Abstract
Although the effects of human leukocyte antigen (HLA) locus matching on clinical outcome in unrelated hematopoietic stem cell transplantations have been characterized, the biologic implications of HLA haplotypes have not been defined. We demonstrated the genetic fixity of Japanese conserved extended haplotypes by multi–single nucleotide polymorphism analysis in 1810 Japanese donor-recipient pairs matching with HLA-A, -B, -C, -DRB1, and -DQB1 alleles. Three major Japanese conserved extended haplotypes (named HP-P1, HP-P2, and HP-P3) were essentially completely conserved at least in the 3.3-Mb HLA region from HLA-A to -DPB1, and extended far beyond HLA-A. The risk of acute graft-versus-host disease (GVHD) of these HLA haplotypes was assessed with multivariate Cox regression in 712 patients transplanted from HLA fully (HLA-A, B, C, DRB1, DQB1, and DPB1) matched unrelated donors. HP-P2 itself reduced the risk of grade 2 to 4 acute GVHD (hazard ratio [HR] = 0.63; P = .032 compared with HP-P2-negative), whereas HP-P3 tended to increase the risk (HR = 1.38; P = .07). Among 381 patients with HP-P1, HP-P1/P3 (HR = 3.35; P = .024) significantly increased the risk of acute GVHD compared with homozygous HP-P1. This study is the first to demonstrate that a genetic difference derived from HLA haplotype itself is associated with acute GVHD in allogeneic hematopoietic stem cell transplantation.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) from a human leukocyte antigen (HLA)–matched unrelated (UR) donor has been established as a mode of curative therapy for hematologic malignancies and other hematologic or immunologic disorders, when an HLA-identical sibling donor is unavailable. Although the effect of donor and recipient HLA locus matching on the clinical outcome of UR-HSCT has been well elucidated,1-4 the biologic implications of HLA haplotype itself have not been explored for HSCT.
HLA-identical sibling shares 2 identical major histocompatibility complex (MHC) haplotypes by descent, including non-HLA polymorphic genes, and it has been generally accepted that transplantation between these related pairs provides a superior outcome. On the other hand, there is no guarantee of matching for non-HLA genes between HLA-allele matched UR donor and recipient pairs, and mismatching of haplotype block in MHC has been suggested to lead to severe acute GVHD5 and an inferior outcome6,7 in UR-HSCT.
In human population, multiple DNA blocks in the MHC are strongly associated with each other, and these relatively long stretches of conserved DNA sequence in the MHC have been named conserved extended haplotypes (CEHs) or ancestral haplotypes.8,9 CEHs are often population-specific and have been investigated as markers for disease susceptibility, particularly in autoimmune diseases.10 However, the relation between CEHs and clinical outcome of UR-HSCT has not been yet reported.
Using the large-scale Japan Marrow Donor Program (JMDP) data, we evaluated the conservation of common HLA haplotypes among a Japanese population and elucidated its impact on acute graft-versus-host disease (GVHD) and other clinical outcomes in UR-HSCT.
Methods
Study population
A total of 5210 donor-recipient pairs who underwent transplantation through the JMDP between January 1993 and January 2006 were retrospectively genotyped for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles as described elsewhere.4 For the genome-wide association studies, 1810 pairs (3620 persons) who matched HLA-A, -B, -C, -DRB1, and -DQB1 alleles and were available for DNA sample were selected from these 5210 pairs. For the analysis of acute GVHD, 712 patients who had received T cell–replete bone marrow from an HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 allele-matched donor were selected from these 5210 pairs. The characteristics of these 712 patients are shown in Table 1. A final clinical survey of the patients was completed by June 2007. Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki, and approval of the study was obtained from the Institutional Review Board of Aichi Cancer Center and JMDP.
Clinical characteristics of patients according to HLA haplotype
| Characteristic . | Haplotype P1 . | Haplotype P2 . | Haplotype P3 . | |||
|---|---|---|---|---|---|---|
| Negative . | Positive . | Negative . | Positive . | Negative . | Positive . | |
| No. of patients | 381 | 331 | 601 | 111 | 608 | 104 |
| Median patient age, y (range) | 33 (0-70) | 33 (1-68) | 33 (0-70) | 35 (1-65) | 33 (1-68) | 36 (0-70) |
| Sex (donor/ patient) | ||||||
| Male/male | 160 | 152 | 250 | 62 | 274 | 38 |
| Male/female | 71 | 63 | 117 | 17 | 112 | 22 |
| Female/male | 66 | 49 | 103 | 12 | 100 | 15 |
| Female/female | 84 | 67 | 131 | 20 | 122 | 29 |
| Disease | ||||||
| ALL | 91 | 71 | 138 | 24 | 144 | 18 |
| ANLL | 90 | 106 | 167 | 29 | 166 | 30 |
| CML | 65 | 54 | 95 | 24 | 101 | 18 |
| Hereditary disease | 5 | 9 | 11 | 3 | 12 | 2 |
| MDS | 52 | 39 | 76 | 15 | 78 | 13 |
| Malignant lymphoma | 39 | 30 | 61 | 8 | 52 | 17 |
| Multiple myeloma | 4 | 4 | 7 | 1 | 7 | 1 |
| Severe aplastic anemia | 24 | 9 | 28 | 5 | 31 | 2 |
| Other | 11 | 9 | 18 | 2 | 17 | 3 |
| Risk of leukemia relapse* | ||||||
| Standard | 137 | 112 | 211 | 38 | 214 | 35 |
| High | 109 | 119 | 189 | 39 | 197 | 31 |
| Disease other than leukemia | 135 | 100 | 201 | 34 | 197 | 38 |
| GVHD prophylaxis | ||||||
| Cyclosporine-based | 199 | 203 | 345 | 57 | 339 | 63 |
| Tacrolimus-based | 182 | 128 | 256 | 54 | 269 | 41 |
| ATG | ||||||
| ATG | 25 | 23 | 38 | 10 | 40 | 8 |
| Non-ATG | 356 | 308 | 563 | 101 | 568 | 96 |
| Preconditioning | ||||||
| TBI regimen | 298 | 241 | 456 | 83 | 463 | 76 |
| Non-TBI regimen | 83 | 90 | 145 | 28 | 145 | 28 |
| Characteristic . | Haplotype P1 . | Haplotype P2 . | Haplotype P3 . | |||
|---|---|---|---|---|---|---|
| Negative . | Positive . | Negative . | Positive . | Negative . | Positive . | |
| No. of patients | 381 | 331 | 601 | 111 | 608 | 104 |
| Median patient age, y (range) | 33 (0-70) | 33 (1-68) | 33 (0-70) | 35 (1-65) | 33 (1-68) | 36 (0-70) |
| Sex (donor/ patient) | ||||||
| Male/male | 160 | 152 | 250 | 62 | 274 | 38 |
| Male/female | 71 | 63 | 117 | 17 | 112 | 22 |
| Female/male | 66 | 49 | 103 | 12 | 100 | 15 |
| Female/female | 84 | 67 | 131 | 20 | 122 | 29 |
| Disease | ||||||
| ALL | 91 | 71 | 138 | 24 | 144 | 18 |
| ANLL | 90 | 106 | 167 | 29 | 166 | 30 |
| CML | 65 | 54 | 95 | 24 | 101 | 18 |
| Hereditary disease | 5 | 9 | 11 | 3 | 12 | 2 |
| MDS | 52 | 39 | 76 | 15 | 78 | 13 |
| Malignant lymphoma | 39 | 30 | 61 | 8 | 52 | 17 |
| Multiple myeloma | 4 | 4 | 7 | 1 | 7 | 1 |
| Severe aplastic anemia | 24 | 9 | 28 | 5 | 31 | 2 |
| Other | 11 | 9 | 18 | 2 | 17 | 3 |
| Risk of leukemia relapse* | ||||||
| Standard | 137 | 112 | 211 | 38 | 214 | 35 |
| High | 109 | 119 | 189 | 39 | 197 | 31 |
| Disease other than leukemia | 135 | 100 | 201 | 34 | 197 | 38 |
| GVHD prophylaxis | ||||||
| Cyclosporine-based | 199 | 203 | 345 | 57 | 339 | 63 |
| Tacrolimus-based | 182 | 128 | 256 | 54 | 269 | 41 |
| ATG | ||||||
| ATG | 25 | 23 | 38 | 10 | 40 | 8 |
| Non-ATG | 356 | 308 | 563 | 101 | 568 | 96 |
| Preconditioning | ||||||
| TBI regimen | 298 | 241 | 456 | 83 | 463 | 76 |
| Non-TBI regimen | 83 | 90 | 145 | 28 | 145 | 28 |
HLA indicates human leukocyte antigen; ALL, acute lymphoblastic leukemia; ANLL, acute nonlymphoblastic leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; GVHD, graft-versus-host disease; ATG, anti–human thymocyte globulin; and TBI, total body irradiation.
Standard risk for leukemia relapse was defined as the status of the first complete remission of ALL and ANLL and the first chronic phase of CML at transplantation. High risk was defined as a more advanced status than standard risk in AML, ANLL, and CML. Disease other than leukemia was defined as other than ALL, ANLL, and CML.
SNP typing and HLA haplotype analysis
The single nucleotide polymorphism (SNP) array experiments were performed according to the standard protocol of Affymetrix GeneChip Mapping 500K Array (Affymetrix). After excluding those SNPs showing less than 95% call rate and deviation from Hardy-Weinberg equilibrium (P < .001), 10.8% of SNPs for the HLA region failed. And 4761 SNPs in the region spanning the MHC (20-46 Mb from the telomere in chromosome 6p) were analyzed to evaluate the conservation of common HLA haplotypes.
Persons who were homozygous for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 were isolated, and the 3 major HLA haplotypes were named HP-P1, HP-P2, and HP-P3. The homozygosity of consecutive SNPs of these HLA haplotypes was analyzed to assess the region of conservation. SNP alleles of the extended homozygous region in each HLA haplotype were analyzed to determine allele frequencies, and a consensus sequence of major alleles in each haplotype was established. Then, the SNP sequence of persons who carried at least one copy of HLA haplotype (shared the same HLA alleles as common HLA haplotype at the HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 loci) was compared with this consensus sequence. Missing alleles in a sample were not accounted for in this analysis.
Statistical analysis
Cumulative incidences of acute GVHD and relapse were assessed by the method described elsewhere to eliminate the effect of competing risk.4 Overall survival was calculated using the Kaplan-Meier method. The competing event regarding acute GVHD was defined as death without acute GVHD. A log-rank test was applied to assess the impact by the factor of interest. Multivariable Cox regression analyses were conducted to evaluate the impact of the specific haplotype on acute GVHD, leukemia relapse, and mortality after transplantation. Confounders considered were sex (donor-recipient pair), patient age (linear), donor age (linear), transplantation year, type of disease, risk of leukemia relapse (standard, high, and diseases other than leukemia), GVHD prophylaxis (cyclosporine-based regimen vs tacrolimus-based regimen), anti–thymocyte globulin (anti–thymocyte globulin vs no anti–thymocyte globulin), and preconditioning (total body irradiation vs non–total body irradiation).
Results
Highly conserved common HLA haplotypes among Japanese
To evaluate for conservation of Japanese common HLA haplotypes, persons who were homozygous HLA haplotype (having homozygous alleles in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 loci) were selected from a total 3620 persons (1810 donor-recipient pairs) for whom genome-wide association study was performed. Among those, 72 persons were homozygous HLA-A*2402 -Cw*1202 -B*5201 -DRB1*1502 -DQB1*0601 -DPB1*0901 (named HP-P1), 10 persons were homozygous HLA-A*3303 -Cw*1403 -B*4403 -DRB1*1302 -DQB1*0604 -DPB1*0401 (named HP-P2), and 8 persons were homozygous HLA-A*2402 -Cw*0702 -B*0702 -DRB1*0101 -DQB1*0501 -DPB1*0402 (named HP-P3).
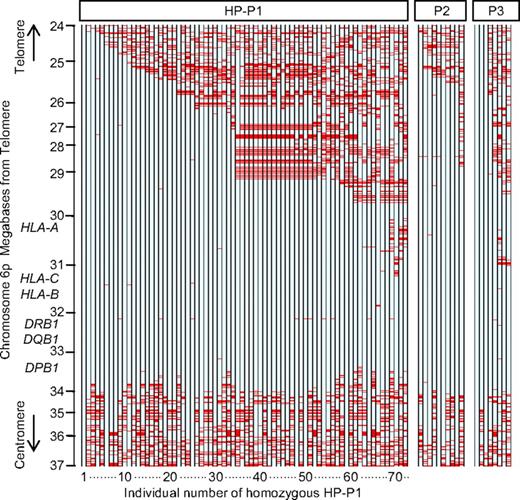
Homozygosity at consecutive SNP loci of persons with homozygous HLA haplotype was shown in Figure 1. The extended homozygous region of HP-P1 gradually broke up the region of HLA-A. The longest homozygous region in persons with HP-P1 was 18.7 Mb. Of 72 persons with homozygous HP-P1, 32 persons (nos. 1-32 of HP-P1 in Figure 1) had more than 99.0% of 1395 consecutive homozygous SNPs throughout the 6.9-Mb region from rs806971 to rs6937061 (nucleotides 26252770-33187790). Although haplotypes of all those 32 persons were identical centromeric rs9257745 (nucleotide 29414635), the telomeric region was clearly divided into 2 different haplotypes (Figure 2). A total of 26 of 32 persons had one of the homozygous haplotypes (named subtype A of HP-P1), and the remaining 6 persons (nos. 3, 9, 19, 23, 24, and 25 of HP-P1 in Figure 1) had another homozygous haplotype (subtype B of HP-P1). A total of 65 of 72 persons with homozygous HP-P1 (nos. 1-65 of HP-P1 in Figure 1) had more than 99.0% homozygous alleles for 888 consecutive SNPs throughout the 3.3-Mb region from rs1610630 to rs6937061 (nucleotides 29837265-33187790). Seven other persons (nos. 66-72 in Figure 1) had an apparently lower conserved region, with 1.0% to 10.0% heterozygous alleles within the 3.3-Mb region.
Representation of genotypes in persons with homozygous HLA haplotype. Data from chromosome 6p (24-37 Mb) of persons with homozygous HLA haplotype are shown. Each column indicates 1 person (72 persons with homozygous HP-P1, 10 persons with homozygous HP-P2, and 8 persons with homozygous HP-P3). Each of the 2389 evenly spaced rows represents 1 SNP locus. Blue row represents homozygous genotype; and red row, heterozygous genotype. Missing genotypes were not counted.
Representation of genotypes in persons with homozygous HLA haplotype. Data from chromosome 6p (24-37 Mb) of persons with homozygous HLA haplotype are shown. Each column indicates 1 person (72 persons with homozygous HP-P1, 10 persons with homozygous HP-P2, and 8 persons with homozygous HP-P3). Each of the 2389 evenly spaced rows represents 1 SNP locus. Blue row represents homozygous genotype; and red row, heterozygous genotype. Missing genotypes were not counted.
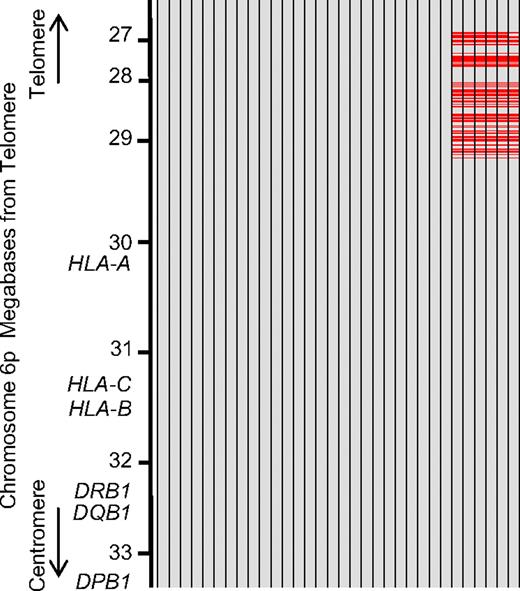
Subtypes of HLA haplotype P1. Data from chromosome 6p (from nucleotides 26252770-33187790) of 32 persons with consecutive homozygous SNPs throughout the 6.9-Mb region. The SNP sequence of persons was compared with consensus sequence across the 6.9-Mb region. Each column indicates 1 person. Each of 1395 evenly spaced rows represents 1 SNP locus. Gray row represents SNPs identical to the consensus alleles; and red row, SNPs different from the consensus alleles. Missing SNPs were not counted. A total of 26 of 32 persons had alleles identical to consensus alleles across 6.9 Mb (subtype A of HP-P1), whereas the remaining 6 persons had apparently different alleles in the telomeric region from nucleotide 29414635 (subtype B of HP-P1). These data indicated that the telomeric region of HP-P1 was clearly divided into 2 different haplotypes.
Subtypes of HLA haplotype P1. Data from chromosome 6p (from nucleotides 26252770-33187790) of 32 persons with consecutive homozygous SNPs throughout the 6.9-Mb region. The SNP sequence of persons was compared with consensus sequence across the 6.9-Mb region. Each column indicates 1 person. Each of 1395 evenly spaced rows represents 1 SNP locus. Gray row represents SNPs identical to the consensus alleles; and red row, SNPs different from the consensus alleles. Missing SNPs were not counted. A total of 26 of 32 persons had alleles identical to consensus alleles across 6.9 Mb (subtype A of HP-P1), whereas the remaining 6 persons had apparently different alleles in the telomeric region from nucleotide 29414635 (subtype B of HP-P1). These data indicated that the telomeric region of HP-P1 was clearly divided into 2 different haplotypes.
All 10 persons with homozygous HP-P2 had more than 99.0% homozygous alleles for consecutive SNPs throughout the 3.3-Mb region. Furthermore, 9 of 10 persons with homozygous HP-P2 showed homozygosity extending across the 7.7-Mb region from rs6912426 to rs6937061 (nucleotides 25517764-33187790), and those 9 persons had identical genotypes in almost all the 1540 consecutive SNPs throughout the 7.7-Mb region.
Among 8 persons with homozygous HP-P3, 5 persons had more than 99.0% homozygous alleles throughout the 3.3-Mb region, and the other 3 persons had 1.7% to 8.0% heterozygous alleles within that region. One person with homozygous HP-P3 showed an extraordinary long stretch of homozygosity across the 25.4-Mb region (nucleotides 20162518-45595922).
Consensus sequence of each haplotype was determined using analysis of persons who were homozygous for almost all the SNPs, ie, 6.9-Mb region in HP-P1 (subtypes A and B), 7.7-Mb region in HP-P2, and 3.3-Mb region HP-P3. The person with the longest homozygous telomeric region of HLA-A served to determine a further extended haplotype (Table 2). These persons had alleles identical to the consensus sequence of major alleles in each haplotype described above in this section.
Longest homozygous region of common HLA haplotype
| Type . | Start of homozygous region . | End of homozygous region . | Region length, Mb . | ||
|---|---|---|---|---|---|
| SNP . | Position, kb . | SNP . | Position, kb . | ||
| HP-P1 | |||||
| Subtype A | rs199026 | 23441.813 | rs2395801 | 42231.646 | 18.8 |
| Subtype B | rs573863 | 24080.365 | rs1536501 | 33835.863 | 9.7 |
| HP-P2 | rs1175427 | 24387.055 | rs1873254 | 34134.467 | 9.7 |
| HP-P3 | rs1688325 | 20162.518 | rs6905847 | 45595.922 | 25.4 |
| Type . | Start of homozygous region . | End of homozygous region . | Region length, Mb . | ||
|---|---|---|---|---|---|
| SNP . | Position, kb . | SNP . | Position, kb . | ||
| HP-P1 | |||||
| Subtype A | rs199026 | 23441.813 | rs2395801 | 42231.646 | 18.8 |
| Subtype B | rs573863 | 24080.365 | rs1536501 | 33835.863 | 9.7 |
| HP-P2 | rs1175427 | 24387.055 | rs1873254 | 34134.467 | 9.7 |
| HP-P3 | rs1688325 | 20162.518 | rs6905847 | 45595.922 | 25.4 |
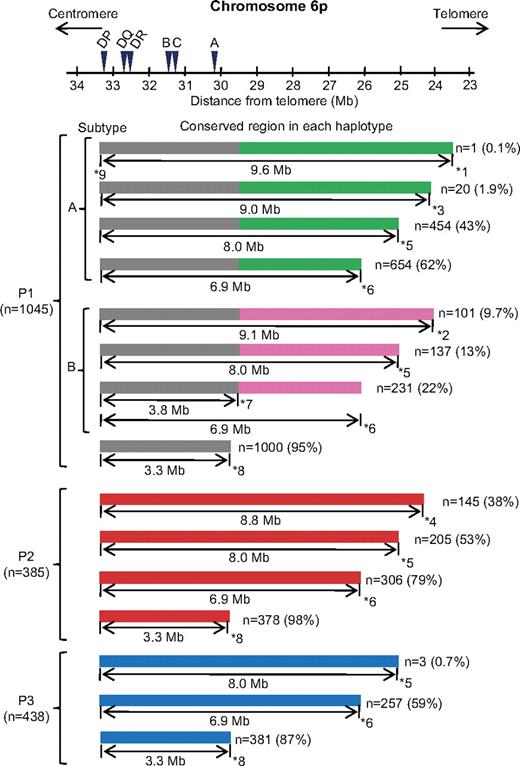
We ascertained whether the consensus sequence of each HLA haplotype was present in the persons carrying at least one copy of HLA haplotype, that is, sharing the same HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles of common HLA haplotypes (Figure 3; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Among 3620 persons analyzed for multi-SNP, 1000 of 1045 persons (95%) with HP-P1 had identical alleles for more than 99.5% of consecutive 888 SNPs as a consensus sequence across 3.3-Mb region from 181 kb telomeric HLA-A to 25 kb centromeric HLA-DPB1. Furthermore, 654 of 1045 (62%) persons with HP-P1 had identical alleles for more than 99.5% of consecutive 1395 SNPs as subtype A of HP-P1 across the 6.9-Mb region, and 231 persons (22%) had identical alleles as subtype B of HP-P1 across the 6.9-Mb region. Fewer persons showed the conserved region extending up to 9.0 Mb. Among 385 persons with HP-P2, 378 (98%) had identical alleles for 888 consecutive SNPs across the 3.3-Mb region. Furthermore, 305 (79%) had identical alleles for more than 99.5% of 1395 consecutive SNPs as a consensus sequence across the 6.9-Mb region, and 205 (53%) also did across the 8.0-Mb region (nucleotides 24111433-33187790). Among 438 persons with HP-P3, 381 (87%) had identical alleles across the 3.3-Mb region, and 257 (59%) had identical alleles for consecutive SNPs as a consensus sequence of HP-P3 across the 6.9-Mb region.
Conservation of common HLA haplotypes. The SNP sequence of persons who carried at least 1 copy of HLA haplotype (shared the same HLA alleles as common HLA haplotype) was compared with consensus sequence of common HLA haplotypes, and conserved regions in each HLA haplotype were illustrated schematically. The majority of persons who share the same HLA alleles in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 as common HLA haplotypes possess at least a 3.3-Mb conserved region from HLA-A to DPB1. HP-P1 splits into the A and B subtypes. The length of conservation in HP-P1, HP-P2, and HP-P3 is more extensive to the telomeric region of HLA-A. ID and position of SNPs are: *1 rs199026 (nucleotide 23551813), *2 rs573863 (nucleotide 24080365), *3 rs1397843 (nucleotide 24111433), *4 rs11754278 (nucleotide 24387055), *5 rs303031 (nucleotide 25144959), *6 rs806971 (nucleotide 26252770), *7 rs9257745 (nucleotide 29414635), *8 rs1610630 (nucleotide 29837265), and *9 rs6937061 (nucleotide 33187790).
Conservation of common HLA haplotypes. The SNP sequence of persons who carried at least 1 copy of HLA haplotype (shared the same HLA alleles as common HLA haplotype) was compared with consensus sequence of common HLA haplotypes, and conserved regions in each HLA haplotype were illustrated schematically. The majority of persons who share the same HLA alleles in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 as common HLA haplotypes possess at least a 3.3-Mb conserved region from HLA-A to DPB1. HP-P1 splits into the A and B subtypes. The length of conservation in HP-P1, HP-P2, and HP-P3 is more extensive to the telomeric region of HLA-A. ID and position of SNPs are: *1 rs199026 (nucleotide 23551813), *2 rs573863 (nucleotide 24080365), *3 rs1397843 (nucleotide 24111433), *4 rs11754278 (nucleotide 24387055), *5 rs303031 (nucleotide 25144959), *6 rs806971 (nucleotide 26252770), *7 rs9257745 (nucleotide 29414635), *8 rs1610630 (nucleotide 29837265), and *9 rs6937061 (nucleotide 33187790).
These results indicate that most of the persons with a common HLA haplotype had a conserved region at least 3.3 Mb from HLA-A to HLA-DPB1. Furthermore, a considerable number of unrelated persons with common HLA haplotype had a more extended conserved telomeric region of HLA-A.
Effect of HLA haplotype on acute GVHD
To elucidate the effect of specific HLA haplotype on acute GVHD, we analyzed 712 patients who underwent transplantation from HLA fully matched (12 of 12 HLA alleles) donor with T cell-replete marrow (Table 1). We excluded HLA-mismatched transplantation to avoid obscuring the relationship between HLA haplotype itself and acute GVHD by powerful allogeneic immune responses caused by HLA allele disparities. Among those patients, 331 (46.4%) had HP-P1, 111 (15.0%) had HP-P2, and 104 (14.6%) had HP-P3.
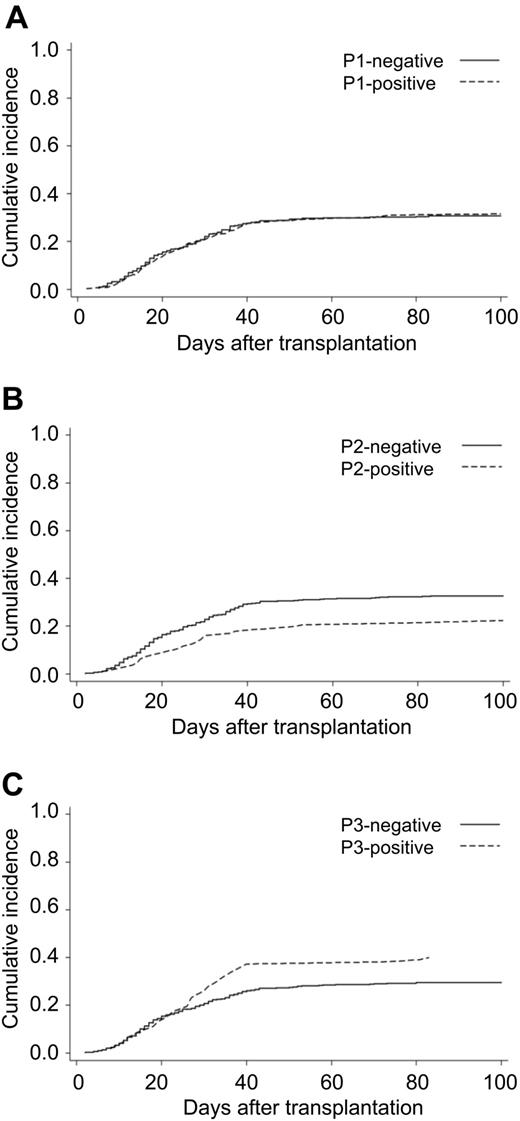
At first, grade 2 to 4 acute GVHD in patients with specific haplotype was compared with those without specific haplotype using multivariate analysis (Table 3). There was no significant difference in the hazard ratio (HR) in grades 2 to 4 acute GVHD between HP-P1-positive and -negative patients, and also no significant difference in the cumulative incidence of acute GVHD between HP-P1-positive and HP-P1-negative patients (31.5% vs 30.7%; Figure 4). Of note, HR of grades 2 to 4 acute GVHD in HP-P2-positive patients was 0.63 (95% confidence interval [CI], 0.41-0.96, P = .032) compared with HP-P2-negative patients, and the cumulative incidence of acute GVHD in patients with HP-P2 was significantly lower than HP-P2-negative patients (22.3% vs 33.7%, P = .031; Figure 4). On the other hand, a trend of increasing risk of acute GVHD was observed in HP-P3-positive patients (HR = 1.38; 95% CI, 0.97-1.95; P = .07). The cumulative incidence of acute GVHD in HP-P3-positive patients was 39.2% and HP-P3-negative patients 29.5% (P = .064; Figure 4).
Hazard ratio of HLA haplotype on acute GVHD (grade 2-4)
| HLA haplotype . | Negative/positive . | No. . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| P1 | Negative | 381 | Referent 1.00 | |
| P1 | Positive | 331 | 1.06 (0.81-1.39) | .665 |
| P2 | Negative | 601 | Referent 1.00 | |
| P2 | Positive | 111 | 0.63 (0.41-0.96) | .032 |
| P3 | Negative | 608 | Referent 1.00 | |
| P3 | Positive | 104 | 1.38 (0.97-1.95) | .07 |
| P1/P1 | 36 | Referent 1.00 | ||
| P1/P2 | 25 | 0.71 (0.17-2.93) | .64 | |
| P1/P3 | 19 | 3.35 (1.18-9.55) | .024 | |
| P1/other | 251 | 2.49 (1.06-5.85) | .036 |
| HLA haplotype . | Negative/positive . | No. . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| P1 | Negative | 381 | Referent 1.00 | |
| P1 | Positive | 331 | 1.06 (0.81-1.39) | .665 |
| P2 | Negative | 601 | Referent 1.00 | |
| P2 | Positive | 111 | 0.63 (0.41-0.96) | .032 |
| P3 | Negative | 608 | Referent 1.00 | |
| P3 | Positive | 104 | 1.38 (0.97-1.95) | .07 |
| P1/P1 | 36 | Referent 1.00 | ||
| P1/P2 | 25 | 0.71 (0.17-2.93) | .64 | |
| P1/P3 | 19 | 3.35 (1.18-9.55) | .024 | |
| P1/other | 251 | 2.49 (1.06-5.85) | .036 |
Multivariate analysis adjusted by clinical factors (see Table 2 and supplemental Table 1).
Cumulative incidence of grade 2 to 4 acute GVHD by common HLA haplotype. The endpoint was the time to diagnosis of grade 2 to 4 acute GVHD with censoring of date of death until 100 days after transplantation. P value was calculated with the log-rank test. (A) Patients with or without HP-P1. (B) Patients with or without HP-P2. (C) Patients with or without HP-P3.
Cumulative incidence of grade 2 to 4 acute GVHD by common HLA haplotype. The endpoint was the time to diagnosis of grade 2 to 4 acute GVHD with censoring of date of death until 100 days after transplantation. P value was calculated with the log-rank test. (A) Patients with or without HP-P1. (B) Patients with or without HP-P2. (C) Patients with or without HP-P3.
A total of 331 patients with HP-P1 in HLA fully matched transplantation made it possible to elucidate the effect of another HLA haplotype on acute GVHD (supplemental Table 1 for patient characteristics). We did not determine unique haplotypes other than HP-P1, HP-P2, and HP-P3 in this study, so all the persons with HP-P1 and unknown haplotype were lumped together with HP-P1/other. The incidence of grade 2 to 4 acute GVHD in patients with HP-P1/P3 (49.9%) was significantly higher than those with homozygous HP-P1 (16.0%), and there was no significant difference between patients with homozygous HP-P1 and those with HP-P1/P2 (12.0%; Figure 5). Multivariate analyses of HR for acute GVHD showed the same results (Table 3). There was no significant difference in the risk of acute GVHD between patients with homozygous HP-P1 and those with HP-P1/P2 (P = .64). Compared with patients with homozygous HP-P1, patients with HP-P1/P3 (HR = 3.35; 95% CI, 1.18-9.55; P = .024) had a significantly higher risk of acute GVHD.
Cumulative incidence of grade 2 to 4 acute GVHD by common HLA haplotype in patients with HP-P1. The endpoint was the time to the diagnosis of grade 2 to 4 acute GVHD with censoring of date of death until 100 days after transplantation. P value was calculated with the log-rank test. A total of 331 patients with HP-P1 were analyzed for the effect of another HLA haplotype by HP-P1, HP-P2, HP-P3, and the other haplotypes, which were lumped together.
Cumulative incidence of grade 2 to 4 acute GVHD by common HLA haplotype in patients with HP-P1. The endpoint was the time to the diagnosis of grade 2 to 4 acute GVHD with censoring of date of death until 100 days after transplantation. P value was calculated with the log-rank test. A total of 331 patients with HP-P1 were analyzed for the effect of another HLA haplotype by HP-P1, HP-P2, HP-P3, and the other haplotypes, which were lumped together.
As for grade 3 to 4 acute GVHD, there were no significant differences between HP-positive and -negative patients. When 331 patients with HP-P1 in HLA fully match transplantation were analyzed, grade 3 to 4 acute GVHD showed the same tendency with grade 2 to 4 acute GVHD. Incidence in patients with homozygous HP-P1 was 2.7%, HP-P1/P2 8.0%, and HP-P1/P3 23.6%.
The cumulative incidence of relapse showed a higher trend in HP-P2-positive patients compared with -negative patients (37.3% vs 29.6%, P = .051), and HR was 1.34 (95% CI, 0.94-1.91, P = .108). There were no significant differences in the relapse rate between HP-P1-positive and -negative patients, and also between HP-P3 -positive and -negative patients.
Overall survival showed no significant differences between HP-positive and -negative patients. When 331 patients with HP-P1 in HLA fully matched transplantation were analyzed, HR of mortality in HP-P1/P3 was 2.03 (95% CI, 0.92-4.49 P = .08) compared with patients with homozygous HP-P1, and HR of HP-P1/P2 1.69 (95% CI, 0.78-3.70, P = .186).
Discussion
First, we demonstrated that Japanese common HLA haplotypes were extraordinarily conserved. Preferential selection of HLA-A–, -B–, and -DR–matched donor through JMDP made it easy to identify a considerable number of persons with homozygous common HLA haplotype extending HLA-DPB1. HP-P1, HP-P2, and HP-P3 have been previously reported as common HLA haplotypes in the Japanese population.11-13 The haplotype frequency of HP-P1 was 0.054 to 0.062, that of HP-P2 was 0.029 to 0.036, and that of HP-P3 was 0.016 to 0.040.
CEHs have been mainly identified by blocks of fragment in the MHC region, such as complement genes, alleles of HLA class I /II gene, and tumor necrosis factor-α gene. Recently, high-density SNP analysis in the HLA region made it possible to confirm the genetic fixity of HLA haplotype.14-16
We determined the consensus sequence of these HLA haplotypes using multi-SNP data of unrelated persons with homozygous HLA haplotype, and the majority of persons who share the same HLA alleles in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 as common HLA haplotypes possess at least a 3.3-Mb conserved region from HLA-A to -DPB1. Furthermore, we showed that those Japanese common HLA haplotypes extend far beyond the HLA-A. We also found, for the first time, that HP-P1 was divided into 2 subtypes based on the telomeric region from nucleotide 29414635 (subtypes A and B). The A1-B8-DR3 CEH, which is one of the most frequent haplotypes in northern European populations, showed that the region of conservation extended 6 Mb telomeric to HLA-A.16,17 Caucasian common HLA haplotypes have often reportedly shown a lack of nonrandom association between HLA-DR, -DQ, and -DP,18 whereas Japanese common HLA haplotypes have been subdivided into haplotypes with only a very limited number of HLA-DPB1.11,12 The highly conserved HLA haplotype that extends to HLA-DPB1 might be attributable to ethnic isolation in the Japanese.
For the analysis of comparison between haplotype-positive and -negative patients (Figure 4; Table 3), patients with HP-P2 significantly reduced the risk of acute GVHD. On the other hand, patients with HP-P3 showed a tendency to increase the risk. Although the relapse rate showed a higher trend in patients with HP-P2 compared with HP-P2-negative patients, the grade 3 to 4 acute GVHD and overall survival did not differ between HP-P2-positive and -negative patients, nor between HP-P3-positive and -negative patients. We suspected the differences with weak power might be attributed to the effects of various other haplotypes combined with a particular haplotype. To confirm the differences in the effects on acute GVHD among a particular HLA haplotype, we analyzed the effect of another haplotype on acute GVHD among patients with HP-P1 (Figure 5). Patients with HP-P1/P3 showed a significantly higher risk of grade 2 to 4 acute GVHD compared with patients with homozygous HP-P1, a tendency to increase the incidence of grade 3 to 4, and also a tendency to decrease overall survival. Patients with homozygous HP-P1 and patients with HP-P1/P2 showed an extremely lower incidence of grade 2 to 4 acute GVHD (16.2% and 12.0%). Therefore, we could not detect any difference between HP-P1 and HP-P2.
Thus, we demonstrated, for the first time, that the HLA haplotype itself affected the occurrence of acute GVHD. These findings suggest that the genetic factor of specific haplotypes would contribute to reducing or increasing the risk of acute GVHD. Alternatively, we should consider the effect of donor and recipient mismatch SNPs because HP-P3 showed more variation than HP-P1 or HP-P2.
There are several possible explanations for the reduced or increased risk of acute GVHD in patients with the specific haplotype. In HSCT, GVHD has been known to result mainly from donor T cells recognizing minor histocompatibility antigens presented on HLA molecules of a recipient's organs.19 Different HLA haplotypes possess a different combination of HLA alleles. Therefore, various HLA alleles in each haplotype might present different immunodominant peptides to T cells and evoke different alloreactivity in HLA-matched UR-HSCT. Presumably, the critical but as yet unidentified minor histocompatibility antigens linked to major histocompatibility antigen should be explored based on HLA haplotype, such as common gene deletion polymorphisms.20 Distinct forms of GVHD were found in different MHC haplotypes in mice, and it has been argued that genes in the MHC locus can dominantly determine the forms of GVHD, probably through MHC-based selection of immunodominant antigens.21
In human retrospective analysis, several single-center studies have shown a reduced incidence of acute GVHD,22 reduced relapse rate,23 and improved overall survival24 for HLA-DR15-positive patients transplanted from HLA-matched donors. HLA-DR15 has been known to be a marker of disease susceptibility and clinical response to immunosuppressive therapy in autoimmune-mediated bone marrow failure,25,26 and it is speculated that immune responses specific to HLA-DR15 are induced. However, our analysis of the effect of HLA-DR15 using the same database showed no effect on acute GVHD (data not shown).
HLA haplotype serves as a model system for studies of disease association, especially in autoimmune disease or infection, and several candidate genes in the HLA region associated with specific haplotype have the potential to modulate immune or inflammatory responses.10 The observed effect of HLA haplotypes on GVHD development could be explained by particular SNPs that are closely associated with those HLA haplotypes. Within the region of conserved HLA haplotypes, there exist several candidate genes whose SNPs may be related to immune responses. Genetic variants of tumor necrosis factor-α gene located in the HLA region might influence the risk of developing GVHD.27,28 In addition, TAP1/TAP2 and LMP2/LMP7 genes encode subunit components of the proteasomes implicated in the processing of class I HLA-bound peptides,29,30 and polymorphisms of these genes may affect antigen presentation on recipient tissues, leading to different susceptibility to GVHD. However, they probably do not explain the observed effects of haplotypes; correlations of each haplotype with known SNPs in these genes are generally weak, although D′ among these alleles is high (> 0.98). Moreover, currently no haplotype-specific non-HLA polymorphisms have been identified in our series, although we could not exclude the possibility that there may exist some nonobserved SNPs that are closely associated with relevant HLA haplotypes.
We showed that Japanese common HLA haplotypes were conserved from HLA-DPB1 to extensively telomeric HLA-A, so it might be possible that the responsible gene is located in the telomeric region of the classic HLA. Interestingly, HP-P1 was divided into 2 subtypes based on the telomeric region from nucleotide 29414635 (subtypes A and B). Although we analyzed the effect of those subtypes on grade 2 to 4 acute GVHD among patients with HP-P1, we could not detect significant differences between patients with HP-P1 subtype A and subtype B. We also could not detect the differences between patients transplanted from a donor with HP-P1 subtype A and subtype B (data not shown).
In conclusion, in the present study, we demonstrated that highly conserved HLA haplotype might contribute to the occurrence of acute GVHD in HSCT. Although the clinical implications of our results should be considered cautiously, these results imply the proof of principle for an association between one or another HLA haplotype and GVHD. Our findings on the conservation of the MHC region may also provide background for exploring not only genetic factors associated with acute GVHD but also genetic disease susceptibility in our population. More extensive studies are warranted to identify specific genes associated with a particular haplotype contributing to acute GVHD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff members of the transplantation centers, donor centers, and the Japan Marrow Donor Program office for their generous cooperation.
This work was supported in part by the Research on Allergic Disease and Immunology (Health and Labor Science Research Grant), the Ministry of Health, Labor, and Welfare of Japan (Grant-in-Aid for Cancer Research), and the Core Research for Evolutional Science and Technology of Japan (Grant-in-Aid).
Authorship
Contribution: S.M., Y.M., S.O., H.S., M.S., H.I., and T.S. participated in the design of this study; S.O., K.K., A.M., and Y.N. performed histocompatibility analysis; Y.M., S.K., and Y.K. organized data collection for transplantation; T.K. performed statistical data analysis; S.M. and Y.M. performed analysis and wrote this paper; and all authors checked the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seishi Ogawa, The 21st Century COE Program, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo Bunkyo-ku, Tokyo, 113-0033 Japan; e-mail: sogawa-tky@umin.net; or Yasuo Morishima, Department of Hematology and Cell Therapy, Aichi Cancer Center Hospital, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan; e-mail: ymorisim@aichi-cc.jp.
References
Author notes
S.M. and S.O. contributed equally to this paper.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal