Abstract
Peripheral T-cell lymphoma (PTCL) is often challenging to diagnose and classify. Gene expression profiling was performed on 144 cases of PTCL and natural killer cell lymphoma and robust molecular classifiers were constructed for angioimmunoblastic T-cell lymphoma (AITL), anaplastic lymphoma kinase-positive (ALK+) anaplastic large-cell lymphoma (ALCL), and adult T-cell leukemia/lymphoma. PTCL-unclassifiable was molecularly heterogeneous, but we were able to identify a molecular subgroup with features of cytotoxic T lymphocytes and a poor survival compared with the remaining PTCL–not otherwise specified cases. Many of the pathologic features and substantial components of the molecular signature of AITL are contributed by the follicular dendritic cells, B-cell, and other stromal components. The expression of Th17-associated molecules in ALK+ ALCL was noted and may represent aberrant activation of Th17-cell differentiation by abnormal cytokine secretion. Adult T-cell leukemia/lymphoma has a homogeneous molecular signature demonstrating high expression of human T-lymphotropic virus type 1–induced genes. These classifiers reflect the biology of the tumor cells as well as their microenvironment. We also constructed a molecular prognosticator for AITL that appears to be largely related to the microenvironmental signature, and the high expression of 2 immunosuppressive signatures are associated with poor outcome. Oncogenic pathways and tumor-host interactions also were identified, and these findings may lead to better therapies and outcome in the future.
Introduction
Peripheral T-cell lymphoma (PTCL) and natural killer–cell lymphomas (NKCLs) represent approximately 10% to 15% of all non-Hodgkin lymphoma in the western world but occur more frequently in Asia.1 The current World Health Organization classification recognizes several distinctive subtypes of PTCL, including angioimmunoblastic T-cell lymphoma (AITL), anaplastic large-cell lymphoma (ALCL), and adult T-cell leukemia/lymphoma (ATLL), as well as several rare entities that are mostly extranodal.2 Some types of PTCL have a disease-defining abnormality, such as the t(2;5)(p23;q35) in ALCL3 or human T-lymphotropic virus 1 (HTLV1) integration in ATLL.4 However, the classification of PTCL remains challenging, with 30% to 50% of cases classified as PTCL unclassifiable (PTCL-NOS [not otherwise specified]), even with current diagnostic approaches. It is also difficult to classify most cases of PTCL according to the normal stages of T-cell differentiation, and the expression of T-cell subset markers is of limited value in distinguishing clinically distinct entities.5,6 With the exception of ALCL, patients with PTCL generally have a poor prognosis with standard chemotherapy.7
We and others8 have shown that gene expression profiling (GEP) can identify biologically and clinically distinctive subgroups of B-cell non-Hodgkin lymphoma. Several recent studies9-14 of T-cell lymphomas, in which the investigators used small numbers of cases, have suggested that some PTCL subtypes have specific molecular profiles or cellular backgrounds. The cell of origin of AITL is now thought to be the follicular helper T cell (TFH),11,12 and PTCL-NOS has multiple molecular subgroups,10 frequent expression of platelet-derived growth factor receptor-α,15 and characteristics of activated peripheral T lymphocytes.13 The association of a high-proliferation gene signature with a shorter survival also was reported recently in nodal PTCL.14 The authors of recent studies16,17 have reported the adhesion molecule TSLC1 as a possible molecular marker for ATLL and the role of TCF-4 in ATLL cell survival. Molecular studies of anaplastic lymphoma kinase-positive ALCL, or ALK+ ALCL, and anaplastic lymphoma kinase-negative ALCL, or ALK− ALCL, have suggested that some pathogenetic mechanisms may be shared by these 2 entities.18,19 Although these preliminary findings are interesting, these studies were limited by the small number of cases, and a more in-depth molecular analysis of a large series of PTCL is warranted.
In this study, we performed GEP on 144 PTCL and NKCL to define molecular classifiers for the more common entities, to identify unique entities within PTCL-NOS, to elucidate unique tumor and microenvironmental interactions and oncogenic pathways in AITL, and to construct a molecular prognosticator for AITL.
Methods
Tumor specimens and cell lines
The International PTCL project included a consortium of 22 institutions that has accessioned 1314 cases of PTCL and NKCL.7 We performed GEP on 144 lymphomas in this study, including AITL (n = 36), ALK+ ALCL (n = 20), ALK− ALCL (n = 8), ATLL (n = 12), T/NKCL (n = 14), PTCL-NOS (n = 44), and other rare PTCL entities (n = 10) by using cryopreserved tissue obtained at the time of diagnosis. The pathology review, diagnostic criteria, and clinical data for these cases have been described.7 We also analyzed 25 of the144 cases for T-cell receptor gamma (TCR-γ) gene rearrangement to estimate the proportion of tumor cells (Table 1).20 The Institutional Review Board of the University of Nebraska Medical Center approved this study. Patients provided informed consent in accordandance with the Declaration of Helsinki.
Clinical characteristics according to their pathologic diagnosis
| Pathologic diagnosis . | No. cases . | Male/female ratio . | Median age, y (range) . | Median transformed (large) T cells, % (range)* . | TCR-γ gene rearrangement by PCR . |
|---|---|---|---|---|---|
| AITL | 36 | 1.8 | 65 (43-87) | 30 (12-59) | 3/3 |
| ALK+ ALCL | 20 | 1.0 | 27 (12-43) | 90 (50-95) | 5/8 |
| ALK − ALCL | 8 | 3.0 | 64 (45-86) | 89 (85-94) | 2/4 |
| ATLL | 12 | 5.0 | 60 (47-80) | 45 (17-80) | ND |
| T/NKCL | 14 | 0.70 | 33 (28-72) | NA | NA |
| PTCL-NOS | 44 | 2.1 | 65 (18-65) | 58 (7-94) | 9/10 |
| Pathologic diagnosis . | No. cases . | Male/female ratio . | Median age, y (range) . | Median transformed (large) T cells, % (range)* . | TCR-γ gene rearrangement by PCR . |
|---|---|---|---|---|---|
| AITL | 36 | 1.8 | 65 (43-87) | 30 (12-59) | 3/3 |
| ALK+ ALCL | 20 | 1.0 | 27 (12-43) | 90 (50-95) | 5/8 |
| ALK − ALCL | 8 | 3.0 | 64 (45-86) | 89 (85-94) | 2/4 |
| ATLL | 12 | 5.0 | 60 (47-80) | 45 (17-80) | ND |
| T/NKCL | 14 | 0.70 | 33 (28-72) | NA | NA |
| PTCL-NOS | 44 | 2.1 | 65 (18-65) | 58 (7-94) | 9/10 |
Not included in this analysis are other 10 cases of PTCL rare entities, including hepatosplenic T-cell lymphoma, mycosis fungoides, enteropathy-associated T-cell lymphoma, and T-cell prolymphocytic leukemia.
AITL indicates angioimmunoblastic T-cell lymphoma; ALCL, anaplastic large-cell lymphoma; ALK, anaplastic lymphoma kinase; ATLL, adult T-cell leukemia/lymphoma; NA, not available; ND, not determined; PCR, polymerase chain reaction; PTCL-NOS, peripheral T-cell lymphoma–not otherwise specified; TCR-γ, T-cell receptor-γ; and T/NKCL, T-/natural killer-cell lymphomas.
Average calculated from 4 pathologists.
We also profiled 9 NK-cell lines, 7 T-cell lines, normal resting and activated T cells (CD4+, CD8+), and NK cells.21 The T-cell subsets were purified through fluorescence-activated cell sorting; stimulated with anti-CD3, anti-CD28, and interleukin-12 (IL-12; BD Biosciences); and harvested after 2, 8, 24, and 48 hours in culture for GEP. NKCL were included for comparative analysis to facilitate the construction of classifiers.
GEP
The methods for isolation and processing of RNA and acquisition of GEP raw data have been described previously.21 We used HG-U133 plus 2 arrays according to the manufacturer's instructions (Affymetrix Inc).
Data analysis
The raw data were uploaded in BRB-ArrayTools (version 3.7.0)22 for normalization and supervised and unsupervised analysis. Consensus clustering (CC)23 by the use of hierarchical clustering (HC), K-mean clustering (KM), and self-organizing map (SOM) was applied to identify a “core” group of cases from pathologically defined PTCL entities. Classifiers for the PTCL entities were constructed by use of the Bayesian algorithm,22,24 which estimated the probability of a case belonging to 1 subtype of PTCL compared with other PTCLs. In our series, genes were selected at significance (P < .001) and a mean fold-difference (> 4) between the 2 groups for Bayesian classification. We arbitrarily chose a 90% or greater probability as the cutoff to classify cases, but also examined 70% or greater as a secondary threshold because of the variable number of neoplastic cells, especially in AITL. Classification precision was evaluated by the use of leave-one-out cross-validation.22,25 Supervised analysis with SAM26 and GSEA27 software programs was used to identify genes and pathways/signatures associated with PTCL subgroups. The microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE19069.
Developing a molecular prognosticator for AITL
A prognosticator was constructed by use of the methodology described by Bair and Tibshirani28 with minor modifications. We first identified differentially expressed genes (P < .05) between cases having an overall survival (OS) less than or greater than 3 years. The selected genes were used in the principal component analysis, and the first 2 principal components were selected for Cox regression. To evaluate the predictive value of the model, leave-one-out cross-validation was used. The OS distributions were estimated by use of the Kaplan-Meier method.
Clinical correlations
The log-rank test was used to compare the survival distributions between PTCL entities.7 The International Prognostic Index (IPI),29 the Prognostic Model for PTCL-NOS (PIT),30 and Ki-67 mRNA expression (measured from HG-U133 plus 2 arrays) also were correlated with overall survival (OS) and event-free survival (EFS). The Fisher exact test was used to analyze categorical data, and the Wilcoxon rank sum test was used to analyze continuous data between groups. Post-hoc tests were adjusted for multiple comparisons by use of the Bonferroni method. SAS software was used for data analysis (SAS Institute Inc).
Evaluation of reclassified cases
The cases reclassified by the molecular classifier were re-reviewed (by D.D.W., T.C.G, and W.C.C.). Additional immunostains, Southern blots for HTLV1 integration, TCR-γ gene rearrangement, and fluorescence in situ hybridization (FISH) analysis for the t(2;5) were performed when feasible to evaluate the diagnoses in these cases.
Results
Patient characteristics
A consensus diagnosis for each case was made by a panel of expert hematopathologists, who also evaluated the cases for the percentage of transformed cells present. The distinct PTCL entities in this study and their clinical characteristics are summarized in Table 1 and supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Approximately one-half of the cases were diagnosed as AITL (n = 36) or PTCL-NOS (n = 44). The median age of patients in these 2 groups was 65 years; however, patients in the ALK+ ALCL and NKCL groups had much younger median ages (26.5 and 33 years, respectively). The OS and EFS are similar for patients with AITL and PTCL-NOS. As expected, patients with ALK+ ALCL had the best 5-year OS (> 70%), and those with ATLL had the worst OS (< 10%; supplemental Figure 1A-B). Similar trends were observed for EFS analysis. High IPI3-5 and PIT2-4 scores were significantly associated with poor OS (P < .001 and P = .008, respectively) and EFS (P < .001 and P = .028, respectively) when PTCL cases were analyzed as a single group and for the PTCL-NOS group (supplemental Figure 1C-J). High Ki-67 mRNA expression also showed marginal association with worse OS (P = .07) when cases were divided into quartiles according to their Ki-67 transcript level (supplemental Figure 1K). The initial therapeutic approaches varied widely in these cases, but the majority (> 60%) with the most common PTCL subtypes received CHOP (ie, cyclophosphamide, doxorubicin, vincristine, and prednisone) or CHOP-like regimens (supplemental Table 1).
Correlation of gene expression with the pathologic diagnosis
In unsupervised analysis in which we used HC,31 we observed that the well-defined subtypes of PTCL each form 1 or 2 main clusters (Figure 1). However, cases of PTCL-NOS were interspersed among the other groups, demonstrating their heterogeneity. Most of the AITL cases were in 2 distinct clusters; with the major cluster including 23 of the 36 cases. The minor PTCL subtypes, including mycosis fungoides, hepatosplenic, and enteropathy-associated T-cell lymphomas, formed distinct clusters but because of low number of cases were not studied further. To assess the stability of these clusters, CC23 with SOM and KM algorithms were run by the use of different numbers of clusters (range, 2-9), and the optimum number of clusters was determined (KM = 4 and SOM = 5) on the basis of the consensus matrix and the corresponding statistic-plots calculated by CC.23 HC generated 3 CCs, and cases that were consistently clustered together by all 3 algorithms were considered a “core” group of cases for an entity and used for derivation of a molecular classifier (supplemental Figure 2). ALK− ALCL and most PTCL-NOS cases did not form unique or tight clusters for core group analysis, aside from approximately one fourth of PTCL-NOS cases (n = 11) that formed a CC (see “A unique PTCL-NOS subgroup with features of cytotoxic cells”).
Normal T and NK cells, cell lines, and PTCL cases classified by unsupervised hierarchical clustering: Major entities of PTCL form tight clusters with cases of PTCL-NOS and other rare entities interspersed. Each column represents a case and each row the expression level of a gene. Gene expression level is depicted according to the color scale shown.
Normal T and NK cells, cell lines, and PTCL cases classified by unsupervised hierarchical clustering: Major entities of PTCL form tight clusters with cases of PTCL-NOS and other rare entities interspersed. Each column represents a case and each row the expression level of a gene. Gene expression level is depicted according to the color scale shown.
Gene expression classifiers and differentially expressed genes in the major PTCL entities
We first focused on the core groups of PTCL defined by CC to derive classifiers and then used the Bayesian algorithm to classify all the cases.
AITL.
The AITL classifier included 3 prominent signatures: a B-cell signature, a follicular dendritic-cell (FDC) signature, and a cytokine signature (Figure 2A; Tables 2,Table 3,Table 4–5). The B-cell signature included B-cell receptor signaling-associated transcripts, members of the immunoglobulin family, and germinal center B cell–associated genes, some of which may also be expressed by the neoplastic T cells (eg, CD10, BCL6). The FDC signature included FDC-associated markers and many components of the complement system. These signatures indicate the consistent presence of significant numbers of B cells and FDCs in the microenvironment of AITL. AITL also showed increased expression of numerous cytokines/receptors and tumor necrosis factor/receptor (TNF/TNFR), including a prominent group of immunosuppressive cytokines/receptors (transforming growth factor β1 [TGF-β1], TGF-βR2, IL-10Rα, and IL-10Rβ). The majority of these cytokines were expressed at a significantly lower level (< 10-fold) in the normal T-cell subsets, suggesting that this signature is largely stromal dependent. The AITL classifier was not entirely stromal in origin because some key genes known to be expressed by neoplastic T cells (CTLA4, ICOS) or normal TFH cells (CXCL13, CXCR5) were noted. Supervised analysis with the use of SAM identified other functional categories that were not specifically included in the classifier. These included many T cell–associated transcripts encoding TCR subunits and coreceptor molecules, components of the TCR signaling pathway, costimulatory and coinhibitory molecules, and molecules critical for bidirectional T- and B-cell interaction, including SH2D1A(SAP) and SAP-associated receptors. Other transcripts were mainly involved in proliferation, angiogenesis, and tumor cell survival (Table 2).
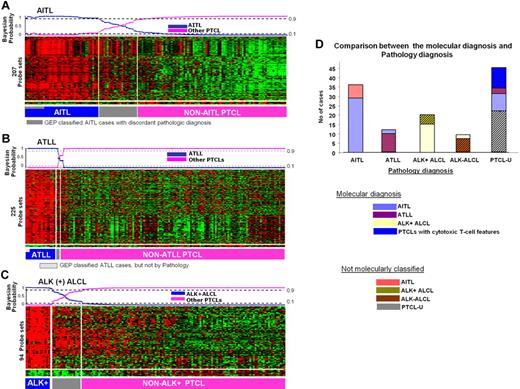
Gene expression–based molecular predictors of the major subgroups of PTCL. (A) AITL, (B) ATLL, and (C) ALK+ ALCL. (D) The correlation between the molecular and the pathology-based diagnoses is illustrated on the right.
Gene expression–based molecular predictors of the major subgroups of PTCL. (A) AITL, (B) ATLL, and (C) ALK+ ALCL. (D) The correlation between the molecular and the pathology-based diagnoses is illustrated on the right.
Differentially expressed genes in the molecularly defined AITL
| . | Genes in classifier . | Other differentially expressed genes* . |
|---|---|---|
| Cytokine/chemokine signature | ||
| Proinflammatory response | LTA (TNFSF1), LTB (TNFSF3) | IFNαR1, IFNγR1, IFNγR2 |
| T- and B-cell activation | TNFRSF4 (OX40/CD134), TNFRSF17(BCM/CD269) TNFSF8 (CD30L/CD153), CSF2RB (CD131), | IL16, IL3RA, IL7 |
| Chemotactic activity for activated T cells | CCL17, CCL19, CCL20, CCL21,CCL22 | CCL18, CCL26, CXCL9, CXCL10 |
| Angiogenesis | CXCL8 (IL8), CXCL5 | IL8, IL23A |
| Homing of B cells and TFH cells to follicles | CXCL13, its receptor (CXCR5) | |
| Monocytes/macrophage activation | CCR2, CSF2RA, CSF1R, CX3CR1, CXCL12 | |
| Granulocyte activation | CSF2RB, CSF3R | |
| TH1 cells | IL-1α, IL21, IL21R, IL23, IL27Rα, IL2Rγ, (LTF) | |
| TH2 cells | IL4, IL6, IL33, IL4R, IL6R, IL6ST | |
| TNF/TNF-R family members (mainly GC-associated) | TNF, TNFRSF11B, TNFRSF1B, TNFSF10, TNFSF11, TNFRSF13C, TNFSF13B, TNFSF13, TNFSF5 (CD40LG) | |
| Immunosuppressive cytokines | TGFβ1, TGFβR2, IL10Rα, IL10Rβ, | |
| Immunosuppressive activities meditated by nontumor cells | VSTM3,VSIG1, LILRB4, LILRB3, LILRB2, LILRB1, LILRA2, LIFR, LIF | |
| B-cell receptor signaling | CD79a, CD19, FCRL5, CD22 | |
| Immunoglobulin family | IgH, IgK, IgL | |
| B-cell–associated/activation | MS4A1 (CD20), CR1 (CD21), CD23, CD24, CD37, FCRLA, AICD, ID3, SpiB | CD19, BLNK, BANK1, BLK, BTK, BTLA, CD79A, CD79B, LYN, SYK, VAV3, SHB, SHCBP1, SOS1 |
| FDC markers | CR1, CR2 (CD21), CD23, CLU, CD200, C4orf7 | FCGR2B (CD32), FDC-M2 (C4b) |
| Complement system | C1S, C3, C4A, C7 | CR1, CR1L, CR2, C1QBP, C1R, C2, C3, C4BPA, C4A, C5AR1, C7, CFB, CFHR1, CFP |
| Antigen presentation | CTSA, CTSC, CTSD, CTSH, CTSK, CTSS, CTSZ | |
| Germinal center–related genes | ||
| B-cell or T-cell | CD10, BCL6, AICDA, GCET1, CXCR5, CD40, POU2AF1 (BOB1), POU2F2 | |
| T cell–specific genes | ||
| TCR subunits, coreceptor molecules | CD28, CD3δ, CD3ϵ, CD3γ, CD4, CD8α, CD8β) | |
| TCR signaling pathway | LCK, FYN, PTPRC (CD45), NFATC1, NFATC2IP,NFATC | |
| Costimulatory | ICOS | CD28, CD27 |
| Coinhibitory | CTLA4 | PCD1/PD1, BTLA |
| SAP/SLAM interaction | SH2D1A(SAP) CD84, LY9 (CD229/SLAM3),LY108 (SLAM6), SLAMF7 (CRACC) | |
| Miscellaneous genes | SOX8, XKR4, GPR64, PTGDS, NTN2L, PLA2G2D, ALPK2, NT5DC4 | |
| Cell-surface receptors, adhesion molecules | ||
| EDG family | EDG1, EDG2, EDG3, EDG4 | |
| Ephrin receptor subfamily | EPHA2, EPHA3, EPHA4, EPHB1, EPHB2, FGFR2I | |
| Protocadherin family | PCDHAC2, PCDH11X, PCDHA10, PCDHA12 | |
| Cell adhesion molecules | ICAM1, ICAM2 ICAM3, ICAM5, VCAM1 | |
| Proliferation-related genes | ||
| G0 to G1/S phase transition | MCM3AP, MCM5, MCM7, NEK3, NEK6 | |
| Cell-cycle progression/regulation | CCNC, CCND2, CCND3, CNDBP1, CCNG2, CCNI, CCNL1, CCNL2, GSPT2, CDK5R1, CDKN1B | |
| Cell division/ regulation | CDC25B, CDC2L1, CDC2L2, CDC37, CDC42, CDC42BPB, CDC42EP4, CDC42EP5, CDC42SE2, CLK1, CDK2AP2, CIZ1 | |
| Transcription factors (oncogenic activities) | CRK, ERG, ETS1, FGR, FOS, JUN, MET1, MYC,MYB, MAF, PVT1 | |
| Antiapoptosis | BCL2, BCL2A1, BCL2L1, FAIM3 |
| . | Genes in classifier . | Other differentially expressed genes* . |
|---|---|---|
| Cytokine/chemokine signature | ||
| Proinflammatory response | LTA (TNFSF1), LTB (TNFSF3) | IFNαR1, IFNγR1, IFNγR2 |
| T- and B-cell activation | TNFRSF4 (OX40/CD134), TNFRSF17(BCM/CD269) TNFSF8 (CD30L/CD153), CSF2RB (CD131), | IL16, IL3RA, IL7 |
| Chemotactic activity for activated T cells | CCL17, CCL19, CCL20, CCL21,CCL22 | CCL18, CCL26, CXCL9, CXCL10 |
| Angiogenesis | CXCL8 (IL8), CXCL5 | IL8, IL23A |
| Homing of B cells and TFH cells to follicles | CXCL13, its receptor (CXCR5) | |
| Monocytes/macrophage activation | CCR2, CSF2RA, CSF1R, CX3CR1, CXCL12 | |
| Granulocyte activation | CSF2RB, CSF3R | |
| TH1 cells | IL-1α, IL21, IL21R, IL23, IL27Rα, IL2Rγ, (LTF) | |
| TH2 cells | IL4, IL6, IL33, IL4R, IL6R, IL6ST | |
| TNF/TNF-R family members (mainly GC-associated) | TNF, TNFRSF11B, TNFRSF1B, TNFSF10, TNFSF11, TNFRSF13C, TNFSF13B, TNFSF13, TNFSF5 (CD40LG) | |
| Immunosuppressive cytokines | TGFβ1, TGFβR2, IL10Rα, IL10Rβ, | |
| Immunosuppressive activities meditated by nontumor cells | VSTM3,VSIG1, LILRB4, LILRB3, LILRB2, LILRB1, LILRA2, LIFR, LIF | |
| B-cell receptor signaling | CD79a, CD19, FCRL5, CD22 | |
| Immunoglobulin family | IgH, IgK, IgL | |
| B-cell–associated/activation | MS4A1 (CD20), CR1 (CD21), CD23, CD24, CD37, FCRLA, AICD, ID3, SpiB | CD19, BLNK, BANK1, BLK, BTK, BTLA, CD79A, CD79B, LYN, SYK, VAV3, SHB, SHCBP1, SOS1 |
| FDC markers | CR1, CR2 (CD21), CD23, CLU, CD200, C4orf7 | FCGR2B (CD32), FDC-M2 (C4b) |
| Complement system | C1S, C3, C4A, C7 | CR1, CR1L, CR2, C1QBP, C1R, C2, C3, C4BPA, C4A, C5AR1, C7, CFB, CFHR1, CFP |
| Antigen presentation | CTSA, CTSC, CTSD, CTSH, CTSK, CTSS, CTSZ | |
| Germinal center–related genes | ||
| B-cell or T-cell | CD10, BCL6, AICDA, GCET1, CXCR5, CD40, POU2AF1 (BOB1), POU2F2 | |
| T cell–specific genes | ||
| TCR subunits, coreceptor molecules | CD28, CD3δ, CD3ϵ, CD3γ, CD4, CD8α, CD8β) | |
| TCR signaling pathway | LCK, FYN, PTPRC (CD45), NFATC1, NFATC2IP,NFATC | |
| Costimulatory | ICOS | CD28, CD27 |
| Coinhibitory | CTLA4 | PCD1/PD1, BTLA |
| SAP/SLAM interaction | SH2D1A(SAP) CD84, LY9 (CD229/SLAM3),LY108 (SLAM6), SLAMF7 (CRACC) | |
| Miscellaneous genes | SOX8, XKR4, GPR64, PTGDS, NTN2L, PLA2G2D, ALPK2, NT5DC4 | |
| Cell-surface receptors, adhesion molecules | ||
| EDG family | EDG1, EDG2, EDG3, EDG4 | |
| Ephrin receptor subfamily | EPHA2, EPHA3, EPHA4, EPHB1, EPHB2, FGFR2I | |
| Protocadherin family | PCDHAC2, PCDH11X, PCDHA10, PCDHA12 | |
| Cell adhesion molecules | ICAM1, ICAM2 ICAM3, ICAM5, VCAM1 | |
| Proliferation-related genes | ||
| G0 to G1/S phase transition | MCM3AP, MCM5, MCM7, NEK3, NEK6 | |
| Cell-cycle progression/regulation | CCNC, CCND2, CCND3, CNDBP1, CCNG2, CCNI, CCNL1, CCNL2, GSPT2, CDK5R1, CDKN1B | |
| Cell division/ regulation | CDC25B, CDC2L1, CDC2L2, CDC37, CDC42, CDC42BPB, CDC42EP4, CDC42EP5, CDC42SE2, CLK1, CDK2AP2, CIZ1 | |
| Transcription factors (oncogenic activities) | CRK, ERG, ETS1, FGR, FOS, JUN, MET1, MYC,MYB, MAF, PVT1 | |
| Antiapoptosis | BCL2, BCL2A1, BCL2L1, FAIM3 |
CT indicates cytotoxic T cells; IFN, interferon; IL, interleukin; NK, natural killer; PTCL, peripheral T-cell lymphoma; TCR, T-cell receptor; TFH, T-follicular helper; TGF, transforming growth factor; and TNF, tumor necrosis factor.
Only up-regulated representative genes are included in each PTCL subtype signature.
Differentially expressed genes in the molecularly defined ATLL
| . | Genes in classifier . | Other differentially expressed genes . |
|---|---|---|
| TCR signaling components and costimulatory/inhibitory molecules | TCRα, TCRβ, CD1ϵ, CTLA4, CD99, TRAC | ICOS, CD28, CD1B, CD1C, CD6, LAT, FYN, FYB |
| T-cell activation or differentiation | TNFRSF25(DR3), CD27, LEF1, NFATC1 | IL2RA (CD25), IL4R, IL18R1, IL23R,IL12RB2, MAL, IL4R, GATA3 |
| Proinflammatory | CCR4, CCL13, IL23R | IL6, IL6ST, CCL26 |
| Melanoma-associated | MAGEA4, MAGEA9, MAGEA4B | MAGEB1, MAGEA3 |
| Immune modulatory enzymes | PGDS (hematopoietic) | PTGER4, PTGER3 |
| B cells | AICDA, MTA3 | |
| Previously published genes associated with ATLL | IKZF2, RGS13, PTHLH, RGS13, CADM1(TSLC1) | TCF4 |
| HTLV-1 Tax target | c-Myc, TIAM1, PDE8B, DOK5, ARNT2, UST, PGDSMYCN, PDE8B, CEBPA, RGS13, PTHLH, SMARCA2, CD99, CADM1(TSLC1),CCR4, RGS2, SPINK2, RORA | IL2RA, CFLAR |
| Treg cell chemoattractant | CCL22, CCL20, CCL17 | |
| Cytotoxic molecules | TIA1 |
| . | Genes in classifier . | Other differentially expressed genes . |
|---|---|---|
| TCR signaling components and costimulatory/inhibitory molecules | TCRα, TCRβ, CD1ϵ, CTLA4, CD99, TRAC | ICOS, CD28, CD1B, CD1C, CD6, LAT, FYN, FYB |
| T-cell activation or differentiation | TNFRSF25(DR3), CD27, LEF1, NFATC1 | IL2RA (CD25), IL4R, IL18R1, IL23R,IL12RB2, MAL, IL4R, GATA3 |
| Proinflammatory | CCR4, CCL13, IL23R | IL6, IL6ST, CCL26 |
| Melanoma-associated | MAGEA4, MAGEA9, MAGEA4B | MAGEB1, MAGEA3 |
| Immune modulatory enzymes | PGDS (hematopoietic) | PTGER4, PTGER3 |
| B cells | AICDA, MTA3 | |
| Previously published genes associated with ATLL | IKZF2, RGS13, PTHLH, RGS13, CADM1(TSLC1) | TCF4 |
| HTLV-1 Tax target | c-Myc, TIAM1, PDE8B, DOK5, ARNT2, UST, PGDSMYCN, PDE8B, CEBPA, RGS13, PTHLH, SMARCA2, CD99, CADM1(TSLC1),CCR4, RGS2, SPINK2, RORA | IL2RA, CFLAR |
| Treg cell chemoattractant | CCL22, CCL20, CCL17 | |
| Cytotoxic molecules | TIA1 |
For abbreviations, see Table 2.
Only up-regulated representative genes are included in each signature.
Differentially expressed genes in the molecularly defined ALK+ ALCL
| . | Genes in classifier . | Other differentially expressed genes . |
|---|---|---|
| Classical ALCL markers | ALK, CD30, MUC1 | |
| Immunoregulatory cytokines | IL26, IL31RA, IL9, IL1R2 | IL20, IL22, IL2RA, IL1RAP, IL1R1 |
| TH17-cell–associated | IL17A, IL17F, RORγ | |
| Proliferation-related | CCNA1, AGT, PDE4DIP, UPK1B | CDC27, FGF5, FOSL2 |
| Genes noted in other tumors | RRAD, RARα, NRCAM, TMEM158, CA12 | VCAN, TNFRSF12A |
| STAT3 regulation targets | SERPINB3, SERPINB4 | SOCS1, SOCS3 |
| Cytotoxic molecules | PRF1, GZMB | |
| Immunosuppressive response | LILRA3 |
| . | Genes in classifier . | Other differentially expressed genes . |
|---|---|---|
| Classical ALCL markers | ALK, CD30, MUC1 | |
| Immunoregulatory cytokines | IL26, IL31RA, IL9, IL1R2 | IL20, IL22, IL2RA, IL1RAP, IL1R1 |
| TH17-cell–associated | IL17A, IL17F, RORγ | |
| Proliferation-related | CCNA1, AGT, PDE4DIP, UPK1B | CDC27, FGF5, FOSL2 |
| Genes noted in other tumors | RRAD, RARα, NRCAM, TMEM158, CA12 | VCAN, TNFRSF12A |
| STAT3 regulation targets | SERPINB3, SERPINB4 | SOCS1, SOCS3 |
| Cytotoxic molecules | PRF1, GZMB | |
| Immunosuppressive response | LILRA3 |
For abbreviations, see Table 2.
Only representative genes are included in each signature.
Differentially expressed genes in the molecularly defined CT-PTCL
| . | Differentially expressed genes . |
|---|---|
| Cytotoxic molecules and related genes | Granzyme-M, -K, -H, -B,-A; cathepsin-A, -B,-C, -D, -K, -L1, -S, -W; PRF1, GNLY, FADD, FASLG |
| Cytotoxic T- or NK-cell markers or associated transcription factors | CD244(2B4), FCGR3B (CD16), FCGR2C (CD32), CD226(DNAM-1), CD8α, CD8β, TBX21, eomesodermin (eomes) |
| Killer cell immunoglobulin-like or lectin-like receptor subfamily | KIR3DL1, KIR3DL2, KIR3DL3, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5A, KIR2DS, KIR2DS2, KIR2DS3, KIR2DS5, KLRB1, KLRC2, KLRC3, KLRC4, KLRD1, KLRF1, KLRK1 |
| Chemokines/receptors associated with either CD8 + T cells or NK cells | CX3CR1, CXCR3, CXCL(-9, -10, -11), CCR1, CCR5, CXCR4, CCL2, CCL5, CCL8,CCL13,CCL18, XCL1, XCL2 |
| Other cytokines (interleukin- and tumor necrosis factor–related) | IFNαR1, IFNγR1, IFNγR1, IFNγR2, IL1RN, IL1R1, IL1R2, IL13Rα1, IL17Rβ, IL18,IL18RAP, IL2Rβ, IL2Rγ, IL21R, IL22,IL27Rα, IL31Rα, IL32, ILF2, ILF3, TNFSF10, TNFSF13, TNFSF14, TNFRSF12α, TNFRSF1A, TNFRSF1B, TNFSF5IP1 |
| Immunosuppression-associated genes | INDO, TGFβ1, TGFβR1, IL10, IL10Rβ, VSIG4, LILRB4, LILRB3, LILRB2, LILRB1,LILRA2, CD47 |
| SAP/SLAM interaction | SH2DIA, LY96, LY6E, SLAM-5, -6, -7, -8 |
| . | Differentially expressed genes . |
|---|---|
| Cytotoxic molecules and related genes | Granzyme-M, -K, -H, -B,-A; cathepsin-A, -B,-C, -D, -K, -L1, -S, -W; PRF1, GNLY, FADD, FASLG |
| Cytotoxic T- or NK-cell markers or associated transcription factors | CD244(2B4), FCGR3B (CD16), FCGR2C (CD32), CD226(DNAM-1), CD8α, CD8β, TBX21, eomesodermin (eomes) |
| Killer cell immunoglobulin-like or lectin-like receptor subfamily | KIR3DL1, KIR3DL2, KIR3DL3, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5A, KIR2DS, KIR2DS2, KIR2DS3, KIR2DS5, KLRB1, KLRC2, KLRC3, KLRC4, KLRD1, KLRF1, KLRK1 |
| Chemokines/receptors associated with either CD8 + T cells or NK cells | CX3CR1, CXCR3, CXCL(-9, -10, -11), CCR1, CCR5, CXCR4, CCL2, CCL5, CCL8,CCL13,CCL18, XCL1, XCL2 |
| Other cytokines (interleukin- and tumor necrosis factor–related) | IFNαR1, IFNγR1, IFNγR1, IFNγR2, IL1RN, IL1R1, IL1R2, IL13Rα1, IL17Rβ, IL18,IL18RAP, IL2Rβ, IL2Rγ, IL21R, IL22,IL27Rα, IL31Rα, IL32, ILF2, ILF3, TNFSF10, TNFSF13, TNFSF14, TNFRSF12α, TNFRSF1A, TNFRSF1B, TNFSF5IP1 |
| Immunosuppression-associated genes | INDO, TGFβ1, TGFβR1, IL10, IL10Rβ, VSIG4, LILRB4, LILRB3, LILRB2, LILRB1,LILRA2, CD47 |
| SAP/SLAM interaction | SH2DIA, LY96, LY6E, SLAM-5, -6, -7, -8 |
For abbreviations, see Table 2.
Only representative genes are included in each signature.
GSEA showed significant enrichment of several pathways (supplemental Table 2) that may be of therapeutic importance, including: (1) nuclear factor (NF)-κB pathway: To validate this observation, we independently selected 10 representative NF-κB target genes in T cells for evaluation. According to this 10-gene signature, AITL cases tended to have an activated NF-kB pathway compared with PTCL-NOS (P < .001; supplemental Figure 3A). (2) Immunosuppressive pathways: Genes that target the function of DCs, directly or indirectly, and ligands secreted by tolerogenic DCs were highly expressed. Furthermore, TGF-β pathway–associated genes and other gene signatures linked with immunosuppression were enriched. (3) IL-6 signaling: IL-6 has diverse functions and is involved in TFH-cell, Epstein-Barr virus (EBV)–infected B-cell proliferation, and T-cell subset differentiation (reviewed by Rose-John et al32 ). The other gene signatures associated with angiogenesis (vascular endothelial growth factor pathway) and DNA damage or genotoxic response also showed significant enrichment in this entity.
Although the pathology review generally showed less than 30% large neoplastic T cells (median = 28.5), gene signatures associated with several T-cell subsets were noted. Consistent with previous findings,11,12,33 the gene signature of TFH cells34,35 was significantly enriched in AITL. The TFH signature was not expressed by other normal T-cell subsets (supplemental Figure 3B), and no enrichment was observed in cases not classified as AITL by the GEP classifier.
The classifier identified 6 of the 13 noncore AITL cases, 9 of 44 PTCL-NOS, and 2 ATLL cases as AITL. Seven cases classified by pathology as AITL were not similarly characterized molecularly (Figure 2D). The reclassified cases of PTCL-NOS were compatible with AITL morphologically upon review in 6 of the 7 cases. The cases were not initially classified pathologically as AITL because they exhibited certain atypical features such as prominent germinal centers, lower vascular proliferation, low Epstein-Barr encoded RNA (EBER) positivity or negative for CD10, and/or CD21 expression. The 2 ATLL cases that were reclassified as AITL did not show HTLV1 integration in Southern blots. The pathology-defined AITL cases that were not characterized molecularly as AITL frequently demonstrated a greater proportion of eosinophils and low EBERs or CD21 positivity (1 of 5 and 0 of 4, respectively). The median number of large transformed cells in these tumors upon morphologic review was lower than the molecularly defined subgroup 16% (range, 10%-25%), suggesting a lower number of tumor cells.
We applied our AITL classifier to a published set of pathologically diagnosed cases of AITL (n = 6) and PTCL-NOS (n = 28).12 By using hierarchical clustering, we observed that 5 of the 6 cases of AITL and 4 of 23 PTCL-NOS cases formed a distinct cluster with high expression of our classifier genes. The number of PTCL-NOS cases reclassified as AITL by our classifier (4 [17%] of 23) was similar to that observed in our cases (9 [20%] of 44). We also compared our classifier with a differentially expressed gene list published by de Leval et al,11 and of the 220 transcripts, 92 (42%) were also in our classifier (P = .003; FDR < 0.025 by GSEA analysis).
ATLL.
The main features of the ATLL classifier were overexpression of a set of genes involved in TCR signaling or activation, a distinct set of genes previously reported to be expressed in HTLV1-induced cell lines or ATLL cases,16,17 and a group of genes associated with melanoma (Figure 2B; Table 3). ATLL did not show an elaborate cytokine profile like AITL, but there was high expression of several cytokines and cytokine receptors involved in T-cell activation and differentiation and recruitment of Treg cells. Of these, CCR4, CCL13, and IL23R were included in the diagnostic signature. Interestingly, CCR4 protein has been shown to be significantly associated with ATLL,36 and it has been proposed as a target for immunotherapy.37 Several inhibitory molecules (TIGIT/VSTM3,38 CTLA4) and immunomodulatory genes involved in the production of prostanoids (PTGER4, PTGER3, PGDS [hematopoietic])39 that promote immunosuppression were also up-regulated. HTLV1 TAX targets such as IRF4, ATF1, CREB1, NF-κB1, and MYB and their target genes showed high expression in ATLL. GSEA revealed the enrichment of TCR signaling genes, a known gene-signature reported in ATLL,17 target genes of the transcription factor retinoic acid receptor-γ, and mature CD4+ T-cell signature but not Treg-related genes (supplemental Table 3).
By use of this classifier, 3 of the PTCL-NOS cases were reclassified as ATLL. These cases were from HTLV1 endemic areas with seropositivity and positive polymerase chain reaction amplification of the viral pX genomic region encoding TAX. However, no clonal integration of HTLV1 DNA was observed by Southern blot.40 These 3 patients died within 3 years from initial diagnosis (range, 0.5-3.0 years) and therefore had a poor prognosis similar to that of ATLL patients in general.7
ALK+ ALCL and ALK− ALCL.
Among the top-ranked genes in our classifier were ALK, TNFRSF8(CD30), MUC1, Th17-cell–associated molecules (IL-17A, IL-17F, retinoic acid receptor–related orphan receptor [ROR]-γ), and a small group of immunoregulatory cytokines/receptors regulating STAT3 (IL-26, IL-31RA) or JAK3 (IL-9) activation (Figure 2C; Table 4). There was low expression of transcripts related to TCR components and TCR signaling or activation but high expression of the cytotoxic molecules GZMB and PRF1. There was low expression of GATA3, which negatively regulates Th17-cell differentiation.41 GSEA revealed significant enrichment of STAT3, BCL3, ESR1, and ETS1 target signatures but marginal enrichment of the ALK signature (52 transcripts) derived from ALCL cell lines by Piva et al42 (supplemental Table 4; supplemental Figure 4).
The ALK+ ALCL classifier identified 15 of 20 pathologically diagnosed ALK + cases with greater than 90% probability. The other 5 ALK+ ALCL cases were either negative for TCR-γ gene rearrangement by polymerase chain reaction (n = 3) or had very low expression of CD30, suggesting that these samples had a low number of neoplastic cells. Two ALK− ALCL cases had a greater than 80% probability of being ALK+ ALCL (supplemental Figure 4B) and showed high expression of CD30, JAK3, and STAT3 and low expression of TCR transcripts. These 2 cases also showed slightly increased (∼ 1.5-fold) expression of ALK transcripts compared with the other PTCLs. However, FISH analysis of the cases did not reveal the t(2;5) translocation or a variant translocation.
The ALK− ALCL cases did not form a unique CC, although GEP analysis showed significant differences from ALK+ ALCL and PTCL-NOS. Compared with PTCL-NOS, genes associated with TCR signaling were expressed at a lower level, whereas 2 cytokines, IL-20, which promotes angiogenesis, and IL-9, which activates Jak3, were highly expressed. Compared with ALK+ ALCL, lower expression of ALK, cytotoxic molecules (PRF1, GZMB), cathepsins (CTS-W, -D, -L1, -B), TH17-cell–associated molecules (IL-17-F, -A, RORγ), and B cell–associated transcripts (Ig-H, -K, -L) was noted. However, ALK− ALCL showed greater expression of a set of cytokine/receptors (CCL1, CCL22, CCR8, CCR4, IL-13RA2, CXCL14, TGF-βR1) and several antiapoptotic factors (BCL2, BIRC6, BIC) but low expression of certain proapoptotic genes (BAX, BCL2L1, BNIP3). GSEA analysis showed no significant enrichment of STAT3 and BCL3 target genes in ALK− ALCL compared with ALK+ ALCL (supplemental Figure 5A-C). Despite these differences, we were not able to derive a robust classifier for ALK− ALCL, most likely related to the limited number of cases available for study.
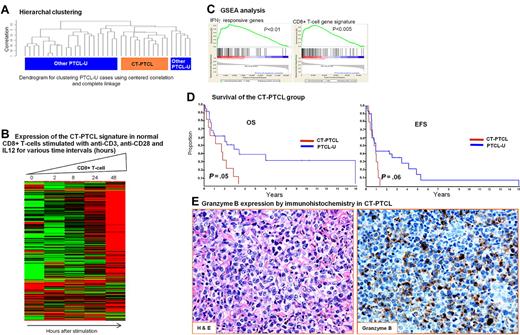
A unique PTCL-NOS subgroup with features of cytotoxic cells.
We have identified a subgroup of PTCL-NOS (n = 11) that formed a CC distinct from the other PTCL-NOS cases (Figure 3). This subgroup expressed a unique repertoire of cytotoxic molecules, including perforin, granulysin, and many granzymes and cathepsins. Other transcripts associated with cytotoxic T- or NK-cell markers, members of the killer cell lectin-like receptor subfamily, and killer cell immunoglobulin-like receptor family members were also highly expressed (Table 5). GSEA analysis revealed significant enrichment of the CD8+ T-cell signature, as well as interferon-α, -β, and -γ responsive genes, genes down-regulated by rapamycin (deregulated mammalian target of rapamycin pathway), and several gene signatures associated with granule secretion. The 2 key transcription factors for CD8+ T cells, T-bet (TBX21) and eomesodermin(eomes), and their known target genes (CXCR3, IL-2RB, CCL3, interferon-γ) also showed greater expression in this subgroup. The overall gene signature associated with this subgroup showed marked up-regulation in normal activated CD8+ T cells compared with their resting counterparts. This subgroup also expressed a distinct set of cytokines/receptors, the majority of which show high expression in normal CD8+ T cells and NK cells. Unexpectedly, these cases also showed an elaborate immunosuppressive gene signature and transcripts associated with SAP/SLAM interaction like AITL. However, the prominent B-cell and FDC signatures were not observed.
A unique subgroup within PTCL-NOS with features of cytotoxic T-cells (CT-PTCL). (A) Hierarchical clustering of the PTCL-NOS group demonstrated a distinct cluster of 11 cases that also clustered together when SOM and KM were used. (B) Gene signature associated with CT-PTCL was highly expressed in activated CD8+ T cells. (C) GSEA analysis identified enrichment of CD8+ T-cell and other gene signatures. The enrichment score curves were obtained from GSEA software. Vertical black lines indicate the position of the enriched genes (Hit) comprising the gene set. The graph on the bottom of each panel shows the ranked list metric (signal-to-noise ratio) for each gene as a function of the rank in the ordered dataset (see Subramanian et al27 for more details). (D) Kaplan-Meier plots of OS and EFS of CT-PTCL and the remaining PTCL-NOS cases indicating poor prognosis of CT-PTCL. (E) A representative CT-PTCL case was immunostained for Granzyme B (original magnification 200× for both panels).
A unique subgroup within PTCL-NOS with features of cytotoxic T-cells (CT-PTCL). (A) Hierarchical clustering of the PTCL-NOS group demonstrated a distinct cluster of 11 cases that also clustered together when SOM and KM were used. (B) Gene signature associated with CT-PTCL was highly expressed in activated CD8+ T cells. (C) GSEA analysis identified enrichment of CD8+ T-cell and other gene signatures. The enrichment score curves were obtained from GSEA software. Vertical black lines indicate the position of the enriched genes (Hit) comprising the gene set. The graph on the bottom of each panel shows the ranked list metric (signal-to-noise ratio) for each gene as a function of the rank in the ordered dataset (see Subramanian et al27 for more details). (D) Kaplan-Meier plots of OS and EFS of CT-PTCL and the remaining PTCL-NOS cases indicating poor prognosis of CT-PTCL. (E) A representative CT-PTCL case was immunostained for Granzyme B (original magnification 200× for both panels).
Immunohistochemical data could be obtained in several of these cases and validated expression of T-cell markers (CD3+ [6 of 6] and TCR-αβ+ [2 of 4]) and cytotoxic molecules, including TIA (4 of 5) and granzyme B (3 of 4; Figure 3E). Interestingly, more cases were CD4+ (6 of 11) than CD8+ (3 of 11), indicating that CD8 expression is not a sufficient marker for identifying these cases. Necrosis was not prominent in these cases. The patients in this group had a worse OS (P = .05) and EFS (P = .06) compared with the other PTCL-NOS cases.
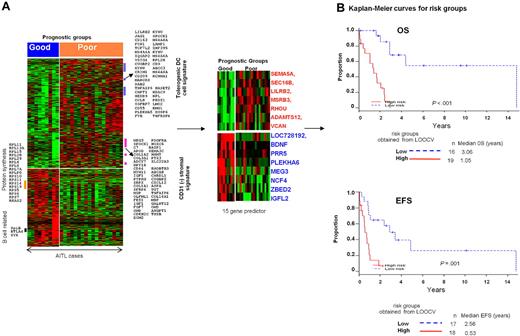
Molecular prognosticator for AITL
We first identified genes that were differentially expressed between AITL cases that had an OS of 3 years or longer (good prognostic group) and those with OS of less than 3 years (poor prognostic group; Figure 4). Genes up-regulated in the poor-prognosis group included 2 gene signatures associated with immunosuppressive functions: tolerogenic DCs43 and CD31− stromal cells (supplemental Table 5). One of the highly expressed genes, VSIG4, is a potent inhibitor of T-cell activation and is secreted by tolerogenic DCs. Cases with a poor prognosis also showed high expression of receptors or cell-adhesion molecules that mediate proliferative signals, including PDGFRα and PDGFRβ. In contrast, transcripts that have inhibitory effects on myeloid cell functions (CD200, MIF, SERPINB1), associated with B cells (SpiB, BTLA4, SYK), or encode members of the ribosomal protein synthesis pathway were highly expressed in the good prognostic group.
Evaluation of a prognosticator in AITL. (A) Differentially expressed genes between the good (≥ 3 years OS) and poor (< 3 years OS) prognostic groups of AITL. (B) Kaplan-Meier plots for AITL risk groups using the 15-gene predictor.
Evaluation of a prognosticator in AITL. (A) Differentially expressed genes between the good (≥ 3 years OS) and poor (< 3 years OS) prognostic groups of AITL. (B) Kaplan-Meier plots for AITL risk groups using the 15-gene predictor.
To evaluate whether a smaller subset of genes can be used as a prognosticator, several gene sets were evaluated to predict the OS and EFS by increasing the stringency in the gene selection criteria. With different iterations and gene selection, a small subset of 15 transcripts was able to predict for the OS and EFS (P < .001), and the genes included in this prognosticator are shown in Figure 4B. Including the IPI and PIT scores as covariates did not improve the predictive power of the molecular prognosticator, demonstrating that it is an independent predictor of outcome.
Discussion
The diagnosis of PTCL is challenging, with significant variability even among expert hematopathologists. Therefore, we have derived molecular classifiers for the common subtypes of PTCL and have identified a new subtype from PTCL-NOS with cytotoxic characteristics. A more in-depth analysis of AITL with the construction of a prognostic model also was performed.
The AITL classifier largely reflects the nonneoplastic cells in the microenvironment with a significant contribution by B cells and FDCs. The complex cytokine milieu may not only recruit various inflammatory cells but also may contribute to a microenvironment that supports malignant T-cell growth and cell survival, as well as angiogenesis and immunosuppression. This classifier allowed us to reclassify a significant number of morphologic PTCL-NOS cases and other PTCLs as AITL. These discrepant cases were rereviewed and demonstrated some morphologic features of AITL but were not considered to be diagnostic because of the absence of certain key features such as numerous EBV-positive B cells, prominent vascular proliferation, or the presence of large sheets of monomorphic tumor cells. The 2 reclassified ATLL cases from Japan had the morphologic features of AITL, and there was no evidence of HTLV1 viral integration in the tumor by Southern blot analysis. Pathologically diagnosed AITL cases that were not confirmed molecularly may represent misdiagnosis, AITL with very low tumor content, or, possibly, nonrepresentative tissue was submitted for GEP studies. Our classifier also classified most of the AITL cases from a previous study,13 reclassified a similar proportion of PTCL-NOS cases (18%), and significantly overlaps the up-regulated genes identified by de Leval et al.11 A similar observation also was reported in another study10 in which 5 (16%) of 32 PTCL-NOS cases clustered tightly with the major AITL cluster. Because of a substantially larger number of cases in our series, we believe this molecular classifier is robust and will allow us to identify cases of AITL in other categories of PTCL, especially PTCL-NOS.
We also were able to generate robust classifiers for ALK+ ALCL and ATLL. As expected in ALK+ ALCL, ALK mRNA was one of the top genes, along with IL-26 and IL-31RA, which are involved in the activation of STAT3.44,45 STAT3 can induce the expression of RORγ which, in turn, promotes the expression of IL-17A, IL-17F, and IL-22. The expression of TH17-associated molecules may represent aberrant activation of TH17-cell differentiation by abnormal cytokine secretion. Interestingly, transcripts characteristic of TH17 cells coexisted with cytotoxic molecules and enrichment of the proto-oncogene ETS-1 target gene signature, indicating a profoundly abnormal differentiation program in ALK+ ALCL. ALK− ALCL could not be accurately classified molecularly, possibly because of the small number of cases available for study. However, ALK− ALCL appeared distinct from PTCL-NOS and ALK+ ALCL, specifically in the expression of TCR signaling-associated genes and the STAT3 target signature, respectively.
Unlike ALK+ ALCL, our ATLL classifier was characterized by high expression of TCR signaling pathway genes, an HTLV1-associated gene signature, and a small subset of MAGE family transcripts that have not been previously reported in ATLL. An intact TCR signaling pathway and several TAX-induced genes/pathways (eg, MYC, NFKB1, and ATF1) may be important for tumor survival and viral propagation. Although GEP indicated an immunosuppressive microenvironment and the expression of chemoattractants for T-reg cells, we were not able to demonstrate enrichment of a Treg-cell signature in ATLL. Consistent with this observation, FOXP3 mRNA was not expressed in ATLL, even though ATLL cells showed significant up-regulation of IL-2Rα (CD25).
Three PTCL-NOS cases were reclassified as ATLL. These cases were serologically positive for HTLV1, but further molecular analysis of 2 cases with available materials demonstrated no evidence of viral integration. It is possible that a combination of molecular events could give rise to a GEP similar to that observed in ATLL, leading to the misclassification, or there could have been partial deletion of HTLV1 during propagation of the lymphoma cells resulting in failure of detection by Southern blot analysis. In general, these classifiers are very specific, and there was little overlap in the molecular diagnosis. However, some cases were missed because of the presence of few tumor cells in the sample analyzed, especially in ALCL, as suggested by negative results from T-cell receptor gene rearrangement analysis or a low CD30 message.
A small subgroup of PTCL-NOS cases identified by CC showed features of cytotoxic T cells with the expression of cytotoxic molecules and many markers related to cytotoxic cells (killer cell immunoglobulin-like receptor and killer cell lectin-like receptor subfamily families) but lacked the expression of CD56 mRNA. GSEA analysis also revealed significant enrichment of the CD8+ T-cell signature, but these cases could not be identified immunophenotypically by the use of the CD8 marker alone. Interestingly, an immunosuppressive gene signature mainly mediated by tolerogenic DCs also was noted in this entity. This group of cases had an inferior survival compared with the other PTCL-NOS cases. There are several reports indicating that expression of cytotoxic markers (TIA, granzymes, and/or perforin) in some PTCLs and these cases are associated with poor clinical outcome.46-49 Some of these cases may correspond to the CT-PTCL defined here, but expression of individual markers is not sufficient to define this subgroup. A comprehensive and refined gene expression signature as presented will be able to separate these cases from PTCL-NOS for further investigations in the future.
The authors of several recent studies6,50-53 also have evaluated cytokines/receptors for diagnosis as well as prognosis. These included the cytokines that correlate with the functional subsets of T cells such as Th1, Th2, and TFH cells.12,54 Although there is preferential expression of chemokine/receptors with PTCL subtypes (eg, CCR4 in ATLL, CCR3 in ALCL, and CXCL13 in AITL),36 individually they are not sufficiently specific for diagnosis, nor can they define Th1 or Th2 differentiation of tumor cells. The association with prognosis reported in few studies may be related to the association of these markers with certain subtypes having different prognosis.
Recent studies have shown the association of a high IPI29 score and PIT30 with poor survival in PTCL30,55 and PTCL-NOS, respectively. Ki-67 expression has also been evaluated in PTCL and showed association with clinical outcome in some studies.6,56 However, in a recent study, Ki-67 expression by itself was not associated with clinical outcome in PTCL.14 In our series, both IP129 and PIT30 had predictive power when PTCL was considered as a single entity, and there was a trend toward worse outcome with greater Ki-67 mRNA expression. Prognosticators based on GEP have been evaluated in PTCL in general14 but not in specific entities. In this study, we have constructed a GEP-derived prognosticator for AITL that is independent of IPI. Notably, the high expression of 2 immunosuppressive signatures was associated with poor survival. We previously demonstrated that host/tumor interactions have a significant impact on survival in follicular lymphoma and diffuse large B-cell lymphoma.57,58 This finding also seems to be true for AITL and suggests that the tumor microenvironment may be an important target for therapy. However, this study was a retrospective one with cases from many centers in which the therapeutic approaches varied widely. Therefore, a prospective study of a large series of well-defined AITL cases is essential to validate and refine this prognosticator in the future.
GEP studies are not only useful in providing robust molecular classifiers but can also illuminate the biology of the tumor and suggest therapeutic targets. Thus, consistent with previous studies,11,12 AITL demonstrates significant enrichment in the TFH-cell gene signature, supporting the contention of a TFH cellular origin. However, there were also obvious differences with normal TFH cells, such as low expression of CD57 and high CD10 mRNA expression.59 SH2DIA (SAP)/SLAM interaction is essential for the localization of TFH cells and their cognate B cells into follicles.60 Although SAP and SLAM molecules were expressed at high levels in AITL, as were molecules that attract B cells, including ICOS, CD40L, OX40, and IL21, follicles are generally poorly developed, and neoplastic T-cells are not concentrated in the follicles in AITL. These findings suggest an abnormality in SAP/SLAM interaction or signaling between the neoplastic T cells and B cells and raise the intriguing possibility that the neoplastic cells may be at a prefollicular stage of development because of their failure to enter follicles, in contrast to the rare follicular T-cell lymphoma.61
Our study found that the NF-κB pathway was highly activated in AITL cases and that this pathway is a strong candidate for therapeutic intervention.62 Inhibitors of the NF-κB pathway may provide significant therapeutic benefit by acting against both the tumor and microenvironmental components. Because of the enrichment of genes related to genotoxic stress, promoting TP53 activity may tip the balance toward apoptosis and enhance the efficacy of therapeutic agents. A small molecular inhibitor of MDM2, Nutlin-3, can enhance TP53 function. Nutlin3 can also suppress EBV-mediated transformation of primary B cells, and its activity is further enhanced by inhibition of NF-κB signaling. A combination of Nutlin 3 and an NF-kB inhibitor may be particularly effective against the neoplastic T cells and EBV-transformed B cells in AITL with intact TP53.
GEP also demonstrated high expression of negative immune response regulators (eg, CTLA4, BTLA, PD1), immunosuppressive cytokines, including IL-10, TGF-β, IL-6, and LIF, and high expression of INDO, which recruits T-reg cells and tolerogenic DCs. Blockade of this immunosuppression may not only suppress the EBV-transformed B cells but also promote antitumor immunity. Angiogenic pathways are highly active in AITL and could be potential targets for therapy. Secretion of IL-6 and IL-21 by malignant TFH cells may have autocrine activities. The potent effects of IL-4, IL-6, and IL-2163 on the differentiation of B cells are likely responsible for the many plasma cells present in AITL and the hypergammaglobulinemia and autoimmune phenomenon often seen in this disease. Many of the pathologic manifestations observed in AITL may be mediated by IL-6 signaling, and targeting this pathway with the humanized anti-human IL-6 receptor antibody tocilizumab64 or with small molecules such as capsacin or SD1008 may be a novel treatment approach for AITL.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Martin Bast for the clinical data collection and Kavita Patel and Lisa Bough for technical assistance.
This work was supported in part by a National Cancer Institute grant (5U01/CA114778) and funds from the International Peripheral T-cell Lymphoma Project and Eppley Core Grant (CA36727 from National Cancer Institute). The UNMC Microarray Core Facility is supported partially by National Institutes of Health grant P20 RR016469 from the INBRE Program of the National Center for Research Resources.
National Institutes of Health
Authorship
Contribution: J.I., T.C.G., and W.C.C. designed and performed the study, supervised all aspects of the research and analysis, and wrote and finalized the manuscript; D.W.W., T.C.G., and W.C.C. were responsible for verifying histology, pathology review, scoring immunohistochemical stains, and for final approval of the manuscript; J.V. and J.A. were involved in the design of the study and reviewed the clinical aspects and edited the manuscript; T.M.K., C.K., H.G., K.D.B., L.S., and K.D. assisted in the design of the study and statistical analysis of the microarray and clinical data; and S.K., M.S., J.D., F.B., F.L., W.B., Y.H.K., and I.S. provided PCTL cases for the study and also scored the immunohistochemical stains and provided FISH and Southern blot data for the cases.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wing C. Chan, MD, Department of Pathology and Microbiology, Co-Director, Center for Research in Lymphoma and Leukemia, 983135 Nebraska Medical Center, Omaha, NE 68198-3135; e-mail: jchan@unmc.edu.
References
Supplemental data
Supplemental figures can be found here.
Differential gene expression (A) ALK (−) ALCL vs PTCL-U (B) ALK (−) vs ALK(+) ALCL (C) Enrichment of STAT3 induced target gene signature in ALK(+)ALCL compared with ALK(−)ALCL and PTCL-U.Supplemental tables can be found here.