In this issue of Blood, Iqbal and colleagues report the results of gene-expression profiling on 144 patients with various T-cell lymphomas from the International Peripheral T Cell Lymphoma Project.1 This report is the largest comprehensive study thus far and provides the molecular signatures of these heterogeneous diseases.
Peripheral T-cell lymphoma (PTCL) is a heterogeneous group of lymphomas. A wide variety of different histologic subtypes have been recognized.2 They constitute approximately 10% to 15% of aggressive lymphomas. Symbolic of the diagnostic issues, the most common subtype is a heterogeneous group of PTCL that has been called unclassifiable (PTCL-U). In terms of frequency, this is followed by anaplastic large cell lymphoma (ALCL) and angioimmunoblastic T-cell lymphoma (AITL). PTCL patients were found to have generally poorer prognoses than patients with B-cell lymphoma when treated with conventional chemotherapy.3 Treating such patients is a challenge as the different disease entities are not easy to distinguish by conventional histology. Even with immunohistochemistry, overlapping diagnoses are frequent. Clinicians want to be confident that the diseases identified by morphologic analysis have distinct biologic etiologies and behaviors. Using gene-profiling technology, Iqbal and colleagues were able to construct robust molecular classifiers for angioimmunoblastic T-cell lymphoma (AITL), ALK-positive anaplastic large cell lymphoma (ALK + ALCL), and adult T-cell leukemia/lymphoma (ATLL). These classifiers reflect the biologic characteristics of the tumor cells as well as their microenvironment. The molecular signature of AITL described previously4 was derived from a peculiar subset of CD4+ T cells normally found in reactive germinal centers (follicular helper T-cells [TFH]) with a significant contribution by B cells and follicular dendritic cells. For ALK + ALCL, TH17 cell–associated molecules were predominant. However, PTCL-U and ALK-negative ALCL are more heterogeneous and not so easy to classify. Nevertheless, Iqbal et al describe a rare entity of PTCLs with cytotoxic T-cell features and a poor prognosis.
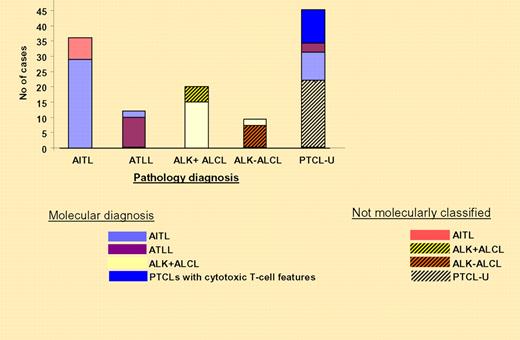
The correlation between the molecular and the pathology-based diagnoses are illustrated on the right. See the complete figure in the article beginning on page 1026.
The correlation between the molecular and the pathology-based diagnoses are illustrated on the right. See the complete figure in the article beginning on page 1026.
It is clear from the data presented by Iqbal et al that gene-expression profiling can classify cases of AITL, ATLL, and ALK + ALCL as distinct pathologic diagnoses. They nicely show the comparison between the classifiers and morphologic characteristics for each subset (see figure). This classifier allowed them to identify a significant number of morphologic PTCL-U cases and other PTCLs as AITL. Unfortunately, the molecular signature was less helpful in the more difficult cases of PTCL-U and ALK-ALCL. The time to establish a microarray with a reasonable number of genes to determine diagnosis has not arrived; however, such an examination will allow direct comparison of compatible data. More important is the need to validate the molecular signatures of these T-cell disorders with more samples.
If the molecular signatures are describing the biology of tumor cells, can they improve the prognostic parameters? The authors stated that this was possible, especially in the group of AITL patients in which they could distinguish a very high-risk versus a low-risk group with a 3-year probability of survival of 50% on the basis of molecular signatures. This analysis was performed on 35 patients and raises methodological questions. Biologists are always enthusiastic to find a correlation between their findings and the outcomes of patients. Numerous prognostic markers have been described in different types of lymphoma. Because of the cost and complexity of their experiments, the number of patients included in each group is limited. These markers have to be correlated with other clinical data in large cohort studies to be sure that they are representative of the disease. There has not been a study of similar magnitude evaluating the International Prognostic Index (IPI) specifically in PTCLs; however, several retrospective studies have demonstrated that the clinical IPI is a useful prognostic tool in some PTCL subtypes. The IPI or modified IPI can be effective in identifying different prognostic risk groups in PTCL-U, ALK + ALCL, and NK/T lymphoma. The IPI was not useful in AITL,5 and with the peculiar presentation of AILT, the finding that host/tumor interactions have a significant impact on survival makes sense. In fact, as T-cell lymphoma generally has a poor prognosis, the distinction in the high-risk population between poor and very poor becomes somewhat meaningless.
From this point of view, the most important point is how the biology can identify therapeutic targets for new drugs. We have not made significant progress in the therapy of T-cell lymphoma with conventional treatment, with only 20% to 30% of patients surviving 5 years. More encouraging survival data have been reported with autologous or allogeneic stem cell transplantation.6 Based on data from this paper, several pathways should be explored in therapy for AITL, such as inhibition of the NF-kB pathway, the anti-angiogenic pathway, microenvironment, and anti-IL6 blockade. Although some clinical studies are ongoing, the road is still long to find the right targets for PTCL. It is time to design prospective studies on T-cell lymphoma that incorporate up-front frozen material to ensure molecular characterization. This study is a step in that direction.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal