Abstract
We investigated the benefit of adding all-trans retinoic acid (ATRA) to chemotherapy for younger patients with nonacute promyelocytic acute myeloid leukemia and high-risk myelodysplastic syndrome, and considered interactions between treatment and molecular markers. Overall, 1075 patients less than 60 years of age were randomized to receive or not receive ATRA in addition to daunorubicin/Ara-C/thioguanine chemotherapy with Ara-C at standard or double standard dose. There were data on FLT3 internal tandem duplications and NPM1 mutations (n = 592), CEBPA mutations (n = 423), and MN1 expression (n = 195). The complete remission rate was 68% with complete remission with incomplete count recovery in an additional 16%; 8-year overall survival was 32%. There was no significant treatment effect for any outcome, with no significant interactions between treatment and demographics, or cytarabine randomization. Importantly, there were no interactions by FLT3/internal tandem duplications, NPM1, or CEBPA mutation. There was a suggestion that ATRA reduced relapse in patients with lower MN1 levels, but no significant effect on overall survival. Results were consistent when restricted to patients with normal karyotype. ATRA has no overall effect on treatment outcomes in this group of patients. The study did not identify any subgroup of patients likely to derive a significant survival benefit from the addition of ATRA to chemotherapy. This study is registered at http://www.controlled-trials.com under ISRCTN17833622.
Introduction
The addition of all-trans retinoic acid (ATRA) to chemotherapy has revolutionized the treatment of acute promyelocytic leukemia (APL),1-3 but preclinical studies have also provided a rationale for the use of ATRA in non-APL acute myeloid leukemia (AML). In vitro sensitivity assays have demonstrated efficacy of single agent ATRA in some cases of non-APL AML,4 but more compelling are the observations by McCulloch and colleagues that cotreatment of leukemic blasts with ATRA makes them more sensitive to cytarabine and anthracycline.5,6 A postulated mechanism is the ATRA-induced shortening of the BCL2 half-life, which has been implicated as a resistance mechanism in AML.7-9
The clinical results of ATRA in non-APL AML have been variable. Venditti et al found that the combination of low-dose Ara-C and ATRA resulted in a remarkably high complete remission (CR) rate in a poor-prognosis AML patient population, particularly those with a lower marrow blast count,10 but in a randomized phase 2 trial Estey et al found no benefit from the addition of ATRA to a fludarabine and cytarabine regimen in newly diagnosed poor prognosis AML and myelodysplastic syndrome.11 Similarly, in a United Kingdom Medical Research Council (MRC) trial, we failed to demonstrate an overall advantage of adding ATRA to standard chemotherapy in patients who were deemed to have high risk AML or who had relapsed,12 or to low-dose cytarabine or hydroxycarbamide as first-line treatment of older patients not considered fit for intensive therapy.13 However, in a randomized trial of older patients conducted by the Austrian-German AML Study Group (AML HD98B), there were improved remission, event-free survival (EFS), and overall survival (OS) in patients receiving ATRA in combination with intensive chemotherapy.14 Of particular interest was their subsequent report that, although the presence of a nucleophosmin (NPM1) mutation was associated with a borderline higher CR rate, improved relapse-free survival and OS were restricted to those patients with a NPM1 mutation who did not have a fms-like tyrosine kinase 3 internal tandem duplication (FLT3/ITD) and were treated with ATRA.15 Furthermore, overexpression of the meningioma 1 gene (MN1), an adverse prognostic marker, is associated with unmutated NPM1 and resistance to ATRA16,17 : in a subset of patients in the AML HD98B trial, coadministration of ATRA led to significantly improved EFS and OS in patients with low MN1 expression.17 Another good prognosis group can be identified by the presence of a mutation in the CCAAT enhancer-binding protein/α (CEBPA) gene, and the response to ATRA is thought to be mediated through this transcription factor.18 The impact of ATRA on outcome in this patient group has not been reported.
In the United Kingdom MRC AML12 trial, we combined ATRA with standard induction chemotherapy in patients, many of whom were molecularly characterized, allowing an assessment of both the effect of the addition of ATRA in younger patients and also the relationship between any treatment benefit and molecular characteristics of patients.
Methods
Patients and treatment protocols
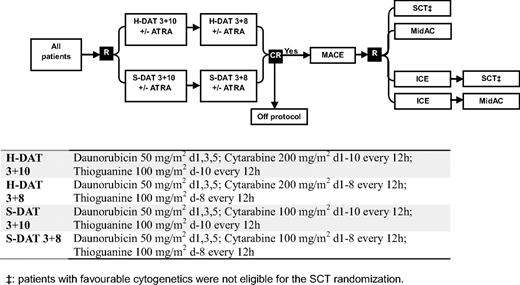
Between November 1998 and May 2002, patients entering the United Kingdom MRC AML12 trial (ICRTN 55678797) were randomized in induction to 2 courses of daunorubicin, Ara-C, and thioguanine (DAT) with Ara-C at a dose of either 100 mg/m2 bid (standard DAT) or 200 mg/m2 bid (high DAT), each with or without ATRA at a dose of 45 mg/m2 from days 1 to 60. In consolidation, all patients received Amsacrine/Ara-C/Etoposide as course 3 and were then randomized between 1 (Mitozantrone/Ara-C) and 2 further courses (Idaribicin/Ara-C/Etoposide, Mitozantrone/Ara-C), and to chemotherapy versus transplant as the final allocated course (ie, as course 4 or 5). Details of the treatment are given in Figure 1.
The trial was open to any patient with de novo or secondary AML and high risk myelodysplastic syndrome, which was defined as marrow blasts greater than 10%. Patients who were in blast transformation of chronic myeloid leukemia, were pregnant or lactating, had other active concurrent malignancy, had received prior cytotoxic therapy for leukemia other than hydroxycarbamide, or who had APL were excluded. Patients were required to give written consent for each of the randomizations separately, in accordance with the Declaration of Helsinki. The trial was approved by the Wales Multicentre Research Ethics Committee and each participating institution's ethics review committee. Archived tissue was stored with informed written consent. The results of the main interventions in this trial have been reported elsewhere.19
Cytogenetics
Cytogenetic analysis was carried out in accredited regional laboratories, and the reports reviewed centrally. The designation of patients as favorable, intermediate, or adverse risk was based on our previously published criteria.20
Molecular characterization
DNA and cDNA were prepared from cells that were banked at the time of diagnosis. In some patients, DNA was extracted retrospectively from diagnostic bone marrow smears using the DNeasy blood and tissue kit (QIAGEN).
FLT3/ITD and NPM1 mutation status using DNA were determined as published previously.21 Using cDNA, methods were as for DNA, except that a nested polymerase chain reaction (PCR) was first performed for FLT3/ITD analysis to circumvent false-positive results due to contaminating DNA, and an alternative exon 11 forward primer was used for the NPM1 analysis (see supplemental Methods, available on the Blood website; see the Supplemental Materials link at the top of the online article). For the CEBPA gene, the entire coding sequence was PCR amplified in 3 overlapping fragments that were analyzed using denaturing high performance liquid chromatography and samples with abnormal chromatograms sequenced (see supplemental Methods).
For the assessment of MN1 expression, LightCycler quantitative PCR (Roche) was performed on cDNA using MN1 and ABL exonic primers (see supplemental Methods). A ratio of MN1 to ABL expression was calculated for each sample, and either the base 10 logarithm of the ratio used as continuous variable, or the dataset dichotomized at the median into low and high expression groups.
Definitions of clinical end points
The protocol defined CR as a normocellular bone marrow aspirate containing less than 5% leukemic blast cells and showing evidence of normal maturation of other marrow elements. The persistence of myelodysplastic features did not exclude the diagnosis of CR; in this report, however, CR patients had to have neutrophil recovery to 1.0 × 109/L and platelets to 100 × 109/L, with no evidence of extramedullary disease. Patients who achieved CR according to the protocol, but who did not exhibit recovery, are denoted in this study as complete remission with incomplete count recovery (CRi).22 Remission failures were classified by the investigating clinician as due either to induction death, that is, death within 30 days of the start of treatment related to treatment and/or hypoplasia, or as resistant disease, that is, related to the failure of therapy to eliminate the disease (including partial remissions with 5%-15% blasts). Where the clinician's evaluation was not available, deaths within 30 days of entry were classified as induction death and deaths at more than 30 days as resistant disease.
The following definitions are also used: OS is the time from randomization to death; for remitters, relapse-free survival is the time from CR/CRi to first event (either relapse or death in CR), the cumulative incidence of relapse is the cumulative probability of relapse with death as a competing risk, and the cumulative incidence of death in first CR is the cumulative probability of dying in CR/CRi with relapse as competing risk. All outcome percentages quoted are given at 8 years.
Statistical analysis
The trial was designed to recruit up to 1200 patients to the ATRA randomization, which would give 95% power to detect a 10% absolute difference in long-term survival from 40% to 50%; 1000 patients recruited would give approximately 90% power to detect the same difference. For time to event endpoints, Kaplan-Meier life tables were constructed and were compared by the log-rank test. Surviving patients were censored on January 1, 2008, when follow-up was up to date for 93% of all patients (the small number of patients lost to follow-up are censored at the date they were last known to be alive). Median follow-up for surviving patients is 8.4 years, with the longest survival at 13.0 years.
Categorical endpoints (eg, CR rates) were compared between arms by Mantel-Haenszel tests, giving Peto odds ratios (OR) and confidence intervals. Continuous variables (eg, nonhematologic toxicity and supportive care requirements) were analyzed by parametric (t test) or nonparametric (Wilcoxon rank sum) tests as appropriate. Interactions between randomized comparisons, and between ATRA and baseline covariates (age, sex, World Health Organization performance status, white blood cell count [WBC], and cytogenetics) and molecular markers (FLT3/ITD, NPM1, CEBPA, and MN1) were investigated by stratified analyses, that is, with each comparison adjusted for the others, using tests for heterogeneity or trend over strata.
Primary analyses of MN1 level were as a continuous variable using logistic and Cox's proportional hazard regression methods, with P values for interaction given using the deviance. OR or hazard ratio (HR) with 95% confidence interval (CI) are quoted for all main endpoints. In all cases, an OR or HR less than 1.0 indicates benefit for ATRA. All P values are 2-tailed. All analyses are performed on the “intention-to-treat” principle with all patients analyzed in their allocated arms, irrespective of whether or not they actually received allocated treatment.
Results
Impact of ATRA on response to therapy and outcome in the total cohort
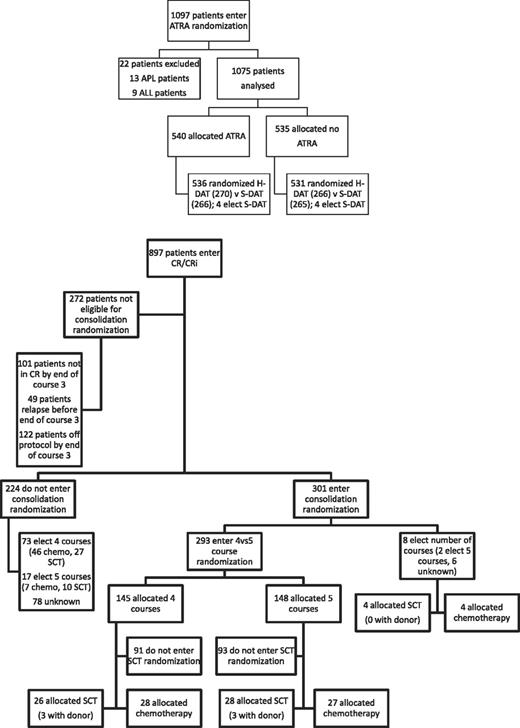
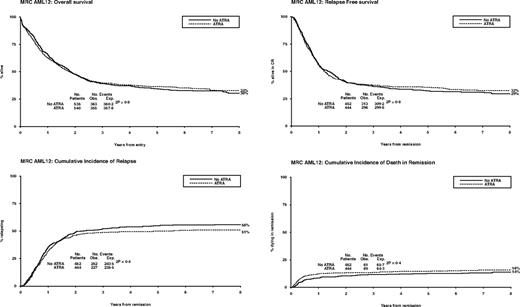
Between November 1998 and May 2002, 1075 patients were randomized to receive ATRA or not with the first 2 courses of induction treatment in the United Kingdom MRC AML12 trial. Patients were randomized between standard DAT and high DAT, and between 4 or 5 courses of total treatment, with the final course either transplant or chemotherapy. There was no difference in outcome for any of these interventions.19 The demographics are shown in Table 1 and the deployment of patients is shown in Figure 2. Compliance with ATRA treatment was 95%. Overall, there was no effect from the addition of ATRA (Figure 3) with equal remission rates (CR + CRi: 83% with vs 84% without ATRA) and OS (33% vs 30%; HR, 0.98 [0.85-1.14]). There were no differences in toxicity profiles, and no significant differences in either hematologic recovery kinetics after each course of treatment or resource usage (supplemental Table 1). Compliance with consolidation therapy was unaffected by ATRA therapy, with 29% versus 27% entering consolidation randomizations (P = .3). In stratified analyses, there were no significant interactions between baseline demographics, or other randomizations and ATRA treatment, with no subgroup showing a benefit for ATRA therapy (supplemental Figure 1).
Characteristics of AML12 patients in the ATRA randomization
| . | Total . | ATRA vs no ATRA, n (%) . | FLT3/ITD status, n (%) . | NPM1 status, n (%) . | CEPBA status, n (%) . | MN1 level, n (%) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATRA . | No ATRA . | P . | WT . | Mutant . | P . | WT . | Mutant . | P . | WT . | Mutant . | P . | Below median . | Above median . | P . | ||
| Overall | 1075 | 540 | 535 | 455 | 137 | 385 | 207 | 388 | 35 | 97 | 98 | |||||
| Age, y | .8* | .6* | .02* | .4* | .4* | |||||||||||
| 0-14 | 1 | 1 (< .5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 15-29 | 157 | 81 (15) | 76 (14) | 78 (17) | 16 (12) | 77 (20) | 17 (8) | 58 (15) | 8 (23) | 18 (19) | 20 (20) | |||||
| 30-39 | 183 | 91 (17) | 92 (17) | 71 (16) | 28 (20) | 63 (16) | 36 (17) | 73 (19) | 2 (6) | 13 (13) | 16 (16) | |||||
| 40-49 | 259 | 129 (24) | 130 (24) | 112 (25) | 34 (25) | 84 (22) | 62 (30) | 100 (26) | 6 (17) | 23 (24) | 26 (27) | |||||
| 50-59 | 432 | 216 (40) | 216 (40) | 174 (38) | 53 (39) | 142 (37) | 85 (41) | 141 (36) | 16 (46) | 36 (37) | 31 (32) | |||||
| 60+ | 43 | 22 (4) | 21 (4) | 20 (4) | 6 (4) | 19 (5) | 7 (3) | 16 (4) | 3 (9) | 7 (7) | 5 (5) | |||||
| Median (range) | 48 (14-68) | 47 (14-68) | 48 (15-67) | 47 (15-67) | 47 (15-68) | 46 (15-68) | 48 (15-64) | 46 (15-68) | 52 (16-67) | 47 (17-64) | 46 (19-68) | |||||
| Sex | .4 | .2 | .2 | .16 | .2 | |||||||||||
| Female | 540 | 264 (49) | 276 (52) | 222 (49) | 75 (55) | 186 (48) | 111 (54) | 203 (52) | 14 (40) | 52 (54) | 44 (45) | |||||
| Male | 535 | 276 (51) | 259 (48) | 233 (51) | 62 (45) | 199 (52) | 96 (46) | 185 (48) | 21 (60) | 45 (46) | 54 (55) | |||||
| Diagnosis | .2 | .1 | < .001 | .8 | .18 | |||||||||||
| De novo | 970 | 481 (89) | 489 (91) | 407 (89) | 129 (94) | 336 (87) | 200 (97) | 350 (90) | 31 (89) | 89 (92) | 84 (86) | |||||
| Secondary | 105 | 59 (11) | 46 (9) | 48 (11) | 8 (6) | 49 (13) | 7 (3) | 38 (10) | 4 (11) | 8 (8) | 14 (14) | |||||
| WBC | .04* | < .001* | < .001* | .5* | .6* | |||||||||||
| < 10 | 494 | 229 (42) | 265 (50) | 219 (49) | 25 (19) | 191 (51) | 53 (27) | 144 (38) | 7 (21) | 31 (33) | 47 (50) | |||||
| 10-49.9 | 333 | 177 (34) | 156 (30) | 142 (32) | 52 (39) | 111 (29) | 83 (42) | 129 (34) | 18 (53) | 42 (44) | 25 (27) | |||||
| 50-99.9 | 120 | 64 (12) | 56 (11) | 44 (10) | 29 (22) | 40 (11) | 33 (17) | 52 (14) | 6 (18) | 15 (16) | 6 (6) | |||||
| 100+ | 103 | 56 (11) | 47 (9) | 39 (9) | 26 (20) | 35 (9) | 30 (15) | 51 (14) | 3 (9) | 7 (7) | 16 (17) | |||||
| Missing | 25 | 14 | 11 | 11 | 5 | 8 | 8 | 12 | 1 | 2 | 4 | |||||
| Median (range) | 12.55 (0.3-559) | 13.95 (0.5-559) | 10.0 (0.3-348) | 10.25 (0.3-502) | 39.15 (0.9-559) | 9.7 (0.3-502) | 28.4 (0.7-559) | 19.45 (0.4-559) | 26.95 (3.7-275) | 21.6 (0.7-310) | 10.8 (0.5-275) | |||||
| Performance status | .8* | .7* | .2* | .03* | .8* | |||||||||||
| WHO 0 | 716 | 361 (67) | 355 (66) | 291 (64) | 87 (64) | 254 (66) | 124 (60) | 228 (59) | 28 (80) | 57 (59) | 56 (57) | |||||
| WHO 1 | 273 | 137 (25) | 136 (25) | 128 (28) | 38 (28) | 101 (26) | 65 (31) | 120 (31) | 6 (17) | 32 (33) | 36 (37) | |||||
| WHO 2 | 36 | 17 (3) | 19 (4) | 17 (4) | 3 (2) | 13 (3) | 7 (3) | 16 (4) | 0 | 3 (3) | 3 (3) | |||||
| WHO 3 | 42 | 21 (4) | 21 (4) | 15 (3) | 8 (6) | 14 (4) | 9 (4) | 20 (5) | 1 (3) | 4 (4) | 2 (2) | |||||
| WHO 4 | 8 | 4 (1) | 4 (1) | 4 (1) | 1 (1) | 3 (1) | 2 (1) | 4 (1) | 0 | 1 (1) | 1 (1) | |||||
| Cytogenetics | .8 | < .001 | < .001 | .1 | < .001 | |||||||||||
| Favorable | 118 | 62 (14) | 56 (13) | 61 (16) | 5 (5) | 66 (20) | 0 | 52 (16) | 0 | 7 (9) | 16 (20) | |||||
| NK | 420 | 218 (51) | 202 (49) | 155 (41) | 78 (73) | 96 (30) | 137 (85) | 153 (48) | 16 (64) | 50 (63) | 26 (33) | |||||
| Other intermediate | 180 | 90 (21) | 90 (22) | 92 (24) | 19 (18) | 92 (28) | 19 (12) | 71 (22) | 7 (28) | 18 (23) | 20 (25) | |||||
| Adverse | 128 | 60 (14) | 68 (16) | 69 (18) | 5 (5) | 69 (21) | 5 (3) | 46 (14) | 2 (8) | 5 (6) | 18 (23) | |||||
| Unknown | 229 | 110 | 119 | 78 | 30 | 62 | 46 | 66 | 10 | 17 | 18 | |||||
| FAB type | .8 | .006 | .03 | .13 | .2 | |||||||||||
| M0 | 60 | 31 (6) | 29 (6) | 36 (9) | 7 (6) | 37 (11) | 6 (3) | 28 (8) | 1 (3) | 0 | 8 (9) | |||||
| M1 | 204 | 107 (22) | 97 (20) | 75 (18) | 30 (24) | 67 (19) | 38 (20) | 66 (19) | 11 (33) | 24 (26) | 20 (22) | |||||
| M2 | 282 | 137 (28) | 145 (31) | 131 (32) | 34 (27) | 108 (31) | 57 (30) | 109 (31) | 15 (45) | 31 (34) | 32 (36) | |||||
| M4 | 192 | 98 (20) | 94 (20) | 80 (20) | 26 (21) | 65 (19) | 41 (22) | 75 (21) | 3 (9) | 17 (18) | 15 (17) | |||||
| M5 | 126 | 71 (14) | 55 (12) | 41 (10) | 25 (20) | 34 (10) | 32 (17) | 48 (14) | 3 (9) | 11 (12) | 8 (9) | |||||
| M6 | 55 | 24 (5) | 31 (7) | 29 (7) | 2 (2) | 22 (6) | 9 (5) | 14 (4) | 0 | 5 (5) | 3 (3) | |||||
| M7 | 23 | 10 (2) | 13 (3) | 6 (1) | 0 | 3 (1) | 3 (2) | 4 (1) | 0 | 1 (1) | 1 (1) | |||||
| Bilineage | 2 | 1 (< .5) | 1 (< .5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| RAEB-t | 22 | 13 (3) | 9 (2) | 12 (3) | 1 (1) | 9 (3) | 4 (2) | 8 (2) | 0 | 3 (3) | 2 (2) | |||||
| Other/unknown | 109 | 48 | 61 | 45 | 12 | 40 | 17 | 36 | 2 | 5 | 9 | |||||
| H-DAT | 536 | 270 (50) | 266 (50) | 1.0 | 246 (54) | 63 (47) | .18 | 210 (55) | 99 (49) | .14 | 205 (54) | 16 (47) | 0.5 | 44 (46) | 58 (60) | 0.05 |
| S-DAT | 531 | 266 (50) | 265 (50) | 207 (46) | 69 (53) | 171 (45) | 105 (51) | 177 (46) | 18 (53) | 52 (54) | 39 (40) | |||||
| FLT3 ITD WT | 455 | 238 (76) | 217 (78) | 0.5 | 337 (88) | 118 (57) | < .001 | 292 (75) | 28 (80) | .5 | 68 (72) | 70 (79) | .3 | |||
| ITD mutant | 137 | 76 (24) | 61 (22) | 48 (12) | 89 (43) | 96 (25) | 7 (20) | 26 (28) | 19 (21) | |||||||
| NPM1 WT | 385 | 201 (64) | 184 (66) | .6 | 337 (74) | 48 (35) | < .001 | 240 (62) | 28 (80) | .03 | 46 (49) | 75 (84) | < .001 | |||
| NPM1 mutant | 207 | 113 (36) | 94 (34) | 118 (26) | 89 (65) | 148 (38) | 7 (20) | 48 (51) | 14 (16) | |||||||
| CEBPA WT | 388 | 214 (91) | 174 (92) | .8 | 292 (91) | 96 (93) | .5 | 240 (90) | 148 (95) | .03 | 74 (91) | 75 (91) | 1.0 | |||
| CEBPA mutant | 35 | 20 (9) | 15 (8) | 28 (9) | 7 (7) | 28 (10) | 7 (5) | 7 (9) | 7 (9) | |||||||
| MN1 low | 97 | 54 (50) | 43 (49) | .9 | 68 (49) | 26 (58) | .3 | 46 (38) | 48 (77) | < .001 | 74 (50) | 7 (50) | 1.0 | |||
| MN1 high | 98 | 54 (50) | 44 (51) | 70 (51) | 19 (42) | 75 (62) | 14 (23) | 75 (50) | 7 (50) | |||||||
| . | Total . | ATRA vs no ATRA, n (%) . | FLT3/ITD status, n (%) . | NPM1 status, n (%) . | CEPBA status, n (%) . | MN1 level, n (%) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATRA . | No ATRA . | P . | WT . | Mutant . | P . | WT . | Mutant . | P . | WT . | Mutant . | P . | Below median . | Above median . | P . | ||
| Overall | 1075 | 540 | 535 | 455 | 137 | 385 | 207 | 388 | 35 | 97 | 98 | |||||
| Age, y | .8* | .6* | .02* | .4* | .4* | |||||||||||
| 0-14 | 1 | 1 (< .5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 15-29 | 157 | 81 (15) | 76 (14) | 78 (17) | 16 (12) | 77 (20) | 17 (8) | 58 (15) | 8 (23) | 18 (19) | 20 (20) | |||||
| 30-39 | 183 | 91 (17) | 92 (17) | 71 (16) | 28 (20) | 63 (16) | 36 (17) | 73 (19) | 2 (6) | 13 (13) | 16 (16) | |||||
| 40-49 | 259 | 129 (24) | 130 (24) | 112 (25) | 34 (25) | 84 (22) | 62 (30) | 100 (26) | 6 (17) | 23 (24) | 26 (27) | |||||
| 50-59 | 432 | 216 (40) | 216 (40) | 174 (38) | 53 (39) | 142 (37) | 85 (41) | 141 (36) | 16 (46) | 36 (37) | 31 (32) | |||||
| 60+ | 43 | 22 (4) | 21 (4) | 20 (4) | 6 (4) | 19 (5) | 7 (3) | 16 (4) | 3 (9) | 7 (7) | 5 (5) | |||||
| Median (range) | 48 (14-68) | 47 (14-68) | 48 (15-67) | 47 (15-67) | 47 (15-68) | 46 (15-68) | 48 (15-64) | 46 (15-68) | 52 (16-67) | 47 (17-64) | 46 (19-68) | |||||
| Sex | .4 | .2 | .2 | .16 | .2 | |||||||||||
| Female | 540 | 264 (49) | 276 (52) | 222 (49) | 75 (55) | 186 (48) | 111 (54) | 203 (52) | 14 (40) | 52 (54) | 44 (45) | |||||
| Male | 535 | 276 (51) | 259 (48) | 233 (51) | 62 (45) | 199 (52) | 96 (46) | 185 (48) | 21 (60) | 45 (46) | 54 (55) | |||||
| Diagnosis | .2 | .1 | < .001 | .8 | .18 | |||||||||||
| De novo | 970 | 481 (89) | 489 (91) | 407 (89) | 129 (94) | 336 (87) | 200 (97) | 350 (90) | 31 (89) | 89 (92) | 84 (86) | |||||
| Secondary | 105 | 59 (11) | 46 (9) | 48 (11) | 8 (6) | 49 (13) | 7 (3) | 38 (10) | 4 (11) | 8 (8) | 14 (14) | |||||
| WBC | .04* | < .001* | < .001* | .5* | .6* | |||||||||||
| < 10 | 494 | 229 (42) | 265 (50) | 219 (49) | 25 (19) | 191 (51) | 53 (27) | 144 (38) | 7 (21) | 31 (33) | 47 (50) | |||||
| 10-49.9 | 333 | 177 (34) | 156 (30) | 142 (32) | 52 (39) | 111 (29) | 83 (42) | 129 (34) | 18 (53) | 42 (44) | 25 (27) | |||||
| 50-99.9 | 120 | 64 (12) | 56 (11) | 44 (10) | 29 (22) | 40 (11) | 33 (17) | 52 (14) | 6 (18) | 15 (16) | 6 (6) | |||||
| 100+ | 103 | 56 (11) | 47 (9) | 39 (9) | 26 (20) | 35 (9) | 30 (15) | 51 (14) | 3 (9) | 7 (7) | 16 (17) | |||||
| Missing | 25 | 14 | 11 | 11 | 5 | 8 | 8 | 12 | 1 | 2 | 4 | |||||
| Median (range) | 12.55 (0.3-559) | 13.95 (0.5-559) | 10.0 (0.3-348) | 10.25 (0.3-502) | 39.15 (0.9-559) | 9.7 (0.3-502) | 28.4 (0.7-559) | 19.45 (0.4-559) | 26.95 (3.7-275) | 21.6 (0.7-310) | 10.8 (0.5-275) | |||||
| Performance status | .8* | .7* | .2* | .03* | .8* | |||||||||||
| WHO 0 | 716 | 361 (67) | 355 (66) | 291 (64) | 87 (64) | 254 (66) | 124 (60) | 228 (59) | 28 (80) | 57 (59) | 56 (57) | |||||
| WHO 1 | 273 | 137 (25) | 136 (25) | 128 (28) | 38 (28) | 101 (26) | 65 (31) | 120 (31) | 6 (17) | 32 (33) | 36 (37) | |||||
| WHO 2 | 36 | 17 (3) | 19 (4) | 17 (4) | 3 (2) | 13 (3) | 7 (3) | 16 (4) | 0 | 3 (3) | 3 (3) | |||||
| WHO 3 | 42 | 21 (4) | 21 (4) | 15 (3) | 8 (6) | 14 (4) | 9 (4) | 20 (5) | 1 (3) | 4 (4) | 2 (2) | |||||
| WHO 4 | 8 | 4 (1) | 4 (1) | 4 (1) | 1 (1) | 3 (1) | 2 (1) | 4 (1) | 0 | 1 (1) | 1 (1) | |||||
| Cytogenetics | .8 | < .001 | < .001 | .1 | < .001 | |||||||||||
| Favorable | 118 | 62 (14) | 56 (13) | 61 (16) | 5 (5) | 66 (20) | 0 | 52 (16) | 0 | 7 (9) | 16 (20) | |||||
| NK | 420 | 218 (51) | 202 (49) | 155 (41) | 78 (73) | 96 (30) | 137 (85) | 153 (48) | 16 (64) | 50 (63) | 26 (33) | |||||
| Other intermediate | 180 | 90 (21) | 90 (22) | 92 (24) | 19 (18) | 92 (28) | 19 (12) | 71 (22) | 7 (28) | 18 (23) | 20 (25) | |||||
| Adverse | 128 | 60 (14) | 68 (16) | 69 (18) | 5 (5) | 69 (21) | 5 (3) | 46 (14) | 2 (8) | 5 (6) | 18 (23) | |||||
| Unknown | 229 | 110 | 119 | 78 | 30 | 62 | 46 | 66 | 10 | 17 | 18 | |||||
| FAB type | .8 | .006 | .03 | .13 | .2 | |||||||||||
| M0 | 60 | 31 (6) | 29 (6) | 36 (9) | 7 (6) | 37 (11) | 6 (3) | 28 (8) | 1 (3) | 0 | 8 (9) | |||||
| M1 | 204 | 107 (22) | 97 (20) | 75 (18) | 30 (24) | 67 (19) | 38 (20) | 66 (19) | 11 (33) | 24 (26) | 20 (22) | |||||
| M2 | 282 | 137 (28) | 145 (31) | 131 (32) | 34 (27) | 108 (31) | 57 (30) | 109 (31) | 15 (45) | 31 (34) | 32 (36) | |||||
| M4 | 192 | 98 (20) | 94 (20) | 80 (20) | 26 (21) | 65 (19) | 41 (22) | 75 (21) | 3 (9) | 17 (18) | 15 (17) | |||||
| M5 | 126 | 71 (14) | 55 (12) | 41 (10) | 25 (20) | 34 (10) | 32 (17) | 48 (14) | 3 (9) | 11 (12) | 8 (9) | |||||
| M6 | 55 | 24 (5) | 31 (7) | 29 (7) | 2 (2) | 22 (6) | 9 (5) | 14 (4) | 0 | 5 (5) | 3 (3) | |||||
| M7 | 23 | 10 (2) | 13 (3) | 6 (1) | 0 | 3 (1) | 3 (2) | 4 (1) | 0 | 1 (1) | 1 (1) | |||||
| Bilineage | 2 | 1 (< .5) | 1 (< .5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| RAEB-t | 22 | 13 (3) | 9 (2) | 12 (3) | 1 (1) | 9 (3) | 4 (2) | 8 (2) | 0 | 3 (3) | 2 (2) | |||||
| Other/unknown | 109 | 48 | 61 | 45 | 12 | 40 | 17 | 36 | 2 | 5 | 9 | |||||
| H-DAT | 536 | 270 (50) | 266 (50) | 1.0 | 246 (54) | 63 (47) | .18 | 210 (55) | 99 (49) | .14 | 205 (54) | 16 (47) | 0.5 | 44 (46) | 58 (60) | 0.05 |
| S-DAT | 531 | 266 (50) | 265 (50) | 207 (46) | 69 (53) | 171 (45) | 105 (51) | 177 (46) | 18 (53) | 52 (54) | 39 (40) | |||||
| FLT3 ITD WT | 455 | 238 (76) | 217 (78) | 0.5 | 337 (88) | 118 (57) | < .001 | 292 (75) | 28 (80) | .5 | 68 (72) | 70 (79) | .3 | |||
| ITD mutant | 137 | 76 (24) | 61 (22) | 48 (12) | 89 (43) | 96 (25) | 7 (20) | 26 (28) | 19 (21) | |||||||
| NPM1 WT | 385 | 201 (64) | 184 (66) | .6 | 337 (74) | 48 (35) | < .001 | 240 (62) | 28 (80) | .03 | 46 (49) | 75 (84) | < .001 | |||
| NPM1 mutant | 207 | 113 (36) | 94 (34) | 118 (26) | 89 (65) | 148 (38) | 7 (20) | 48 (51) | 14 (16) | |||||||
| CEBPA WT | 388 | 214 (91) | 174 (92) | .8 | 292 (91) | 96 (93) | .5 | 240 (90) | 148 (95) | .03 | 74 (91) | 75 (91) | 1.0 | |||
| CEBPA mutant | 35 | 20 (9) | 15 (8) | 28 (9) | 7 (7) | 28 (10) | 7 (5) | 7 (9) | 7 (9) | |||||||
| MN1 low | 97 | 54 (50) | 43 (49) | .9 | 68 (49) | 26 (58) | .3 | 46 (38) | 48 (77) | < .001 | 74 (50) | 7 (50) | 1.0 | |||
| MN1 high | 98 | 54 (50) | 44 (51) | 70 (51) | 19 (42) | 75 (62) | 14 (23) | 75 (50) | 7 (50) | |||||||
Test for trend.
Overall outcomes for ATRA versus not randomization: OS, relapse-free survival, cumulative incidence of relapse, and cumulative incidence of death in first CR.
Overall outcomes for ATRA versus not randomization: OS, relapse-free survival, cumulative incidence of relapse, and cumulative incidence of death in first CR.
Impact of ATRA on response to therapy by molecularly defined subgroups
Data were available on FLT3/ITD and NPM1 mutations in 592 patients, CEBPA mutations in 423 patients, and MN1 expression level in 195 patients. Patients with FLT3/ITD, NPM1, and CEBPA data tended to have a higher presenting WBC (P = .005 for FLT3/NPM1; P < .001 for CEBPA), and patients with CEBPA data a worse performance status (P = .002). There were no significant differences in outcome in those with/without FLT3/NPM1 data; patients with CEBPA data tended to have marginally better OS (36% vs 31%; P = .04). There were no significant differences in demographics in those with/without MN1 data, although survival was slightly better in those with data (39% vs 31%; P = .04). In all cases, there was no significant heterogeneity in the effect of ATRA by whether or not molecular data were available (OS by FLT3/NPM1, P = .8; by CEBPA, P = .5; by MN1, P = .4; supplemental Figure 2).
The relationship of molecular characteristics is shown in Table 1, with a significant association between NPM1 mutation and low MN1 expression as well as the known association of FLT3/ITD and NPM1 mutations. There were no significant associations between treatment received and FLT3/ITD, NPM1, CEBPA, or MN1 status. The level of MN1 did not affect clinical outcome.
Table 2 shows the outcomes by treatment in each of the molecularly defined subgroups.
Outcomes by molecular characteristics and treatment
| Outcome/treatment . | FLT3/ITD status, % . | NPM1 status, % . | ITD/NPM1 status, % . | CEBPA status, % . | MN1 expression, % . | Treatment . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT . | Mutant . | WT . | Mutant . | NPM1 mutant, ITD WT . | Others . | WT . | Mutant . | Below median . | Above median . | Overall, % . | HR/OR, 95% CI . | P . | |
| CR | 1.02 (0.79-1.32) | .9 | |||||||||||
| ATRA | 71 | 67 | 67 | 77 | 80 | 68 | 72 | 75 | 70 | 65 | 68 | ||
| No ATRA | 68 | 79 | 65 | 81 | 83 | 68 | 70 | 93 | 81 | 61 | 68 | ||
| CR/CRi | 1.16 (0.84-1.61) | .4 | |||||||||||
| ATRA | 84 | 84 | 81 | 89 | 89 | 82 | 84 | 90 | 79 | 82 | CRi 15 | ||
| No ATRA | 86 | 89 | 85 | 89 | 94 | 84 | 85 | 93 | 91 | 83 | CRi 16 | ||
| RD | 1.29 (0.84-1.98) | .2 | |||||||||||
| ATRA | 9 | 9 | 13 | 3 | 2 | 11 | 11 | 5 | 2 | 16 | 9 | ||
| No ATRA | 8 | 5 | 9 | 4 | 2 | 8 | 9 | 7 | 7 | 13 | 7 | ||
| ID | 1.02 (0.65-1.58) | .9 | |||||||||||
| ATRA | 7 | 7 | 7 | 8 | 9 | 6 | 6 | 5 | 19 | 4 | 8 | ||
| No ATRA | 6 | 7 | 7 | 6 | 4 | 7 | 6 | 0 | 2 | 2 | 8 | ||
| 30-day mortality | |||||||||||||
| ATRA | 6 | 5 | 5 | 7 | 8 | 6 | 4 | 5 | 15 | 4 | 7 | ||
| No ATRA | 6 | 7 | 5 | 6 | 4 | 6 | 5 | 0 | 5 | 2 | 7 | ||
| 8-week mortality | |||||||||||||
| ATRA | 9 | 7 | 8 | 8 | 9 | 8 | 7 | 5 | 17 | 6 | 9 | ||
| No ATRA | 8 | 8 | 9 | 7 | 4 | 9 | 8 | 7 | 5 | 5 | 10 | ||
| 8-year OS | 0.98 (0.85-1.14) | .8 | |||||||||||
| ATRA | 35 | 31 | 27 | 47 | 56 | 28 | 38 | 35 | 44 | 44 | 33 | ||
| No ATRA | 34 | 31 | 30 | 39 | 40 | 32 | 33 | 47 | 36 | 30 | 30 | ||
| 8-year RFS | 0.98 (0.83-1.14) | .8 | |||||||||||
| ATRA | 33 | 30 | 26 | 42 | 52 | 26 | 36 | 28 | 45 | 40 | 32 | ||
| No ATRA | 28 | 31 | 25 | 37 | 33 | 28 | 31 | 29 | 33 | 28 | 29 | ||
| 8-year CIR | 0.93 (0.78-1.11) | .4 | |||||||||||
| ATRA | 50 | 53 | 54 | 46 | 37 | 55 | 48 | 61 | 38 | 49 | 51 | ||
| No ATRA | 59 | 52 | 61 | 49 | 51 | 59 | 53 | 64 | 54 | 53 | 56 | ||
| 8-year CIDCR | 1.16 (0.82-1.63) | .4 | |||||||||||
| ATRA | 17 | 17 | 20 | 12 | 10 | 19 | 16 | 11 | 17 | 11 | 16 | ||
| No ATRA | 13 | 17 | 14 | 14 | 16 | 13 | 16 | 7 | 14 | 19 | 14 | ||
| Outcome/treatment . | FLT3/ITD status, % . | NPM1 status, % . | ITD/NPM1 status, % . | CEBPA status, % . | MN1 expression, % . | Treatment . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT . | Mutant . | WT . | Mutant . | NPM1 mutant, ITD WT . | Others . | WT . | Mutant . | Below median . | Above median . | Overall, % . | HR/OR, 95% CI . | P . | |
| CR | 1.02 (0.79-1.32) | .9 | |||||||||||
| ATRA | 71 | 67 | 67 | 77 | 80 | 68 | 72 | 75 | 70 | 65 | 68 | ||
| No ATRA | 68 | 79 | 65 | 81 | 83 | 68 | 70 | 93 | 81 | 61 | 68 | ||
| CR/CRi | 1.16 (0.84-1.61) | .4 | |||||||||||
| ATRA | 84 | 84 | 81 | 89 | 89 | 82 | 84 | 90 | 79 | 82 | CRi 15 | ||
| No ATRA | 86 | 89 | 85 | 89 | 94 | 84 | 85 | 93 | 91 | 83 | CRi 16 | ||
| RD | 1.29 (0.84-1.98) | .2 | |||||||||||
| ATRA | 9 | 9 | 13 | 3 | 2 | 11 | 11 | 5 | 2 | 16 | 9 | ||
| No ATRA | 8 | 5 | 9 | 4 | 2 | 8 | 9 | 7 | 7 | 13 | 7 | ||
| ID | 1.02 (0.65-1.58) | .9 | |||||||||||
| ATRA | 7 | 7 | 7 | 8 | 9 | 6 | 6 | 5 | 19 | 4 | 8 | ||
| No ATRA | 6 | 7 | 7 | 6 | 4 | 7 | 6 | 0 | 2 | 2 | 8 | ||
| 30-day mortality | |||||||||||||
| ATRA | 6 | 5 | 5 | 7 | 8 | 6 | 4 | 5 | 15 | 4 | 7 | ||
| No ATRA | 6 | 7 | 5 | 6 | 4 | 6 | 5 | 0 | 5 | 2 | 7 | ||
| 8-week mortality | |||||||||||||
| ATRA | 9 | 7 | 8 | 8 | 9 | 8 | 7 | 5 | 17 | 6 | 9 | ||
| No ATRA | 8 | 8 | 9 | 7 | 4 | 9 | 8 | 7 | 5 | 5 | 10 | ||
| 8-year OS | 0.98 (0.85-1.14) | .8 | |||||||||||
| ATRA | 35 | 31 | 27 | 47 | 56 | 28 | 38 | 35 | 44 | 44 | 33 | ||
| No ATRA | 34 | 31 | 30 | 39 | 40 | 32 | 33 | 47 | 36 | 30 | 30 | ||
| 8-year RFS | 0.98 (0.83-1.14) | .8 | |||||||||||
| ATRA | 33 | 30 | 26 | 42 | 52 | 26 | 36 | 28 | 45 | 40 | 32 | ||
| No ATRA | 28 | 31 | 25 | 37 | 33 | 28 | 31 | 29 | 33 | 28 | 29 | ||
| 8-year CIR | 0.93 (0.78-1.11) | .4 | |||||||||||
| ATRA | 50 | 53 | 54 | 46 | 37 | 55 | 48 | 61 | 38 | 49 | 51 | ||
| No ATRA | 59 | 52 | 61 | 49 | 51 | 59 | 53 | 64 | 54 | 53 | 56 | ||
| 8-year CIDCR | 1.16 (0.82-1.63) | .4 | |||||||||||
| ATRA | 17 | 17 | 20 | 12 | 10 | 19 | 16 | 11 | 17 | 11 | 16 | ||
| No ATRA | 13 | 17 | 14 | 14 | 16 | 13 | 16 | 7 | 14 | 19 | 14 | ||
CRi indicates complete remission with incomplete count recovery; RD, resistant disease; ID, induction death; RFS, relapse-free survival; CIR, cumulative incidence of relapse; and CIDCR, cumulative incidence of death in CR/CR.
FLT3/ITD and NPM1
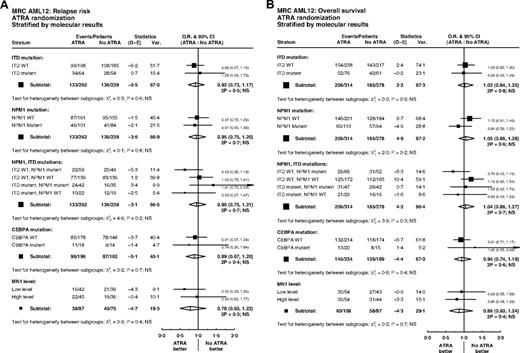
Overall, of 592 patients with data, FLT3/ITD mutations were present in 137 (23%) and NPM1 mutations in 207 (35%). Patients with FLT3 and NPM1 mutations were equally distributed between treatment groups (P = .5, P = .6; Table 1). There were no significant interactions between ATRA and FLT3/ITD or NPM1 status, either individually or combined, for remission (ITD, P = .7; NPM1, P = .6; ITD/NPM1, P = .1), relapse (ITD, P = .6; NPM1, P = .8; ITD/NPM1, P = .2), or OS (ITD, P = .9; NPM1, P = .2; ITD/NPM1, P = .3) (Figure 4). In particular, the effect of ATRA was not significantly different in any of the 4 subgroups defined by combinations of FLT3 and NPM1 status (Figure 5). In the group of patients with FLT3/wild-type and NPM1 mutant disease, 8-year survival was 56% when treated with ATRA, and 40% in the no ATRA group, but the difference was not statistically significant (P = .2). This lack of heterogeneity of treatment effect was maintained when analyses were restricted to patients with normal karyotype (supplemental Figure 3) and whether results were censored at the time of transplantation (data not shown).
Stratified analyses of ATRA randomization by molecular markers FLT3/ITD, NPM1, CEBPA, and MN1: (A) relapse; (B) overall survival.
Stratified analyses of ATRA randomization by molecular markers FLT3/ITD, NPM1, CEBPA, and MN1: (A) relapse; (B) overall survival.
Overall survival for ATRA randomization, split by FLT3/ITD and NPM1 status.
CEBPA
Limited DNA was obtained from bone marrow smears, and therefore, CEBPA analysis was only performed on samples from 423 patients. Of these, 35 (8%) had a CEBPA mutation. There was no significant association between CEBPA mutation status and randomized treatment (P = .8; Table 1). Tests for interaction with ATRA treatment were performed, but found no significant heterogeneity of treatment effect on remission (P = .8), relapse (P = .7), or OS (P = .4) (Figure 4).
MN1
MN1 expression was available in 195 patients; there were no differences in MN1 level by randomized treatment (Table 1). In multivariable analyses adjusted for age, WBC, type of AML, cytogenetics, performance status, FLT3/ITD, or NPM1 mutant status, there were no interactions between MN1 level as a continuous variable and treatment for CR (P = .8) or OS (P = .5). There was a suggestion that ATRA was beneficial for relapse in patients with lower MN1 expression levels (P = .03 for interaction). However, this was not seen in the less sensitive unadjusted analysis dichotomized at the median (P = .4; Figure 4A). Similar results were found when restricting attention to patients with a normal karyotype, with a significant interaction between MN1 level as a continuous variable and ATRA for cumulative incidence of relapse in multivariable analysis (P = .02), but no significant heterogeneity of treatment effect in either CR or OS, and no significant interaction in unadjusted analyses dichotomized at the median (supplemental Figure 3).
Discussion
In this randomized trial of more than 1000 younger adults with previously untreated non-APL AML, we were unable to demonstrate a higher CR rate, reduced relapse risk, or improved OS associated with the addition of ATRA to standard intensive chemotherapy. This contrasts with the results from the Austrian-German AML Study Group in which the addition of ATRA to intensive chemotherapy, albeit in older patients (median age 67 years), resulted in an increase in the CR rate from 39% to 52% (P = .05), with a significant increase in EFS (P = .03) and OS (P = .01).14 One possibility for this difference is that our study was carried out in younger patients (median age, 47 years) in which the CR/CRi rate was already high without ATRA (83%), and further improvements in the CR rate may be difficult to demonstrate. However, the M.D. Anderson trial, in an older population with a median age of 65 years, had a CR rate of 51% with no benefit from the addition of ATRA.11
There are differences in the way ATRA was administered between these trials. In our trial, it was given from days 1 to 60 at a dose of 45 mg/m2. In the M.D. Anderson trial, ATRA was commenced on day −2 for low WBC cases or on day 1 of chemotherapy for high WBC cases, and continued until 3 days after the end of the course of treatment.11 In the Austrian-German trial, ATRA was given from days 3 to 6 of chemotherapy at the dose of 45 mg/m2 and from days 3 to 28 at 15 mg/m2.14 It is unlikely that these results could be explained by the dose of ATRA used, although the possibility that the timing of ATRA administration might be influential cannot be discounted. There are also differences in the chemotherapy used. Our trial used a DAT combination; the M. D. Anderson trial used fludarabine, Ara-C, and idarubicin; and the Austrian-German trial used idarubicin, Ara-C, and etoposide. It has recently been suggested that the ATRA benefit is attributable to the inclusion of etoposide, based on an observation that a subsequent experience in which etoposide was not used showed no benefit,23 but the rationale for this is weak.
Another possible reason for the discrepancy in overall results relates to possible genotypic differences between patients in the different trials, as an analysis of the Austrian-German AMLSG trial suggested that the improved CR rate was only found in patients with a NPM1 mutation and the OS benefit was restricted to patients with a NPM1 mutation and no FLT3/ITD,15 which comprise 15% of the Austrian-German patients and 20% in our trial. We were unable to replicate their findings, or indeed to demonstrate any heterogeneity of response or survival dependent upon the genotype. The Austrian-German study had a relatively small number of patients (169 patients analyzed for OS), with only 26 patients with a NPM1 mutation in the absence of a FLT3/ITD, the group in which the benefit of ATRA was apparent, leading to the speculation that this result may be an outlier. Our data do raise the possibility that any benefit of ATRA may be in patients with lower MN1 expression, but given the multiple analyses and the lack of evidence of heterogeneity on OS, the evidence is not convincing. However, our data do not rule out an effect of ATRA, and together with the Austrian-German result indicate that any definitive answer awaits the results of randomized trials of ATRA in addition to etoposide-containing regimens. Pending these results, we believe that the addition of ATRA to any subgroup of patients with non-APL AML is premature.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the staff of the Clinical Trials Service Unit, Oxford, for support during recruitment; Birmingham Clinical Trials Unit for patient follow-up; the many research nurses and pharmacists who supported the trial; and Drs D. Swirsky, M. Griffiths, and P. Evans for patient material.
This work was supported by the United Kingdom Medical Research Council. Research was supported by Leukemia Research (United Kingdom), Cancer Research (United Kingdom), and the United Kingdom Medical Research Council.
Authorship
Contribution: A.K.B., K.W., A.H.G., and D.W.M. devised the trial; A.K.B. and A.H.G. were principal investigators; A.K.B., A.H.G., D.W.M., and A.G.P. were trial coordinators; D.C.L., R.E.G., and A.G. coordinated tissue collection and assays; assays were performed by C. Green, S.J., K.K., Y.P., C. Guy, and A.G.; R.K.H. analyzed the data; and A.K.B., D.C.L., R.E.G., and R.K.H. wrote the manuscript, which was reviewed by all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan K. Burnett, Department of Haematology, School of Medicine, Cardiff University, Heath Park, Cardiff, CF14 4XN, United Kingdom; e-mail: BurnettAK@Cardiff.ac.uk.
References
Author notes
Presented in part at the 46th annual meeting of the American Society of Hematology, Philadelphia, PA, December 9, 2002, and the 50th annual meeting of the American Society of Hematology, San Francisco, CA, December 8, 2008.