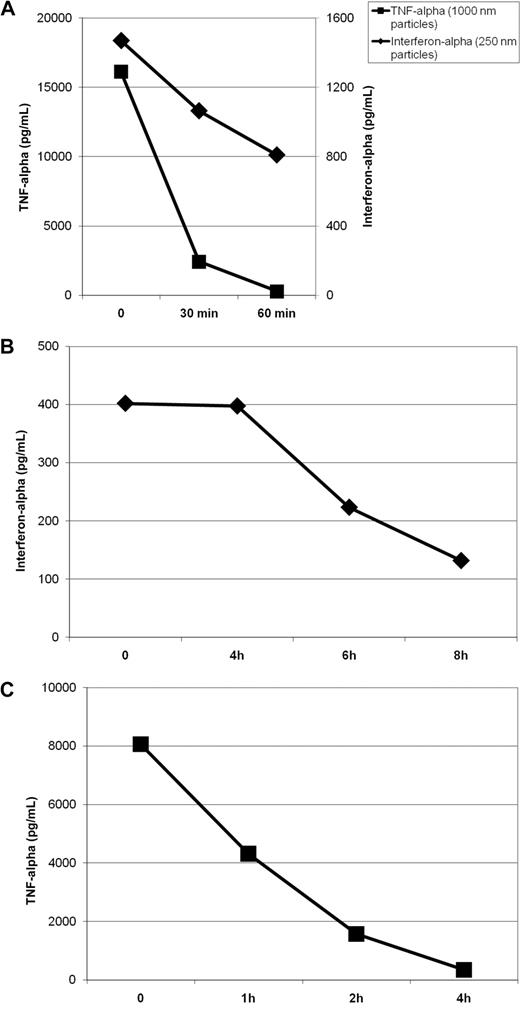
In the described experiments (Figure 3), particles were formulated and immediately put into cell culture. To test the stability of the preparations, we investigated their immunostimulating capacity after incubation at room temperature for 0, 30, and 60 minutes. Particles were prepared at the described time points before fresh human PBMCs were added to the wells. As shown in Figure 4A, the immunostimulating activity of nanoparticles decreased slowly over time. After 1 hour at room temperature, over 50% of maximal activity was still observed. In contrast, the immunostimulating capacity of micrometric particles dropped more quickly: nearly 90% of activity was lost within 30 minutes. This effect could reflect the spontaneous aggregation of microparticles into a solid pellet that may limit the availability of free particles and thus stimulation of immune cells.
Stability of protamine-RNA particles. Protamine 5000 diluted 14 times with water or with Ringer lactate solution was mixed 1-to-1 with RNA18 diluted to 1 mg/mL with water or Ringer lactate solution to generate nano- or microparticles, respectively. The formulation was repeated at several time intervals. (A) Particles were diluted with 9 volumes of PBS and incubated at room temperature. Particles of 220 nm (B) or 1200 nm (C) were diluted with 9 volumes of fresh human serum and incubated at 37°C. Fresh human PBMCs were added to the particles after various incubation times as indicated. After overnight incubation, the interferon-α (for stimulation with nanoparticles) or TNF-α (for stimulation with microparticles) concentrations in the culture supernatants were measured by ELISA. The data shown are representative of 5 independent experiments.
Stability of protamine-RNA particles. Protamine 5000 diluted 14 times with water or with Ringer lactate solution was mixed 1-to-1 with RNA18 diluted to 1 mg/mL with water or Ringer lactate solution to generate nano- or microparticles, respectively. The formulation was repeated at several time intervals. (A) Particles were diluted with 9 volumes of PBS and incubated at room temperature. Particles of 220 nm (B) or 1200 nm (C) were diluted with 9 volumes of fresh human serum and incubated at 37°C. Fresh human PBMCs were added to the particles after various incubation times as indicated. After overnight incubation, the interferon-α (for stimulation with nanoparticles) or TNF-α (for stimulation with microparticles) concentrations in the culture supernatants were measured by ELISA. The data shown are representative of 5 independent experiments.
To estimate particle half-life in vivo, we studied the immunostimulating capacity of protamine-RNA complexes incubated in 90% human serum at 37°C. Using data from several donors, we estimated the half-life of nanoparticles to be approximately 6 hours (Figure 4B), whereas the half-life of microparticles was approximately 1 hour (Figure 4C). The extended half-life of microparticles in serum at 37°C versus in simple buffer at room temperature (∼ 1 hour vs < one-half hour) could be due to serum viscosity preventing fast precipitation of the microparticles, leaving them free longer to stimulate immune cells.
Deciphering the mechanisms of differential immunostimulation by nano- and microparticles
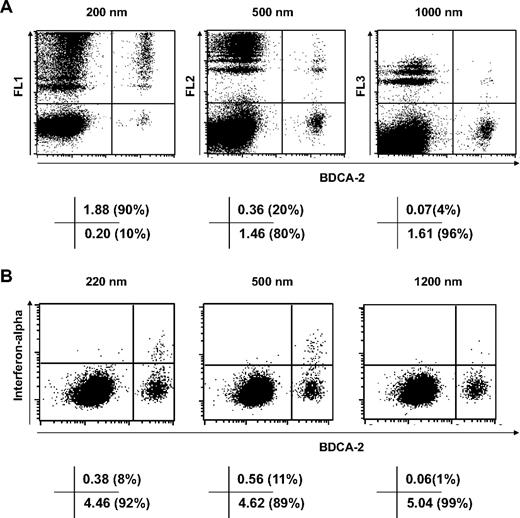
We studied which populations of cells in PBMCs take up protamine-RNA particles using fluorescent nanospheres of 200, 500, and 1000 nm. The results shown in Figures 5 and 6 demonstrate that pDCs, the main producers of interferon-α, efficiently phagocytose nanoparticles but take up microparticles far less efficiently (Figure 5A). Meanwhile, CD11c+ cells, which make up over 85% of the monocyte population, take up particles of all tested sizes with similar efficiency (Figure 6A).
Selective phagocytosis of nanoparticles by pDCs. Fresh human PBMCs were incubated for 4 hours with 10 μg/mL fluorescent nanospheres (A) or for 6 hours with 20 μg/mL protamine-RNA particles (B), then stained with antibody specific for BDCA-2 and, in panel B, anti–interferon-α fluorescein isothiocyanate antibody. Cells were analyzed by fluorescence-activated cell sorting. The value in each quadrant indicates the percentage of cells as a percentage of the parent gate. The data shown are representative of 4 independent experiments.
Selective phagocytosis of nanoparticles by pDCs. Fresh human PBMCs were incubated for 4 hours with 10 μg/mL fluorescent nanospheres (A) or for 6 hours with 20 μg/mL protamine-RNA particles (B), then stained with antibody specific for BDCA-2 and, in panel B, anti–interferon-α fluorescein isothiocyanate antibody. Cells were analyzed by fluorescence-activated cell sorting. The value in each quadrant indicates the percentage of cells as a percentage of the parent gate. The data shown are representative of 4 independent experiments.
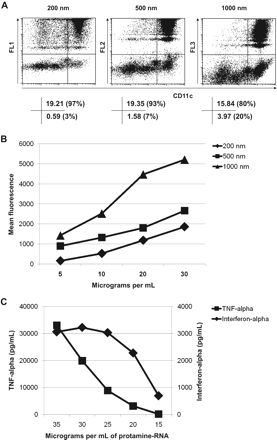
The activation threshold is higher for monocytes than for pDCs. Fresh human PBMCs were incubated for 4 hours with 10 μg/mL fluorescent spheres (A) or 5, 10, 20, or 30 μg/mL fluorescent spheres (B) stained by APC-labeled CD11c and analyzed by fluorescence-activated cell sorting. The dot plots presented in panel A and analyzed in panel B were gated on monocytes on the basis of forward-side light scatter plot (gate r9, supplemental Figure 6A, or gate r3, supplemental Figure 6B). The value in each quadrant in panel A indicates the percentage of cells as a percentage of total cells. In panel B, mean fluorescence (FL1 for 200 nm, FL2 for 500 nm, and FL3 for 1000 nm) of CD11c+ cells at different particle concentrations is shown. (C) TNF-α and interferon-α present in the supernatant of fresh human PBMCs incubated 24 hours with 15, 20, 25, 30, or 35 μg/mL protamine RNA nanoparticles (220 nm) was tested by ELISA. The data shown are representative of 3 independent experiments.
The activation threshold is higher for monocytes than for pDCs. Fresh human PBMCs were incubated for 4 hours with 10 μg/mL fluorescent spheres (A) or 5, 10, 20, or 30 μg/mL fluorescent spheres (B) stained by APC-labeled CD11c and analyzed by fluorescence-activated cell sorting. The dot plots presented in panel A and analyzed in panel B were gated on monocytes on the basis of forward-side light scatter plot (gate r9, supplemental Figure 6A, or gate r3, supplemental Figure 6B). The value in each quadrant in panel A indicates the percentage of cells as a percentage of total cells. In panel B, mean fluorescence (FL1 for 200 nm, FL2 for 500 nm, and FL3 for 1000 nm) of CD11c+ cells at different particle concentrations is shown. (C) TNF-α and interferon-α present in the supernatant of fresh human PBMCs incubated 24 hours with 15, 20, 25, 30, or 35 μg/mL protamine RNA nanoparticles (220 nm) was tested by ELISA. The data shown are representative of 3 independent experiments.
Although more than 90% of pDCs uptake one or (for the majority of cells) several 200-nm particles, only 4% of pDCs cells take up 1000-nm particles (Figure 5A). The 500-nm fluorescent particles are phagocytosed by pDCs less efficiently than 200-nm particles but more efficiently than 1000-nm particles. This selective uptake depending on particle size is mirrored by the intracellular detection of interferon-α in pDCs pulsed with protamine-RNA nano- or microparticles (Figure 5B). Thus the production of interferon-α by PBMCs incubated with nanoparticles but not microparticles of protamine-RNA is dictated by the selective uptake of nanoparticles by pDCs.
In contrast, phagocytosis by monocytic CD11c+ cells is unselective in relation to particle size (Figure 6A). Thus, the higher production of TNF-α triggered by microparticles compared with nanoparticles is not a matter of size-selective phagocytic capacity. The cargo of microparticles is much higher than the cargo of nanoparticles: one 1200-nm particle contains approximately 100 times more material than one 220-nm particle. Thus, we tested whether high load of nanoparticles could mimic the activity of microparticles. As shown in Figure 6B, it is possible to increase the amount of particles taken up by monocytic CD11c+ cells by increasing the amount of fluorescent spheres in the cell culture. This increase is mirrored by the TNF-α production induced by high amounts of nanoparticles (Figure 6C). At a final concentration of 20 μg/mL, the production of interferon-α is nearly maximal, whereas TNF-α production is low. Increasing the concentration of 220-nm particles increases TNF-α production to the level observed using microparticles (> 20 000 pg/mL). Thus the activation threshold of monocytes is higher than the activation threshold of pDCs. The former requires a greater quantity of danger signal (RNA) than the latter for full activation.
RNA characteristics needed for optimal activation by protamine-RNA particles
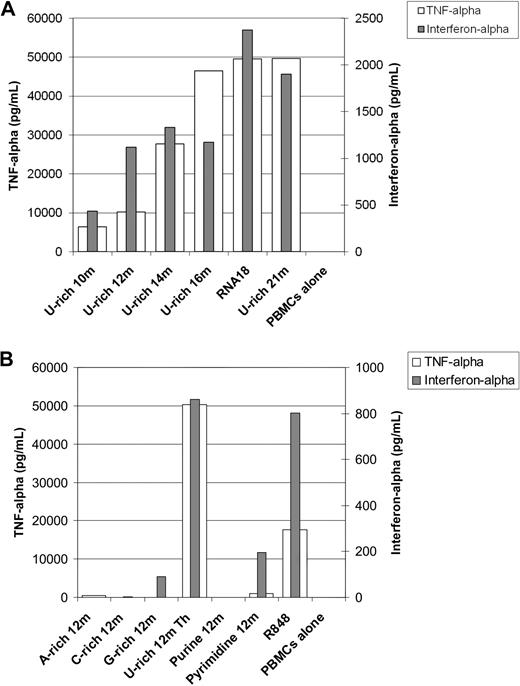
Using 500-nm nanoparticles, we studied the impact of different RNA sequences on both TNF-α and interferon-α production simultaneously in fresh human PBMCs. As shown in Figure 7A, although a short oligonucleotide of 10 residues rich in U bases can induce TNF-α and interferon-α production by PBMCs after it is mixed with protamine, optimal activation was seen with longer oligonucleotides. The 18 residues were enough to stimulate maximum TNF-α and interferon-α production: 21-residue RNA oligonucleotides as well as long in vitro–transcribed RNA (data not shown) did not induce greater production of TNF-α or interferon-α. Using oligonucleotides of 12 residues, we studied the effect of the sequence on immunostimulation. As shown in Figure 7B, U-residues are important for both TNF-α and interferon-α production. However, a polyU RNA oligonucleotide consisting of 12 U-residues alone barely stimulated immune cells (data not shown). G-residues may mediate some immunostimulation: the G-rich 12m oligonucleotide that does not contain U-residues reproducibly induced some detectable cytokine production. However an AG purine oligonucleotide, Purine 12m, did not stimulate human immune cells as measured by cytokine production. The Pyrimidine 12m containing UC repeats induced weak but detectable TNF-α and interferon-α production. In summary, uraciles are the most immunostimulating bases in RNA formulated with protamine. G-residues may be somewhat immunostimulating, whereas A- and C-residues are not immunostimulating.
RNA sequence requirements for immunostimulation by protamine-RNA particles. Large nanoparticles were produced using different RNA oligonucleotides at 1 mg/mL in water, and Protamine 1000 diluted 2.5 times in water. Resulting particles were approximately 500 nm in diameter. Fresh human PBMCs were added and cultured for 18 hours. Supernatants were then tested for TNF-α and interferon-α content by ELISA. (A) The effect of the RNA length on immunostimulation was tested. (B) The effect of the RNA sequence on immunostimulation was tested. Each set of experiments was repeated 3 times. Representative data are shown.
RNA sequence requirements for immunostimulation by protamine-RNA particles. Large nanoparticles were produced using different RNA oligonucleotides at 1 mg/mL in water, and Protamine 1000 diluted 2.5 times in water. Resulting particles were approximately 500 nm in diameter. Fresh human PBMCs were added and cultured for 18 hours. Supernatants were then tested for TNF-α and interferon-α content by ELISA. (A) The effect of the RNA length on immunostimulation was tested. (B) The effect of the RNA sequence on immunostimulation was tested. Each set of experiments was repeated 3 times. Representative data are shown.
Discussion
The capacity of ssRNA and dsRNA to induce nonspecific immune responses including interferon production was described long before TLRs were characterized.17 The potentiation of such phenomena by cationic molecules such as protamine has also been previously demonstrated.7 Having shown in 2004 that protamine-mRNA complexes activate immune cells through receptors of the IL-1 receptor family,8 we also tested the potential for use of those products in the clinic.18,19 However, the reagents and formulation conditions10 resulted in the production of macroscopic heterogenous aggregates (Figure 1). Here, we show that RNA oligonucleotides can be used to formulate homogenous microparticles (Figure 1C). We further show that using RNA and Protamine 5000 (the highest concentration available for pharmaceutical protamine) both diluted to 1 mg/mL in water, we can generate homogenous nanoparticles (Figure 1A). One advantage of particles of less than 500 nm is that they remain polydispersed, ie they do not precipitate spontaneously into a pellet. Thus, they are more readily usable for clinical applications: preparation, short-term storage, uptake in a syringe, and injection. A protamine-RNA mass ratio of between 4:1 and 1:2 does not affect the size of the nanoparticles, but the ratio does affect the resulting surface charge; it is negative at a ratio of 1:2 and slightly positive at a 1:1 ratio or higher. It is preferable for pharmaceutical applications that the charge of a particle is not too highly positive because positively charged particles bind to negatively charged serum protein and generate large aggregates that cause transient embolism in the lung capillaries.16 Microparticles formulated with different protamine-RNA ratios have surface charges similar to those of nanoparticles (data not shown) but, in contrast to nanoparticles, tend to get smaller in size at higher protamine-RNA ratios (Figure 1C). From these data, it can be concluded that for nanoparticles, the 1:1 mass ratio of protamine-RNA may provide complexes with ideal biophysical characteristics. As well as exhibiting favorable immunostimulating capacities (induction of therapeutically relevant cytokines such as interferon-α) and adequate stability and half-life of activity (Figure 4), nanoparticles also activated pDCs from all advanced cancer patients who we tested (data not shown). This suggests the possibility that protamine-RNA nanoparticles could be used as an adjuvant to anticancer vaccines or as a nonspecific immunostimulation regimen for patients suffering from advanced malignant diseases.
Interestingly, we demonstrate here that nanoparticles and microparticles formed with the same RNA and same protamine at equivalent concentrations and ratios, but in different electrolyte conditions (low salt to form nanoparticles, isotonic salts to form microparticles), have different immunostimulating characteristics: nanoparticles but not microparticles induce interferon-α production in human blood cells (Figure 2A). Using fluorescent particles, we could show (Figure 5) that pDCs selectively take up nanoparticles. This size-selective characteristic of phagocytosis in pDCs explains the observed cytokine production pattern induced in these cells by immunostimulating nano- and microparticles.
In contrast, monocytes take up nano- and microparticles similarly (Figure 6A). However, we could show that the threshold of activation of monocytes is higher than the threshold of activation of pDCs (Figure 6C) in terms of the amount of danger signal needed for activation. Thus, monocytes require a higher number of nanoparticles than microparticles for full activation. As a result, at a concentration of 20 μg/mL, microparticles of protamine-RNA but not 220-nm nanoparticles activate maximal TNF-α production by human monocytes.
The immunomodulating capacities of danger signals have not previously been linked to the size of the associated structure. Purified, soluble lipopolysaccharide, CpG ODN, or dsRNA are usually used for in vitro and in vivo experiments. However, these danger signals are physiologically within structures of different sizes: nanostructures (compare, viruses) in the case of dsRNA, nano- or microstructures (compare, viruses and bacteria) in the case of CpG DNA or ssRNA, and microstructures (compare, bacteria) in the case of lipopolysaccharide. For CpG DNA and ssRNA, the structure-related function of the danger signal was largely overlooked, all focus being on how the sequence and modifications of these nucleic acids modulates their immunostimulating activity. Of note, however, the packaging of CpG DNA oligonucleotides in nanoparticles using condensation on protamine20 or adsorption on preformed polystyrene nanoparticles21 was shown to render CpG-B sequences capable of inducing interferon-α production by pDCs, whereas soluble CpG-B sequences did not induce production. In addition, the particularity of CpG-A sequences to induce interferon-α by pDCs was found to be associated with the capacity of this oligonucleotide to spontaneously form nanoparticles.21 According to those studies and our present results, we suggest that the immune system has developed the capacity to specifically react to pathogens according to their size: pDCs selectively take up nanoparticles (viruses), whereas monocytes require a larger amount of danger signal than pDCs to be fully activated. In this way, viruses (typically 5-300 nm) would trigger the potent antivirus interferon-α production, whereas bacteria (typically 500 nm to micrometers) would trigger the potent antibacterial monocytic activation. Both pathogens stimulate mammalian immune systems by the same danger signals, their nucleic acids: CpG motifs and/or unmodified RNA. However, the precise type of immune response is dictated by both the size of the pathogen and its correlate, ie, the amount of danger signal. Aside from pathogens, physiologic DNA- and RNA-containing nanoparticles (exosomes) and microparticles22 are present in plasma. In particular, microparticles shed from the plasma membranes of activated and dying cells may play significant roles in the pathophysiology of thromboses, neuropathologies, inflammation (rheumatic diseases), and cancers.23,24 In light of our results, the particular immunostimulating activities of different disease-associated microparticles can be re-evaluated.
In general, our data invite revision of the activities of danger signals and associated pathogens in view of the size of the original physiologic structure. This revision would lead to a better understanding of the immunomodulating capacities of pathogens or natural nano- and micrometric structures as well as facilitating better design of customized immunotherapy approaches.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Ludwig Institute for Cancer Research, Atlantic Philanthropies, and the Helmut Horten Stiftung.
Authorship
Contribution: L.R. performed in vitro immunologic experiments; S.P.H. performed in vitro immunologic experiments; A.G.B. performed the electron microscopy studies; L.v.B. performed the Multiplex; A.C. obtained informed consent and drew blood; S.D.K. supervised the studies with Zetasizer; A.K. supervised the study; and S.P. designed and supervised the study and performed in vitro experiments.
Conflict-of-interest disclosure: A.K. and S.P. are inventors of a filed patent related to the utilization of protamine-RNA particles for pharmaceutical use. The remaining authors declare no competing financial interests.
The current affiliation for S.P.H. is Department for Hematology, Oncology, Immunology, Rheumatology, and Pulmonology, University Hospital of Tübingen, Tübingen, Germany.
Correspondence: Steve Pascolo, Department of Oncology, University Hospital of Zürich, Haeldeliweg 4, 8044 Zürich, Switzerland; e-mail: steve.pascolo@usz.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal