Abstract
High VEGFC mRNA expression of acute myeloid leukemia (AML) blasts is related to increased in vitro and in vivo drug resistance. Prognostic significance of VEGFC on long-term outcome and its associated gene expression profiles remain to be defined. We studied effect of VEGFC on treatment outcome and investigated gene expression profiles associated with VEGFC using microarray data of 525 adult and 100 pediatric patients with AML. High VEGFC expression appeared strongly associated with reduced complete remission rate (P = .004), reduced overall and event-free survival (OS and EFS) in adult AML (P = .002 and P < .001, respectively). Multivariable analysis established high VEGFC as prognostic indicator independent of cytogenetic risk, FLT3-ITD, NPM1, CEBPA, age, and white blood cell count (P = .038 for OS; P = .006 for EFS). Also, in pediatric AML high VEGFC was related to reduced OS (P = .041). A unique series of differentially expressed genes was identified that distinguished AML with high VEGFC from AML with low VEGFC, that is, 331 up-regulated genes (representative of proliferation, vascular endothelial growth factor receptor activity, signal transduction) and 44 down-regulated genes (eg, related to apoptosis) consistent with a role in enhanced chemoresistance. In conclusion, high VEGFC predicts adverse long-term prognosis and provides prognostic information in addition to well-known prognostic factors.
Introduction
Vascular endothelial growth factor-C (VEGFC) is a (lymph)angiogenic growth factor and signals through kinase insert domain receptor (KDR, ie, VEGF receptor-2) and fms-related tyrosine kinase 4 receptor (FLT4; ie, VEGF receptor-3).1,2 In general, by VEGF stimulation, VEGFRs become phosphorylated and transmit intracellular signals, resulting in cell proliferation and survival.3,4
Acute myeloid leukemia (AML) blasts express VEGFC and its receptors KDR and FLT4.5 Exogenously added VEGFC promotes in vitro cell survival by activation of the FLT4 and KDR heterodimeric receptor, as shown in 2 AML cell lines and in 5 cases of primary AML.6 Recently, we described that endogenous VEGFC mRNA expression levels of primary AML cells were related to increased in vitro resistance for 6 AML-related drugs.7 In addition, a relation was shown between high VEGFC mRNA and slow AML blast disappearance during induction treatment in vivo. This was apparent from the higher blast counts in the bone marrow on day 15 after the start of induction chemotherapy and a prolonged time to achieve complete remission.7 Currently, insight into molecular mechanisms responsible for the unfavorable treatment response and possible relationships of VEGFC levels with other biologic factors with prognostic significance is lacking. Furthermore, to date, the effect of VEGFC on long-term outcome has not been assessed in a large series of cases. We set out to study the effect of VEGFC on overall and event-free survival (OS and EFS) both in pediatric as well as in adult AML and considered the effect of VEGFC in relation to other established cytogenetic and gene mutation prognostic markers. Finally, we investigated gene expression profiles associated with VEGFC mRNA expression with the use of Affymetrix HGU133Plus2.0 gene expression data of 525 adult and 100 pediatric AML cases.
Methods
Patients
Gene expression profiling (GEP) has been performed on 525 consecutive adult patients with AML who have been treated according to sequential Dutch-Belgian Hemato-Oncology Cooperative Group and the Swiss Group for Clinical Cancer Research (HOVON/SAKK) AML-04, -04A, -29, -32, -42, and -43 protocols (available at http://www.hovon.nl).8-10 The adult and pediatric patients with AML have been included in previous GEP studies.11-13 An independent second series of cell specimens from 100 pediatric patients with newly diagnosed AML, who have been treated according to subsequent Dutch Childhood Oncology Group (DCOG) AML DCOG-BFM-87, DCOG-92/94, DCOG-97 protocols (available at http://www.skion.nl), was used for validation of the data of the adult AML cohort. All adult as well as pediatric patients in this study were newly diagnosed with AML and established according to World Health Organization criteria. All patients provided written informed consent in accordance with the Declaration of Helsinki, and the study was approved by all participating institutional review boards. Cell specimens were collected at the time of diagnosis. All subjects provided written informed consent. Cytogenetic risk group distinction (favorable, intermediate, and unfavorable) is according to HOVON/SAKK and DCOG protocols.14-16 Favorable cytogenetic risk was defined as t(8;21)(q22;q22), inv(16)(p13.1;q22), or t(16;16)(p13.1;q22) and t(15;17). Unfavorable cytogenetic risk was defined as complex cytogenetic abnormalities (ie, 3 or more distinct clonal abnormalities), −7, −5, del 5q, or del 7q; abnormalities of the long arm of chromosome 3 (abn 3q); t(6;9)(q23;q34); t(9;22)(q34;q11); or abnormalities of the long arm of chromosome 11 (abn 11q23). All other cytogenetic abnormalities as well as AML without cytogenetic abnormalities were considered to indicate an intermediate cytogenetic risk. In pediatric AML, 11q23 and t(6;9)(q23;q34) abnormalities were considered as intermediate cytogenetic risk.
Isolation and quality control of RNA, GEP, and quality control
The samples for GEP were obtained and analyzed as described previously.11,12 Detailed clinical, cytogenetic, and molecular information is available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession no. GSE6891 for adult patients with AML and accession no. GSE22056 for pediatric patients with AML).
Class comparison
Differentially expressed probes were identified for 262 AML samples with high VEGFC mRNA (ie, above the median VEGFC mRNA level) versus 262 AML samples with low VEGFC (ie, below the median VEGFC mRNA level) mRNA expression levels with the use of a multivariable permutation test in Biometric Research Branch ArrayTools (BRB ArrayTools) Version 3.8.0. BRB ArrayTools has been developed by the Biometric Research Branch of the US National Cancer Institute (http://linus.nci.nih.gov/BRB-ArrayTools.html). Differential expression was considered significant at P less than .001. A random variance t test was selected to permit the sharing of information among probe sets within class variation without assuming that all of the probe sets possess the same variance.
Gene ontology analysis
To investigate the biologic significance of the gene lists, we used gene ontology (GO) analysis (http://www.geneontology.org). After mapping each gene to the GO tree structure, the number of genes was determined at or below any given node in the GO hierarchy and the amount of statistically enrichment (Fisher exact test and false discovery rate adjustment) for each GO node relative to chance observation, using a previously developed procedure (GeneTrail).17
Statistical analysis
Statistical analyses were performed with SPSS software (SPSS Inc), release 16.0. Actuarial probabilities of OS (with death resulting from any cause) as well as EFS (with failure in case of no complete remission or relapse or death) were estimated according to the Kaplan-Meier method. For quantitative parameters overall differences between the cohorts were evaluated with an F test (or Student t test in case of 2 groups) for normally distributed variables or a Kruskal-Wallis test (or Mann-Whitney U test in case of 2 groups) for skewed distributed variables. For qualitative parameters, overall group differences were evaluated with a χ2 test (or Fisher exact in 2 × 2 setting). Correlations were calculated with the Spearman rank correlation coefficient (ρ). The association between VEGFC and OS and EFS was tested in univariate Cox models. Cox regression analysis was applied to determine the association of VEGFC expression and OS/EFS with adjustment for risk factors such as age, white blood cell (WBC) count, cytogenetic risk group (ie, favorable, intermediate, or unfavorable), Fms-like tyrosine kinase 3 gene internal tandem duplication (FLT3-ITD), mutations in genes encoding nucleophosmin 1 (NPM1) and the transcription factor CCAAT/enhancer binding protein α (CEBPA). The proportional hazard assumption was checked with log-log survivor functions (parallel curves). In addition the presence of time dependence, indicating violation of the proportional hazard assumption, was assessed. All tests were 2 tailed, and a P value of less than .05 was considered statistically significant.
Results
High VEGFC expression level is related to reduced OS and EFS in adult AML
Five hundred twenty-five adult patients with AML were analyzed for mRNA expression of VEGFC by GEP. Clinicopathologic, demographic, and molecular data, including age at diagnosis, baseline cytogenetics, initial WBC count, percentage blasts, platelet count, FLT3-ITD, and NPM1 and CEBPA mutation status are included in Table 1. The median OS for the total cohort was 16.3 months, and the median follow-up for survivors was 61.4 months.
Adult AML patient characteristics
| Characteristics . | All patients . | Low VEGFC . | High VEGFC . |
|---|---|---|---|
| No. of patients | 525 | 262 | 262 |
| Median age, y (range) | 46.6 (15.2-77.2) | 43.7 (15.2-75.5) | 49.7 (15.8-77.2) |
| Median WBC, ×109/L (range) | 26 (0.3-510) | 34 (0.6-510) | 20 (0.3-349) |
| Median blasts, % (range) | 65 (1-99) | 70 (2-99) | 62 (1-99) |
| Median platelets, ×109/L (range) | 56 (3-998) | 53 (6-494) | 59 (3-998) |
| Cytogenetic risk | |||
| Favorable | 89 (17%) | 57 (22%) | 32 (12%) |
| t(8;21) | 34 | 23 | 11 |
| t(15;17) | 20 | 12 | 8 |
| inv16 | 35 | 22 | 13 |
| Intermediate | 331 (63%) | 164 (63%) | 166 (63%) |
| Normal karyotype | 218 | 111 | 107 |
| + 8 | 25 | 8 | 16 |
| −9q | 7 | 4 | 3 |
| Other | 81 | 41 | 40 |
| Unfavorable | 85 (16%) | 34 (13%) | 51 (19%) |
| 11q23 | 11 | 6 | 5 |
| Complex | 20 | 7 | 13 |
| −5(q)/−7(q) | 42 | 16 | 26 |
| abn(3q) | 2 | 1 | 1 |
| t(6;9) | 6 | 3 | 3 |
| t(9;22) | 2 | 1 | 1 |
| Other | 2 | 0 | 2 |
| Not available | 20 (4%) | 7 (3%) | 13 (5%) |
| FLT3-ITD vs no FLT3-ITD | 143/382 | 68/194 | 74/188 |
| NPM1 wild type vs mutant | 366/159 | 184/78 | 181/81 |
| CEBPA wild type vs mutant* | 486/38 | 240/22 | 245/16 |
| Allogeneic SCT | 140† | 66‡ | 74§ |
| Autologous SCT | 68 | 40 | 28 |
| Cycles to CR | |||
| 1 | 297 (57%) | 149 (57%) | 147 (56%) |
| 2 | 111 (21%) | 67 (25%) | 44 (17%) |
| 3 | 8 (2%) | 6 (2%) | 2 (1%) |
| More than 3 | 5 (1%) | 2 (1%) | 3 (1%) |
| No CR | 104 (20%) | 38‖ (15%) | 66‖ (25%) |
| Relapse | 202 (39%) | 97 (37%) | 105 (40%) |
| Dead vs alive | 316/209 | 141/121‖ | 174/88‖ |
| Characteristics . | All patients . | Low VEGFC . | High VEGFC . |
|---|---|---|---|
| No. of patients | 525 | 262 | 262 |
| Median age, y (range) | 46.6 (15.2-77.2) | 43.7 (15.2-75.5) | 49.7 (15.8-77.2) |
| Median WBC, ×109/L (range) | 26 (0.3-510) | 34 (0.6-510) | 20 (0.3-349) |
| Median blasts, % (range) | 65 (1-99) | 70 (2-99) | 62 (1-99) |
| Median platelets, ×109/L (range) | 56 (3-998) | 53 (6-494) | 59 (3-998) |
| Cytogenetic risk | |||
| Favorable | 89 (17%) | 57 (22%) | 32 (12%) |
| t(8;21) | 34 | 23 | 11 |
| t(15;17) | 20 | 12 | 8 |
| inv16 | 35 | 22 | 13 |
| Intermediate | 331 (63%) | 164 (63%) | 166 (63%) |
| Normal karyotype | 218 | 111 | 107 |
| + 8 | 25 | 8 | 16 |
| −9q | 7 | 4 | 3 |
| Other | 81 | 41 | 40 |
| Unfavorable | 85 (16%) | 34 (13%) | 51 (19%) |
| 11q23 | 11 | 6 | 5 |
| Complex | 20 | 7 | 13 |
| −5(q)/−7(q) | 42 | 16 | 26 |
| abn(3q) | 2 | 1 | 1 |
| t(6;9) | 6 | 3 | 3 |
| t(9;22) | 2 | 1 | 1 |
| Other | 2 | 0 | 2 |
| Not available | 20 (4%) | 7 (3%) | 13 (5%) |
| FLT3-ITD vs no FLT3-ITD | 143/382 | 68/194 | 74/188 |
| NPM1 wild type vs mutant | 366/159 | 184/78 | 181/81 |
| CEBPA wild type vs mutant* | 486/38 | 240/22 | 245/16 |
| Allogeneic SCT | 140† | 66‡ | 74§ |
| Autologous SCT | 68 | 40 | 28 |
| Cycles to CR | |||
| 1 | 297 (57%) | 149 (57%) | 147 (56%) |
| 2 | 111 (21%) | 67 (25%) | 44 (17%) |
| 3 | 8 (2%) | 6 (2%) | 2 (1%) |
| More than 3 | 5 (1%) | 2 (1%) | 3 (1%) |
| No CR | 104 (20%) | 38‖ (15%) | 66‖ (25%) |
| Relapse | 202 (39%) | 97 (37%) | 105 (40%) |
| Dead vs alive | 316/209 | 141/121‖ | 174/88‖ |
Cytogenetic risk group distinction (favorable, intermediate, and unfavorable) is described in “Patients.”
Low VEGFC indicates a VEGFC mRNA expression level below the median VEGFC level; high VEGFC, a VEGFC mRNA expression level above the median VEGFC level; WBC, white blood cell count; NPM1, nucleophosmin 1; FLT3, fms-related tyrosine kinase 3; ITD, internal tandem duplication; CEBPA, CCAAT/enhancer binding protein α; SCT, stem cell transplantation; and CR, complete remission.
For 1 patient, the CEBPA status is unknown.
Twelve of 140 patients with an allogeneic SCT after nonmyeloablative conditioning.
Four of 66 patients with an allogeneic SCT after nonmyeloablative conditioning.
Eight of 74 patients with an allogeneic SCT after nonmyeloablative conditioning.
A significant difference when AML patients with a low versus high VEGFC expression level were compared (P < .01).
First, we tested the association of VEGFC expression as a continuous variable with patient survival. Higher expression levels of VEGFC were significantly associated with reduced OS and EFS (HR: 1.66, 95% CI: 1.16-2.37, P = .006 for OS; HR: 1.66, 95% CI: 1.19-2.32, P = .003 for EFS). We then defined patient subgroups on the basis of high and low VEGFC expression. In an initial step, within the entire cohort of 525 patients, the association between different levels of VEGFC expression and survival was evaluated with quartiles of VEGFC expression to assess appropriateness of VEGFC expression as a continuous variable (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Because the risks in the first and second quartiles were highly comparable and significantly different from those in the third and fourth quartiles, the median VEGFC level was chosen as the appropriate cutoff (hereafter referred to as high and low VEGFC). The latter cutoff corresponds with the threshold used in most previous studies on VEGFC mRNA/protein in various malignancies, including AML.7,18-20
Table 1 lists the clinical characteristics of the entire cohort as well as of patient subgroups distinguished by high versus low mRNA expression of VEGFC. No significant differences in VEGFC mRNA expression were observed among different cytogenetic risk groups or the genotypic NPM1/FLT3-ITD subgroups or CEBPA mutant subsets. Also age at diagnosis, WBC count, percentage of bone marrow blasts, platelet count, and the number of patients who had received an allogeneic stem cell transplantation were not different among patients with high versus low VEGFC.
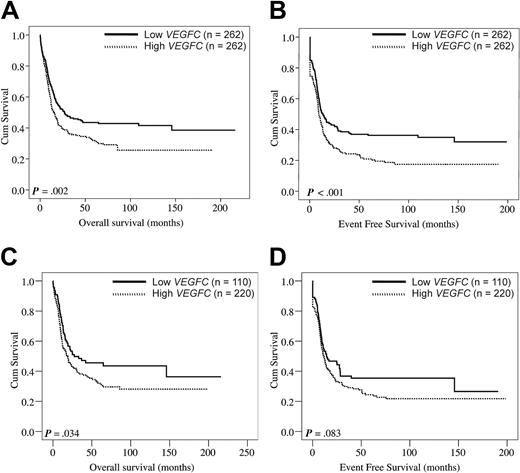
The complete remission rate was significantly lower among patients with AML with high VEGFC compared with patients with low VEGFC (Table 1; P = .004). Interestingly, the patients with high VEGFC expression levels showed a significantly reduced OS (Figure 1A; P = .002) and a reduced EFS (Figure 1B; P < .001). Patients with high and low VEGFC transcript levels had median OS of 13 and 19 months and estimated 5-year OS rates of 35% and 47%, respectively. In addition, we specifically evaluated the effect of VEGFC on OS and EFS among cytogenetically intermediate-risk AML, representing the largest prognostically distinct subgroup (63% of cases studied). First, we tested the association of VEGFC expression as a continuous variable with patient survival in AML cases with intermediate cytogenetic risk. Higher expression levels of VEGFC tend to associate with reduced OS and EFS (HR: 1.45, 95% CI: 0.95-2.22, P = .085 for OS; HR: 1.40, 95% CI: 0.94-2.08, P = .100 for EFS). Furthermore, within the cohort of intermediate cytogenetic risk cases, patients with low VEGFC compared favorably with high VEGFC in terms of OS (P = .034) and EFS (P = .083) (Figure 1C-D). Although numbers were relatively small, we next studied the effect of VEGFC on OS and EFS within specific cytogenetic subgroups (supplemental Figure 2). No difference in OS was found among core binding factor AMLs [ie, t(8;21) and inv16)] with low versus high VEGFC (P = .193). Interestingly, core binding factor AMLs with high VEGFC showed a significantly reduced EFS (P = .017). In addition, patients harboring t(15;17) with high VEGFC expression showed no difference in OS (P = .582) but, interestingly, tend to show a reduced EFS (P = .084). Hereafter, the 2 largest specific cytogenetic subgroups of patients with AML with poor risk cytogenetics within our cohort (ie, complex cytogenetics and −5(q)/−7(q) abnormalities) were compared for OS and EFS among patients with low versus high VEGFC. In patients with complex cytogenetics no effect of VEGFC on OS and EFS was evident (P = .676 and P = .851, respectively). Finally, patients with −5(q)/−7(q) cytogenetic abnormalities and high VEGFC tend to show a reduced OS (P = .067), but no effect was evident on EFS (P = .320).
VEGFC in relation to OS and EFS in adult AML. Kaplan-Meier plots show the OS (A) and EFS (B) of adult patients with AML subgroups with high (n = 262) versus low (n = 262) VEGFC expression. (C-D) Evaluation of the effect of VEGFC on OS and EFS among patients with cytogenetically intermediate-risk AML (n = 331). First, within the 331 patients with cytogenetically intermediate-risk AML the association between VEGFC expression and survival was evaluated with the use of tertiles of VEGFC expression. Because the risks in the second and third tertile were comparable (data not shown), the first versus the combination of the second and third VEGFC tertiles were compared with regard to OS and EFS. Kaplan-Meier plots show the OS (C) and EFS (D) in patients with cytogenetically intermediate risk AML with high (ie, second and third tertiles combined) versus low (ie, first tertile) transcript levels of VEGFC. Intermediate cytogenetic risk is defined in “Patients.”
VEGFC in relation to OS and EFS in adult AML. Kaplan-Meier plots show the OS (A) and EFS (B) of adult patients with AML subgroups with high (n = 262) versus low (n = 262) VEGFC expression. (C-D) Evaluation of the effect of VEGFC on OS and EFS among patients with cytogenetically intermediate-risk AML (n = 331). First, within the 331 patients with cytogenetically intermediate-risk AML the association between VEGFC expression and survival was evaluated with the use of tertiles of VEGFC expression. Because the risks in the second and third tertile were comparable (data not shown), the first versus the combination of the second and third VEGFC tertiles were compared with regard to OS and EFS. Kaplan-Meier plots show the OS (C) and EFS (D) in patients with cytogenetically intermediate risk AML with high (ie, second and third tertiles combined) versus low (ie, first tertile) transcript levels of VEGFC. Intermediate cytogenetic risk is defined in “Patients.”
Prognostic value of VEGFC expression level in the context of other risk factors in adult AML
Univariate analysis showed that besides high VEGFC expression values (HR: 1.41, 95% CI: 1.13-1.76, P = .003 for OS; HR: 1.44, 95% CI: 1.17-1.78, P = .001 for EFS), also age, WBC count, NPM1 mutation, FLT3-ITD, CEBPA, and cytogenetic risk group (ie, unfavorable risk, intermediate risk, and favorable risk) significantly affected OS and EFS (data not shown). When we subsequently considered the latter variables in a multivariable analysis, high VEGFC maintained its independent prognostic value for both OS as well as EFS (HR: 1.29, 95% CI: 1.02-1.63, P = .038 for OS; HR: 1.38, 95% CI: 1.09-1.71, P = .006 for EFS; details in Table 2).
Multivariable analysis of VEGFC[/bi] as prognostic marker for overall and event-free survival
| Variable . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| VEGFC* | 1.29 (1.02-1.63) | .038 | 1.37 (1.09-1.71) | .006 |
| Intermediate† | 2.14 (1.43-3.20) | < .001 | 1.69 (1.19-2.41) | .004 |
| Poor† | 3.79 (2.46-5.84) | < .001 | 3.01 (2.04-4.43) | < .001 |
| Age, decades | 1.12 (1.03-1.22) | .008 | 1.06 (.98-1.15) | .170 |
| WBC‡ | 1.35 (1.06-1.73) | .016 | 1.27 (1.01-1.60) | .038 |
| FLT3-ITD§ | 1.74 (1.32-2.28) | < .001 | 1.59 (1.23-2.06) | < .001 |
| NPM1 mutation‖ | .57 (.43-.77) | < .001 | .60 (.45-.79) | < .001 |
| CEBPA mutation¶ | .51 (.30-.84) | .008 | .58 (.36-.91) | .018 |
| Variable . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| VEGFC* | 1.29 (1.02-1.63) | .038 | 1.37 (1.09-1.71) | .006 |
| Intermediate† | 2.14 (1.43-3.20) | < .001 | 1.69 (1.19-2.41) | .004 |
| Poor† | 3.79 (2.46-5.84) | < .001 | 3.01 (2.04-4.43) | < .001 |
| Age, decades | 1.12 (1.03-1.22) | .008 | 1.06 (.98-1.15) | .170 |
| WBC‡ | 1.35 (1.06-1.73) | .016 | 1.27 (1.01-1.60) | .038 |
| FLT3-ITD§ | 1.74 (1.32-2.28) | < .001 | 1.59 (1.23-2.06) | < .001 |
| NPM1 mutation‖ | .57 (.43-.77) | < .001 | .60 (.45-.79) | < .001 |
| CEBPA mutation¶ | .51 (.30-.84) | .008 | .58 (.36-.91) | .018 |
HR indicates hazard ratio; CI, confidence interval; intermediate, intermediate cytogenetic risk as defined in “Patients”; poor, poor cytogenetic risk as defined in “Methods”; WBC, white blood cell count; FLT3, fms-related tyrosine kinase 3; ITD, internal tandem duplication; NPM1, nucleophosmin 1; and CEBPA, CCAAT/enhancer binding protein α.
High VEGFC (ie, above the median VEGFC level) versus low VEGFC (ie, below the median VEGFC level).
Cytogenetic risk versus good cytogenetic risk.
WBC greater than 20 × 109/L.
FLT3-ITD versus no FLT3-ITD.
NPM1 mutation versus no NPM1 mutation.
CEBPA mutation versus no CEBPA mutation.
Moreover, we evaluated patients with AML with intermediate-risk cytogenetics and considered VEGFC transcript level, FLT3-ITD, and NPM1 gene mutation status separately. Patients with low VEGFC expression in the absence of a FLT3-ITD constituted a favorable subset of patients with a significantly improved OS and EFS compared with the other 3 groups combined (P = .026 for OS; P = .032 for EFS; Figure 2A-B). For patients with low VEGFC levels and absent FLT3-ITD, the estimated 5-year OS was 50% and the estimated EFS at 5 years was 40%. In contrast, the outcome of patients with both high VEGFC expression and FLT3-ITD were worse, because the 5-year OS and EFS rates were only 30% and 26%, respectively. Of note, also within cytogenetically normal karyotype AML, patients with low VEGFC in the absence of a FLT3-ITD tend to have a better prognosis than patients with the other cytogenetically normal karyotype AML (P = .11 for OS; P = .04 for EFS; supplemental Figure 3A and B, respectively). Furthermore, patients with the combination of a high VEGFC level without a NPM1 mutation showed a nonsignificant trend as regards a reduced OS (P = .098; supplemental Figure 4A) and a significantly reduced EFS compared with the other 3 groups combined (P = .012; supplemental Figure 4B).
Evaluation of the combination of FLT3-ITD and VEGFC expression among AML with intermediate-risk cytogenetics. Evaluation of the combination of FLT3-ITD and VEGFC expression level among patients with AML with intermediate-risk cytogenetics. (A) OS and EFS (B) are shown for the 4 subgroups divided by FLT3-ITD and VEGFC status. High VEGFC indicates a VEGFC expression level above the median VEGFC level in the specific patient group, whereas low VEGFC indicates a VEGFC expression level below the median VEGFC level in the specific patient group. The low and high VEGFC without FLT3-ITD subgroups consist of 108 patients per group and the low and high VEGFC with FLT3-ITD subgroups consist of 57 patients per group. P values are given for the overall comparison across all 4 groups.
Evaluation of the combination of FLT3-ITD and VEGFC expression among AML with intermediate-risk cytogenetics. Evaluation of the combination of FLT3-ITD and VEGFC expression level among patients with AML with intermediate-risk cytogenetics. (A) OS and EFS (B) are shown for the 4 subgroups divided by FLT3-ITD and VEGFC status. High VEGFC indicates a VEGFC expression level above the median VEGFC level in the specific patient group, whereas low VEGFC indicates a VEGFC expression level below the median VEGFC level in the specific patient group. The low and high VEGFC without FLT3-ITD subgroups consist of 108 patients per group and the low and high VEGFC with FLT3-ITD subgroups consist of 57 patients per group. P values are given for the overall comparison across all 4 groups.
Consistence of VEGFC as a prognostic factor in pediatric AML
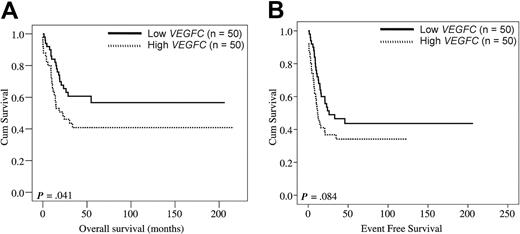
We wanted to study the effect of VEGFC on outcome with the use of an independent cohort of pediatric patients with AML (Table 3) and defined patient subgroups on the basis of high and low VEGFC expression with the use of the identical cutoff (ie, median) as applied to the above adult series of AML. As in adult patients with AML, the complete remission rate was significantly reduced in pediatric patients with AML with high VEGFC (Table 3; P = .048). Univariate analysis showed that high VEGFC significantly affected OS in pediatric AML (HR: 1.81, 95% CI: 1.02-3.21, P = .044). For EFS a nonsignificant trend was seen (HR: 1.57, 95% CI: 0.94- 2.62, P = .087). Figure 3 shows the Kaplan-Meier plots for pediatric patients with AML with high versus low VEGFC with regard to OS (Figure 3A; P = .041) and EFS (P = .084; Figure 3B). Thus, the VEGFC prognostic data in pediatric AML appear in general agreement with those in adult AML.
Pediatric AML patient characteristics
| Characteristics . | All patients . | Low VEGFC . | High VEGFC . |
|---|---|---|---|
| No. of patients | 100 | 50 | 50 |
| Median age, y (range) | 9.0 (0.0-16.7) | 10 (0.0-15.0) | 8.0 (0.6-16.7) |
| Median WBC, ×109/L (range) | 36.5 (2.3-483) | 33.8 (2.5-320) | 46.7 (2.3-483) |
| Cytogenetic risk | |||
| Favorable | 25 (25%) | 15 (30%) | 10 (20%) |
| t(8;21) | 14 | 9 | 5 |
| t(15;17) | 4 | 2 | 2 |
| inv16 | 7 | 4 | 3 |
| Intermediate | 58 (58%) | 27 (54%) | 31 (62%) |
| Normal karyotype | 16 | 10 | 6 |
| Other | 39 | 16 | 23 |
| +8 | 3 | 1 | 2 |
| Unfavorable | 12 (12%) | 6 (12%) | 6 (12%) |
| Complex | 10 | 6 | 4 |
| −5(q)/−7(q) | 2 | — | 2 |
| Not available | 5 (5%) | 2 (4%) | 3 (6%) |
| CR vs no CR | 79/21 | 44/6* | 35/15* |
| Relapse | 40 | 23 | 17 |
| Dead vs alive | 48/52 | 20/30 | 28/22 |
| Characteristics . | All patients . | Low VEGFC . | High VEGFC . |
|---|---|---|---|
| No. of patients | 100 | 50 | 50 |
| Median age, y (range) | 9.0 (0.0-16.7) | 10 (0.0-15.0) | 8.0 (0.6-16.7) |
| Median WBC, ×109/L (range) | 36.5 (2.3-483) | 33.8 (2.5-320) | 46.7 (2.3-483) |
| Cytogenetic risk | |||
| Favorable | 25 (25%) | 15 (30%) | 10 (20%) |
| t(8;21) | 14 | 9 | 5 |
| t(15;17) | 4 | 2 | 2 |
| inv16 | 7 | 4 | 3 |
| Intermediate | 58 (58%) | 27 (54%) | 31 (62%) |
| Normal karyotype | 16 | 10 | 6 |
| Other | 39 | 16 | 23 |
| +8 | 3 | 1 | 2 |
| Unfavorable | 12 (12%) | 6 (12%) | 6 (12%) |
| Complex | 10 | 6 | 4 |
| −5(q)/−7(q) | 2 | — | 2 |
| Not available | 5 (5%) | 2 (4%) | 3 (6%) |
| CR vs no CR | 79/21 | 44/6* | 35/15* |
| Relapse | 40 | 23 | 17 |
| Dead vs alive | 48/52 | 20/30 | 28/22 |
Cytogenetic risk group distinction (favorable, intermediate, and unfavorable) is described in “Patients.”
“Low VEGFC” indicates a VEGFC mRNA expression level below the median VEGFC level; high VEGFC, a VEGFC mRNA expression level above the median VEGFC level; and CR, complete remission.
A significant difference when AML patients with a low versus high VEGFC expression level were compared (P < .01).
VEGFC in relation to OS and EFS in pediatric AML. The 100 pediatric patients with AML were split in a group with high VEGFC (ie, above the median VEGFC level; n = 50) and a group with low VEGFC (ie, below the median VEGFC level; n = 50) mRNA expression levels. Kaplan-Meier plots show the OS (A) and EFS (B) of pediatric AML patient subgroups with high versus low transcript levels of VEGFC.
VEGFC in relation to OS and EFS in pediatric AML. The 100 pediatric patients with AML were split in a group with high VEGFC (ie, above the median VEGFC level; n = 50) and a group with low VEGFC (ie, below the median VEGFC level; n = 50) mRNA expression levels. Kaplan-Meier plots show the OS (A) and EFS (B) of pediatric AML patient subgroups with high versus low transcript levels of VEGFC.
Distinct gene expression profiles between AML samples with high versus low VEGFC expression level
To extend the observation that clinical parameters differ between AML samples with high versus low VEGFC, the transcriptome of 262 adult AML samples with high VEGFC was compared with the transcriptome of 262 adult AML samples with low VEGFC. This comparison showed that 459 unique genes (represented by 778 probe sets) were higher expressed (“Up-with-VEGFC” group) and that 192 unique genes (represented by 250 probe sets) were lower expressed (“Down-with-VEGFC” group) with elevated VEGFC at the significance level of P less than .001 (supplemental Table 1). With regard to the differential expression of VEGFC itself, as expected, the VEGFC gene was the highest ranked among the list of differentially expressed genes. Of note, when a more extreme cutoff for VEGFC was chosen (ie, first vs fourth quartile), a highly comparable list of differentially expressed genes was found; 648 of 651 genes were similar between both methods.
Validation of VEGFC-dependent differences in gene expression profiles
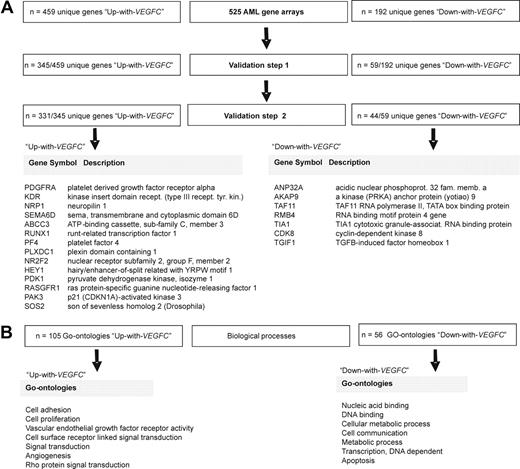
To validate the list of differently expressed genes between AML samples with high versus low VEGFC, 2 independent GEP cohorts were used; ie, the pediatric AML cohort (n = 100) and a publicly available adult AML GEP cohort (n = 180).21 The 459 Up-with-VEGFC genes and the 192 Down-with-VEGFC genes were individually validated as a continuous variable dependent on VEGFC. Hence, 345 of 459 Up-with-VEGFC genes (75%) as well as 59 of 192 Down-with-VEGFC genes (31%) could be confirmed (ie, validation step 1′ in Figure 4A) in the pediatric AML set. Subsequently, a third independent adult AML cohort (n = 180) described by Tomasson et al21 was used for an additional validation (ie, validation step 2′ in Figure 4A); 331 of 345 Up-with-VEGFC genes and 44 of 59 Down-with-VEGFC genes were found. In summary, of the differentially expressed genes on VEGFC in the original dataset, 331 Up-with-VEGFC genes and 44 Down-with-VEGFC genes could be confirmed to be significantly differently expressed as a continuous variable depending on VEGFC in 2 independent AML cohorts (Figure 4A). The entire list of differentially expressed genes after 2 validation steps is presented in supplemental Table 1.
Genes and GOs distinguishing AML samples with a high versus low VEGFC mRNA expression level. (A) The transcriptome of 262 adult AML samples with high VEGFC was compared with the transcriptome of 262 adult AML samples with low VEGFC mRNA expression level. This comparison showed 459 unique genes that were higher expressed in the high VEGFC group (Up-with-VEGFC group), and 192 unique genes that were lower expressed in the high VEGFC group (Down-with-VEGFC group) at the significance level of P < .001. The GEP cohort of 100 pediatric AML samples was used to validate the differentially expressed unique genes between samples with high versus low VEGFC (ie, validation step 1). Hereafter, an independent publicly available cohort of 180 adult AML samples was used (ie, validation step 2).20 After 2 validation steps 331 unique genes were found to be higher expressed in patients with AML with high compared with low VEGFC, and 44 unique genes were found to be lower expressed in patients with AML with high VEGFC compared with patients with AML with low VEGFC. (B) Biologic processes (represented by GOs) enriched among the 331 (Up-with-VEGFC) and 44 (Down-with-VEGFC) differentially expressed unique genes showed 105 significantly Up-with-VEGFC and 56 significantly Down-with-VEGFC GOs at the significance level of P < .05.
Genes and GOs distinguishing AML samples with a high versus low VEGFC mRNA expression level. (A) The transcriptome of 262 adult AML samples with high VEGFC was compared with the transcriptome of 262 adult AML samples with low VEGFC mRNA expression level. This comparison showed 459 unique genes that were higher expressed in the high VEGFC group (Up-with-VEGFC group), and 192 unique genes that were lower expressed in the high VEGFC group (Down-with-VEGFC group) at the significance level of P < .001. The GEP cohort of 100 pediatric AML samples was used to validate the differentially expressed unique genes between samples with high versus low VEGFC (ie, validation step 1). Hereafter, an independent publicly available cohort of 180 adult AML samples was used (ie, validation step 2).20 After 2 validation steps 331 unique genes were found to be higher expressed in patients with AML with high compared with low VEGFC, and 44 unique genes were found to be lower expressed in patients with AML with high VEGFC compared with patients with AML with low VEGFC. (B) Biologic processes (represented by GOs) enriched among the 331 (Up-with-VEGFC) and 44 (Down-with-VEGFC) differentially expressed unique genes showed 105 significantly Up-with-VEGFC and 56 significantly Down-with-VEGFC GOs at the significance level of P < .05.
Biologic processes enriched for Up- and Down-with-VEGFC genes
Then biologic processes (represented by GOs) enriched among AML samples with high versus low VEGFC were analyzed. For this analysis, those genes that were significantly up-regulated or down-regulated after 2 validation steps were used. These 331 (Up-with-VEGFC) and 44 (Down-with-VEGFC) differentially expressed genes consisted of 105 significantly Up-with-VEGFC and 56 significantly Down-with-VEGFC GOs (supplemental Table 2). This analysis showed, for instance, that genes involved in biologic processes as proliferation, signal transduction (eg, PAK3 and SOS2, which are both upstream activators of mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase/ERK,22-25 and PDK1, which is a direct activator of Akt/protein kinase B (PKB)26,27 ), cell adhesion, vascular endothelial growth factor receptor activity (eg, KDR, PDGFRA, and NRP1), angiogenesis, and wnt-protein binding were up-regulated with increasing VEGFC. Genes involved in biologic processes as apoptosis (TIA1 and ANP32A), cellular metabolic process, cell communication, and DNA-dependent transcription were found to be down-regulated with increasing VEGFC (Figure 4B).
Discussion
In this study we show that adult as well as pediatric patients with AML with a high VEGFC transcript level have an inferior disease outcome. To further improve the insight into the molecular mechanisms associated and possibly responsible for the differences in VEGFC-related poor outcome in AML, we have analyzed expression profiles of 525 patients with AML in relation to VEGFC. This analysis showed, after validation in 2 independent AML cohorts of 100 children and 180 adults, distinct gene expression profiles in AML with high VEGFC versus low VEGFC expression.
VEGFC is predominantly known for its ability to promote the formation of new lymphatic vessels by inducing proliferation, migration, and sprout formation of existing lymphatic endothelial cells (ie, lymphangiogenesis).28-31 The spread of cancer cells from a primary tumor to the lymphatics and blood stream is now understood to be due to active recruitment of new lymphatics by tumor-derived VEGFC.29,31,32 However, several studies reported other functions for VEGFC, which are independent of lymphangiogenesis, and instead are important for cancer progression. For example, an autocrine VEGFC loop promoting the invasion and metastasis of lung, breast, and gastric cancer cells and stimulating survival of Kaposi sarcoma, malignant mesothelioma, and prostate cancer cells has been described.28,33-39 Furthermore, in vivo, several studies have linked VEGFC to the progression of diverse cancer types (eg, lung adenocarcinoma, head and neck carcinomas, and breast, prostate, and colorectal cancers).38-43
A possible role for VEGFC in the pathogenesis of AML was for the first time suggested by Fiedler et al5 who detected VEGFC and KDR/FLT-4 expression of AML blasts at protein and/or mRNA levels. In addition, VEGFC/FLT4 expression was found to be significantly higher in patients with AML compared with healthy controls with the use of immunohistochemical staining.44 In vitro studies of Dias et al6 identified a role for exogenously added VEGFC (as a model for paracrine signaling) in survival and proliferation of leukemic cells and protection against chemotherapy-induced apoptosis by blc-2 induction through FLT4 signaling. Furthermore, AML cells are also able to express VEGFRs themselves; therefore, autocrine signaling has been described in AML.45,46 We found KDR, but not FLT4, to be up-regulated with VEGFC in this study. The autocrine VEGF signaling is found to be primarily mediated by KDR.45,46 To our knowledge, autocrine VEGFC signaling by FLT4 has not been described in AML. However, this clearly needs further studies.
Interestingly, in a relatively small cohort of 90 adult patients with AML, subgroup analysis suggested that patients with a low VEGFC expression level (in combination with a high ANG2 expression level) had a significantly better long-term prognosis than did patients with a high VEGFC expression level.47 Our study shows for the first time that a high VEGFC expression level is associated with reduced complete remission rate, reduced OS, as well as EFS in AML. The prognostic effect of VEGFC expression was also apparent in the subset of AML with intermediate prognostic risk cytogenetics. Furthermore, within the subgroup of patients with AML with intermediate cytogenetic risk, it appeared that patients with low VEGFC without FLT3-ITD had a better prognosis. Of note, comparable results were found among patients with cytogenetically normal karyotype AML with low VEGFC and without FLT3-ITD in terms of treatment outcome. Although numbers were relatively small, our results suggest that patients with favorable cytogenetic risk AML with high VEGFC showed reduced EFS. Finally, patients with −5(q)/−7(q) cytogenetic abnormalities with high VEGFC tend to show reduced OS. However, further and larger studies are needed to generate more conclusive results in (these) specific cytogenetic subgroups regarding VEGFC expression and treatment outcome.
Most of the patients of this study were fully characterized with regard to FLT3-ITD, NPM1, CEBPA, cytogenetics, and clinical characteristics such as age and WBC count, which thus allowed consideration of a comprehensive panel of gene mutation and cytogenetic and clinical prognostic markers. In multivariable analyses, high VEGFC was identified as an independent risk indicator for both OS and EFS. Of note, our analysis relied on relative expression differences between samples and not on absolute expression levels. Nonetheless, before quantitative measures of VEGFC expression can be used for clinical decision making, additional standardization of the methods used to determine VEGFC expression levels in combination with prospective trials is necessary.
GEP showed a unique signature associated with (high) VEGFC. As one might expect, the biologic process VEGFR activity (representing genes such as KDR, PDGFRA, and NRP1) was elevated with high VEGFC. Notably, no correlation was found between VEGFA and VEGFC. Furthermore, the GEP signature showed genes involved in proliferation (eg, PAK3 and SOS2, known activators of the mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/ERK pathway22-25 and PDK1, which is a direct activator of Akt/PKB26,27 ) to be up-regulated with high VEGFC. In endothelial cells the ERK and Akt pathways are reported to be activated by VEGFC and to be important for proliferation and survival signals, respectively.28,32 Of note, it has been suggested by Su et al43 that signal transduction in response to VEGFC varies with cell type, because VEGFC did not activate the ERK pathway in lung adenocarcinoma cells.
The GEP signature showed up-regulation of a set of genes whose overexpression has been found to negatively correlate with prognosis of AML; for example, ABCC3 (also known as MRP3) gene and certain other members of the adenosine triphosphate–binding cassette (ABC) family (ie, ABCB9, ABCC8, and CFTR) were found to be associated with elevated VEGFC. Many ABC-family members have been shown to be able to efflux cytostatic drugs.48,49 In AML, as well as in acute lymphoblastic leukemia, ABCC3 is associated with a lower chance of survival, both in adults and in children.50,51 In vitro, it was shown that MRP3 causes resistance against etoposide, teniposide, and methotrexate.52,53
In conclusion, our study provides evidence to indicate that VEGFC expression predicts outcome in both pediatric and adult patients with AML. In multivariable analysis, high VEGFC emerged as a prognostic indicator that independently predicted shorter survival. Finally, GEP showed a unique signature associated with elevated VEGFC, which we established in 3 independent AML cohorts. The interruption of VEGFC signaling in AML might offer potential therapeutic targets for antileukemic treatment interventions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially supported by the Foundation for Pediatric Oncology Research Groningen (H.J.M.d.J.) and the Dutch Cancer Society (grant 3661; E.S.J.M.d.B.).
Authorship
Contribution: H.J.M.d.J. designed research, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript; P.J.M.V. analyzed and interpreted data, collected data, and wrote the manuscript; N.J.G.M.V. analyzed and interpreted data and performed statistical analysis; A.t.E. analyzed and interpreted data; M.L.d.B., J.C., V.d.H., M.M.v.d.H.-E., G.J.L.K., C.M.Z., and W.A.K. collected data; B.L. analyzed and interpreted data, collected data, and wrote the manuscript; E.S.J.M.d.B. collected data, designed research, analyzed and interpreted data, and wrote the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eveline S. J. M. de Bont, Division of Pediatric Oncology/Hematology, Department of Pediatrics, Beatrix Children's Hospital, University Medical Center Groningen, University of Groningen, PO Box 30.001, 9700 RB Groningen, The Netherlands; e-mail: e.de.bont@bkk.umcg.nl.