Abstract
Patients with systemic lupus erythematosus (SLE) have a markedly increased risk to develop cardiovascular disease, and traditional cardiovascular risk factors fail to account for this increased risk. We used microarray to probe the platelet transcriptome in patients with SLE and healthy controls, and the gene and protein expression of a subset of differentially expressed genes was further investigated and correlated to platelet activation status. Real-time PCR was used to confirm a type I interferon (IFN) gene signature in patients with SLE, and the IFN-regulated proteins PRKRA, IFITM1 and CD69 (P < .0001) were found to be up-regulated in platelets from SLE patients compared with healthy volunteers. Notably, patients with a history of vascular disease had increased expression of type I IFN-regulated proteins as well as more activated platelets compared with patients without vascular disease. We suggest that interferogenic immune complexes stimulate production of IFNα that up-regulates the megakaryocytic type I IFN-regulated genes and proteins. This could affect platelet activation and contribute to development of vascular disease in SLE. In addition, platelets with type I IFN signature could be a novel marker for vascular disease in SLE.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by inflammation in many different organ systems such as skin, joints, kidney, nervous system, serosal membranes and blood elements. B-cell abnormalities, autoantibodies, complement activation and an ongoing type I interferon (IFN) production are all of importance in the pathogenesis of SLE.1 The IFNα production in SLE is detectable in serum,2 especially in certain phenotypes of active disease. Furthermore, microarray analyses of peripheral blood mononuclear cells (PBMCs) from SLE patients demonstrate an over-expression of IFNα-regulated genes, termed the type I IFN signature.3 Due to diagnosis earlier and better treatment options in SLE, the survival has increased substantially over the past decades. A bimodal pattern of mortality was described already 35 years ago with early mortality being caused by uncontrolled disease activity and infections while late mortality mainly was due to cardiovascular complications.4 Cardiovascular disease occurs even in young females with SLE and a 50-fold increased risk of myocardial infarction has been reported in this group of SLE patients.5 Traditional Framingham risk factors can only partly explain the cardiovascular morbidity in SLE, and the disease itself seems to be an independent risk factor for cardiovascular events.5 Pathogenetic mechanisms driven by SLE are not clear but systemic inflammation with damaging effects on vessels are most likely involved in which immune complexes, autoantibodies and complement play a role together with lymphocytes and monocyte derived cells. Endothelial dysfunction, which can be an early stage in the development of atherosclerosis, has also been described in SLE. It has been proposed that there is an imbalance between vascular damage and repair. Interestingly, this seems to be controlled by IFNα, a key cytokine in the pathogenesis of SLE.6 There is also a substantially increased risk for venous thrombosis in SLE, often associated with antiphospholipid (aPL) antibodies and coagulation abnormalities.7 Approximately 10% to 20% of SLE patients have secondary aPL antibody syndrome (APS) with venous thrombosis, stroke, recurrent spontaneous abortions, thrombocytopenia and aPL antibodies. In summary, large portions of patients with SLE have significant clinical problems involving both arteries and veins, which we will refer to collectively as vascular disease (VD).
The possible role of aberrant platelet activation in the pathogenesis of VD in SLE is suggested in a few studies, but has not been extensively investigated. Increased platelet expression of P-selectin and phosphatidylserine (PS) has been demonstrated8 but has not, however, been reproduced.9 Furthermore, platelet-monocyte complexes, which could accelerate the production of tissue factor,10 are found more often in SLE.9 Markers of sustained platelet activation such as extracellular phosphorylation of the plasma proteins fibrinogen and C3 has been found in SLE patients and may be associated with venous thrombosis.11,12 Even though the function of platelets is central in coagulation and VD, the platelet contribution to the increased risk of VD in SLE remains unknown. In the present study, platelet gene expression was probed using microarray, and the altered expression in SLE of selected genes was confirmed on both mRNA and protein level and related to platelet activation status. We could demonstrate an increased expression of type I IFN-regulated genes and proteins and increased platelet activation in SLE. Furthermore, SLE patients with a previous history of VD had clearly increased levels of type I IFN-regulated proteins and increased platelet activation compared with SLE patients without a previous history of VD.
Methods
Patients
Patients were recruited in 2 sets: exploratory cohort I including 10 SLE patients for the gene expression analyses, and the confirmatory cohort II including 69 SLE patients for the protein expression analyses. Patients were recruited at routine clinical visits (to the Department of Rheumatology, Lund University Hospital, Lund, Sweden). A selection of patients with a known history of VD was done in the confirmatory cohort II in which 45% of the patients had a history of VD compared with 24% in the previously described unselected SLE cohort.13-16 In this study, VD is defined as previous episodes of either myocardial infarction (MI), arterial thrombosis (12/13 with cerebrovascular incidents), and venous thrombosis (pulmonary embolism or deep venous thrombosis) as defined by the Systemic Lupus International Collaborative Clinics/American College of Rheumatology Damage Index.17 The median time since the last VD event was 8 years for the SLE patients (Table 1). Healthy age- and sex-matched volunteers, without a history of VD were used as controls. An overview of clinical characteristics and treatment at the time of blood sampling is presented in Tables 1 and 2. All patients fulfilled at least 4 American College of Rheumatology (ACR) classification criteria for SLE.18 Auto-antibodies and complement levels in serum were measured according to routine analyses at the Department of Clinical Immunology, Lund University Hospital, Lund, Sweden. The study was approved by the regional ethics board (LU 378-02).
Clinical characteristics of the SLE patients according to ACR criteria and presence of cardiovascular events at any time during disease
| . | Cohort I (n = 10) . | Cohort II (n = 69) . |
|---|---|---|
| Malar rash, % | 30 | 61 |
| Discoid rash, % | 20 | 30 |
| Photosensitivity, % | 40 | 64 |
| Oral ulcers, % | 10 | 30 |
| Arthritis, % | 50 | 81 |
| Serositis, % | 40 | 65 |
| Renal disease, % | 50 | 42 |
| Neurologic disorder, % | 10 | 9 |
| Hematologic manifestations, % | 70 | 52 |
| Leukopenia, % | 70 | 39 |
| Lymphopenia, % | 60 | 28 |
| Thrombocytopenia, % | 20 | 20 |
| Immunology, % | 80 | 67 |
| ANA, % | 100 | 100 |
| Anti-DNA antibodies, % | 90 | 71 |
| Anti-cardiolipin antibodies, % | 50 | 45 |
| Anti-phospholipid antibody syndrome, % | 20 | 29 |
| Vascular disease, % | 10 | 45 |
| Median time since event, y (range) | 24 | 8 (2-36) |
| Venous thrombosis, % | 10 | 29 |
| Median time since event, y (range) | 24 | 16 (3-36) |
| Arterial thrombosis, % | 0 | 19 |
| Median time since event, y (range) | 8 (1-18) | |
| Myocardial infarction, % | 0 | 15 |
| Median time since event, y (range) | 7 (2-30) |
| . | Cohort I (n = 10) . | Cohort II (n = 69) . |
|---|---|---|
| Malar rash, % | 30 | 61 |
| Discoid rash, % | 20 | 30 |
| Photosensitivity, % | 40 | 64 |
| Oral ulcers, % | 10 | 30 |
| Arthritis, % | 50 | 81 |
| Serositis, % | 40 | 65 |
| Renal disease, % | 50 | 42 |
| Neurologic disorder, % | 10 | 9 |
| Hematologic manifestations, % | 70 | 52 |
| Leukopenia, % | 70 | 39 |
| Lymphopenia, % | 60 | 28 |
| Thrombocytopenia, % | 20 | 20 |
| Immunology, % | 80 | 67 |
| ANA, % | 100 | 100 |
| Anti-DNA antibodies, % | 90 | 71 |
| Anti-cardiolipin antibodies, % | 50 | 45 |
| Anti-phospholipid antibody syndrome, % | 20 | 29 |
| Vascular disease, % | 10 | 45 |
| Median time since event, y (range) | 24 | 8 (2-36) |
| Venous thrombosis, % | 10 | 29 |
| Median time since event, y (range) | 24 | 16 (3-36) |
| Arterial thrombosis, % | 0 | 19 |
| Median time since event, y (range) | 8 (1-18) | |
| Myocardial infarction, % | 0 | 15 |
| Median time since event, y (range) | 7 (2-30) |
SLE indicates systemic lupus erythematosus; and ACR, American College of Rheumatology.
Clinical characteristics and treatment of the SLE patients and healthy controls at the time point of blood sampling. Items included in SLEDAI are shown
| . | Cohort I (n = 10) . | Cohort II (n = 69) . | Controls I (n = 10) . | Controls II (n = 69) . |
|---|---|---|---|---|
| Median age, y (range) | 36 (22-70) | 51 (20-84) | 36 (22-70) | 53 (19-79) |
| Female, % | 100 | 87 | 100 | 87 |
| Male, % | 0 | 13 | 0 | 13 |
| Median disease duration, y (range) | 4 (1-20) | 13 (0-49) | ||
| Median SLEDAI score (range) | 2 (0-14) | 2 (0-14) | ||
| Seizure | 0 | 0 | ||
| Psychosis | 0 | 0 | ||
| Organic brain syndrome | 0 | 0 | ||
| Visual disturbance | 0 | 1 | ||
| Cranial nerve disorder | 0 | 0 | ||
| Lupus headache | 0 | 0 | ||
| Cerebrovascular accident | 0 | 0 | ||
| Vasculitis | 2 | 2 | ||
| Arthritis | 0 | 8 | ||
| Myositis | 1 | 0 | ||
| Kidney involvement (urinary cast, hematuria, proteinuria or pyuria) | 2 | 12 | ||
| Mucocutaneous activity (rash, alopecia or mucosal ulcers) | 3 | 10 | ||
| Pleurisy | 0 | 0 | ||
| Pericarditis | 0 | 0 | ||
| Low complement (C3 or C4) | 3 | 19 | ||
| Anti-DNA antibodies | 1 | 15 | ||
| Fever | 0 | 1 | ||
| Thrombocytopenia | 0 | 1 | ||
| Leukopenia | 0 | 3 | ||
| Hydroxychloroquine | 4 | 46 | ||
| Azathioprine | 0 | 19 | ||
| Mycophenolatmofetil | 1 | 6 | ||
| Rituximab (within 12 mo) | 0 | 1 | ||
| Methotrexate | 0 | 6 | ||
| Cyclophosphamide | 1 | 3 | ||
| Cyclosporin A | 1 | 2 | ||
| NSAIDs | 0 | 12 | ||
| Acetylsalicylic acid | 2 | 13 | ||
| Warfarin | 1 | 24 |
| . | Cohort I (n = 10) . | Cohort II (n = 69) . | Controls I (n = 10) . | Controls II (n = 69) . |
|---|---|---|---|---|
| Median age, y (range) | 36 (22-70) | 51 (20-84) | 36 (22-70) | 53 (19-79) |
| Female, % | 100 | 87 | 100 | 87 |
| Male, % | 0 | 13 | 0 | 13 |
| Median disease duration, y (range) | 4 (1-20) | 13 (0-49) | ||
| Median SLEDAI score (range) | 2 (0-14) | 2 (0-14) | ||
| Seizure | 0 | 0 | ||
| Psychosis | 0 | 0 | ||
| Organic brain syndrome | 0 | 0 | ||
| Visual disturbance | 0 | 1 | ||
| Cranial nerve disorder | 0 | 0 | ||
| Lupus headache | 0 | 0 | ||
| Cerebrovascular accident | 0 | 0 | ||
| Vasculitis | 2 | 2 | ||
| Arthritis | 0 | 8 | ||
| Myositis | 1 | 0 | ||
| Kidney involvement (urinary cast, hematuria, proteinuria or pyuria) | 2 | 12 | ||
| Mucocutaneous activity (rash, alopecia or mucosal ulcers) | 3 | 10 | ||
| Pleurisy | 0 | 0 | ||
| Pericarditis | 0 | 0 | ||
| Low complement (C3 or C4) | 3 | 19 | ||
| Anti-DNA antibodies | 1 | 15 | ||
| Fever | 0 | 1 | ||
| Thrombocytopenia | 0 | 1 | ||
| Leukopenia | 0 | 3 | ||
| Hydroxychloroquine | 4 | 46 | ||
| Azathioprine | 0 | 19 | ||
| Mycophenolatmofetil | 1 | 6 | ||
| Rituximab (within 12 mo) | 0 | 1 | ||
| Methotrexate | 0 | 6 | ||
| Cyclophosphamide | 1 | 3 | ||
| Cyclosporin A | 1 | 2 | ||
| NSAIDs | 0 | 12 | ||
| Acetylsalicylic acid | 2 | 13 | ||
| Warfarin | 1 | 24 |
SLE indicates systemic lupus erythematosus; SLEDAI, SLE Disease Activity Index; and NSAIDs, non steroidal anti-inflammatory drugs.
Gene expression analysis
Isolation and purification of platelets, cDNA synthesis, microarray and real-time PCR of CD58 and CD69 in cohort I was performed as described by Amisten et al.19 All microarray data are available at GEO, accession number GSE22132. Cluster-analysis was carried-out using the Ingenuity Pathways Analysis software (Ingenuity Systems Inc). Real-time PCR of IFITM1 and PRKRA (QIAGEN Sciences) was performed according to the manufacturer's instructions and run in a Rotor-Gene 2000 (Corbett Life Science), and all gene expression is presented relative to the expression of the house-keeping gene Glyceraldehyde 3-phosphate dehydrogenase.
Detection of surface molecules on platelets
Blood was collected in sodium-citrate tubes (BD Biosciences Pharmingen) and used within 15 minutes. For direct surface staining, 5 μL of whole blood was incubated with fluorescein isothiocyanate (FITC)–conjugated annexin V, FITC- or phycoerythrin (PE)–conjugated antibodies against CD58 and P-selectin, respectively, (BD Biosciences Pharmingen) in HS-buffer (145mM NaCl, 5mM KCl, 10mM HEPES, pH 7.4) in a total volume of 50 μL. After 15 minutes of incubation at room temperature, 500 μL of 0.2% paraformaldehyde (PFA) was added and the incubation continued for 15 minutes at 4°C. The cells were diluted 1/20 in phosphate-buffered saline (PBS) pH 7.2 and analyzed by flow cytometry (Epics XL-MCL; Beckman-Coulter). An antibody isotype control was used as a negative control with a cutoff value of 1% positive platelets.
For detection of CD69, platelet rich plasma (PRP) obtained by centrifugation of whole blood for 10 minutes at 280g, was used. The PRP was mixed with 10mM EDTA and the platelets centrifuged at 1125g for 10 minutes and resuspended in 500 μL of HS-buffer. Platelet suspension (0.4 μL) was then incubated with antibodies against CD69 (Santa Cruz Biotechnology), in HS-buffer at a total volume of 50 μL for 40 minutes at room temperature, washed once and incubated with FITC-conjugated rabbit anti–mouse Ig antibodies (Dako) for an additional 30 minutes at 4°C. The incubation was ended with addition of 500 μL of 0.2% PFA. The platelets were diluted 1/5 in PBS before analyzed by flow cytometry. An antibody isotype control was used as a negative control with a cutoff value of 2% positive platelets.
Detection of intracellular proteins
Platelets obtained as described in the previous paragraph were incubated with 4% PFA for 10 minutes, washed with PBS and permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) for 15 minutes at room temperature. The platelets were washed, resuspended in 500 μL of HS-buffer and incubated for 30 minutes at room temperature with antibodies against PRKRA or IFITM1 (Santa Cruz Biotechnology). The platelets were then washed and incubated for an additional 30 minutes at room temperature with a FITC-conjugated mouse anti–goat Ig antibody (Santa Cruz Biotechnology). The incubation was stopped by the addition of 500 μL of 0.2% PFA before the samples were diluted 1/5 in PBS and analyzed by flow cytometry. An antibody isotype control was used with a cut-off value of 2% positive platelets.
Detection of platelet-monocyte complexes
For the detection of platelet-monocyte complexes, 5 μL of blood was incubated with PE-conjugated anti-CD45 antibodies and FITC-conjugated anti-CD42a antibodies (BD Biosciences Pharmingen) for 15 minutes at room temperature in HS-buffer in a total volume of 50 μL. The incubation was ended by the addition of 750 μL of 0.5% formaldehyde, after which the cells were analyzed by flow cytometry.
Serum IFNα detection
Serum IFNα was measured according to the manufacturers' instructions using the Procarta cytokine assay kit (Panomics Srl) and analyzed using a Luminex 200 (Luminex Corporation). Serum IFNα concentration was defined as increased if the concentration was higher than 0.13 pg/mL (ie, median + 2 SDs for 70 healthy volunteers).
Western blot analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed using Mini Protean II cell equipment (Bio-Rad). The serum samples, diluted 1/500 in PBS, were mixed 1:1 (vol/vol) with nonreducing sample buffer and boiled for 5 minutes before loading 0.5 μL onto the 6% polyacrylamide protein mini gels. PageRuler Protein Ladder Plus (Fermentas) was used as the high-molecular-mass standard. The gels were transferred to polyvinylidenefluoride (PVDF) membranes (Immobilon P; Millipore) as described by Matsudaira.20 For detection of IgG immune complexes (ICs), membranes were blocked in 5% (wt/vol) skim milk (DIFCO) dissolved in PBS containing 0.05% (vol/vol) Tween 20 for 1 hour at room temperature. The blots were incubated with peroxidase-conjugated goat anti–human IgG, (AbD Serotec) diluted 1/4000 for 1 hour at 37°C. After the subsequent washing step in PBS containing 0.05% (vol/vol) Tween 20, the membranes were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Densitometric analysis of protein bands was performed using the public software ImageJ 1.41o developed by Wayne Rasband at the National Institutes of Health.
Enzyme-linked immunosorbent assay measurement of immune complexes
Microtiter plates (Greiner Bio-one) were coated with 100 μL of human C1q (10 μg/mL), prepared as described by Tenner et al,21 and kept at 4°C overnight. The plates were washed 3 times in PBS and blocked for 2 hours at room temperature with 1% (wt/vol) gelatin in PBS. Sera were pre-diluted 1/3 in 0.2M EDTA and incubated for 30 minutes at 37°C. This was further diluted 1/10 in veronal buffered saline containing 10mM EDTA and 0.05% (vol/vol) Tween (VBS-ET) and incubated at 37°C for 1 hour and then at 4°C for 20 hours. As a positive standard, heat-aggregated IgG was used. After the subsequent wash step in VBS-ET, an alkaline phosphatase conjugated goat anti–human antibody (Sigma-Aldrich) was added at a dilution of 1/10 000 and incubated for 1 hour at 4°C. After a wash step, the phosphatase substrate (Sigma-Aldrich) was added. The absorbance at 405 nm was read using a Wallac 1420 Multilabel Counter (PerkinElmer).
Induction of type I IFN-regulated proteins in MEG-01
MEG-01 cells (CRL-2021; ATCC) were cultured in RPMI-1640 supplemented with 10% fetal calf serum, 1% non essential amino acids (vol/vol), 1 mM sodium pyruvate (PAA Laboratories GmbH) and 100 μg/mL gentamicin (Invitrogen) for 4 days with or without the addition of 2000 U/mL IFNα (Intron A; SP) after which the cells were analyzed for expression of type I IFN-regulated proteins.
Statistical analysis
Correlations were determined by Spearman's correlation test and the Mann-Whitney U test was used for group comparisons. Values are expressed as the mean plus or minus 1 SD, median with 95% coincidence interval or median with range. All P values were considered significant at P less than .05.
Results
The overall strategy was to identify platelet genes of interest by microarray analysis and to demonstrate the expression of those genes by real-time PCR in cohort I. In cohort II, corresponding platelet protein expression and platelet function was examined. Correlations with clinical data were performed only in cohort II due to the small number of patients in cohort I.
Identification of genes differentially expressed in SLE patients
Using gene cluster-analysis several clusters of genes were identified that were differentially expressed in SLE patients in cohort I compared with the healthy controls (supplemental Table 1 [available on the Blood Web site; see the Supplemental Materials link at the top of the online article] and GEO, accession number GSE22132). In general, platelets from SLE patients had increased expression of genes encoding cytokines, chemokines and proteins involved in apoptosis, and decreased expression of genes encoding several complement proteins. Interestingly, many type I IFN-regulated genes were highly up-regulated (Table 3) and CD58, PRKRA, CD69, and IFITM1 were selected for further analyses. Real-time PCR revealed that the median mRNA expression of CD58 in SLE patients and controls were 3.80 (1.85-5.72) and 1.31 (0.91-1.55), respectively (P = .06) and the corresponding figures for PRKRA were 0.002 (0.001-0.005) and 0.001 (0-0.005), respectively (P = .72). Thus, mRNA expression of CD58 and PRKRA did not differ significantly between SLE patients and healthy controls. However, the median relative mRNA expression of IFITM1 was 1.27 (0.75-2.94) in SLE and 0.42 (0.03-0.99) in controls, which was statistically significant (P = .008). Also CD69 was statistically significant increased (P = .04) in SLE with a relative mRNA expression of 6.07 (1.97-34.06) in SLE and 2.04 (1.12-2.79) in controls. Thus, we could confirm the presence of a type I IFN transcriptional signature in platelets from SLE patients.
Expression of type I IFN-regulated genes in platelets from SLE patients in comparison with healthy controls given as fold change
| Gene symbol . | Gene title . | Fold change . |
|---|---|---|
| IFI27 | interferon, alpha-inducible protein 27 | 23.12 |
| CD58* | CD58 antigen, (lymphocyte function-associated antigen 3) | 13.04 |
| PRKRA* | protein kinase, interferon-inducible double stranded RNA dependent activator | 6.34 |
| G1P3 | interferon, alpha-inducible protein (clone IFI-6-16) | 6.27 |
| CD69* | CD69 antigen (p60, early T-cell activation antigen) | 5.25 |
| IFITM1* | interferon induced transmembrane protein 1 (9-27) | 3.42 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | 2.87 |
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 2.58 |
| LY6E | lymphocyte antigen 6 complex, locus E | 2.43 |
| IFNGR1 | interferon gamma receptor 1 | 2.18 |
| IFRD1 | interferon-related developmental regulator 1 | 2.10 |
| IFI44L | interferon-induced protein 44-like | 2.04 |
| OAS3 | 2′-5′-oligoadenylate synthetase 3, 100kDa | 1.98 |
| G1P2 | interferon, alpha-inducible protein (clone IFI-15K) | 1.98 |
| PLSCR1 | phospholipid scramblase 1 | 1.97 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71kDa | 1.92 |
| IRF2BP2 | interferon regulatory factor 2 binding protein 2 | 1.92 |
| GBP1 | guanylate binding protein 1, interferon-inducible, 67kDa | 1.87 |
| IFI16 | interferon, gamma-inducible protein 16 | 1.83 |
| IFITM3 | interferon induced transmembrane protein 3 (1-8U) | 1.80 |
| IRF2 | interferon regulatory factor 2 | 1.70 |
| Gene symbol . | Gene title . | Fold change . |
|---|---|---|
| IFI27 | interferon, alpha-inducible protein 27 | 23.12 |
| CD58* | CD58 antigen, (lymphocyte function-associated antigen 3) | 13.04 |
| PRKRA* | protein kinase, interferon-inducible double stranded RNA dependent activator | 6.34 |
| G1P3 | interferon, alpha-inducible protein (clone IFI-6-16) | 6.27 |
| CD69* | CD69 antigen (p60, early T-cell activation antigen) | 5.25 |
| IFITM1* | interferon induced transmembrane protein 1 (9-27) | 3.42 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | 2.87 |
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 2.58 |
| LY6E | lymphocyte antigen 6 complex, locus E | 2.43 |
| IFNGR1 | interferon gamma receptor 1 | 2.18 |
| IFRD1 | interferon-related developmental regulator 1 | 2.10 |
| IFI44L | interferon-induced protein 44-like | 2.04 |
| OAS3 | 2′-5′-oligoadenylate synthetase 3, 100kDa | 1.98 |
| G1P2 | interferon, alpha-inducible protein (clone IFI-15K) | 1.98 |
| PLSCR1 | phospholipid scramblase 1 | 1.97 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71kDa | 1.92 |
| IRF2BP2 | interferon regulatory factor 2 binding protein 2 | 1.92 |
| GBP1 | guanylate binding protein 1, interferon-inducible, 67kDa | 1.87 |
| IFI16 | interferon, gamma-inducible protein 16 | 1.83 |
| IFITM3 | interferon induced transmembrane protein 3 (1-8U) | 1.80 |
| IRF2 | interferon regulatory factor 2 | 1.70 |
The genes were selected based on type I IFN-regulated genes reported up-regulated in SLE patients' peripheral blood mononuclear cells and also several other type I IFN-associated genes as determined by the open access database INTERFEROME (http://www.interferome.org). All microarray data are available in supplemental Table 1 and at GEO under accession number GSE22132.
IFN indicates interferon; and SLE, systemic lupus erythematosus.
Expression of genes was further validated by real-time polymerase chain reaction and flow cytometry.
Increased type I IFN-regulated protein expression in SLE patients
Because SLE patients in cohort I displayed increased expression of type I IFN-regulated genes as demonstrated by microarray and real-time PCR, we investigated if the expression of the corresponding proteins in platelets were also elevated. The protein expression of CD58, CD69, IFITM1 and PRKRA was validated in cohort II and all 4 proteins were found to be up-regulated in platelets from SLE patients (P = .004, P < .0001, P < .0001, and P < .0001, respectively, Figure 1). Thus, the elevated expression of the CD58, CD69, IFITM1 and PRKRA proteins confirmed the gene expression results and demonstrated the presence of a type I IFN protein signature in platelets from SLE patients.
Increased platelet protein expression of CD58, CD69, IFITM1 and PRKRA in SLE patients. Surface (CD58, CD69) or intracellular (IFITM1, PRKRA) protein levels were measured on platelets from healthy controls and systemic lupus erythematosus (SLE) patients by flow cytometry. The values are expressed as percentage of cells being positive compared with an antibody isotype control. The lines represent the median-value in each group.
Increased platelet protein expression of CD58, CD69, IFITM1 and PRKRA in SLE patients. Surface (CD58, CD69) or intracellular (IFITM1, PRKRA) protein levels were measured on platelets from healthy controls and systemic lupus erythematosus (SLE) patients by flow cytometry. The values are expressed as percentage of cells being positive compared with an antibody isotype control. The lines represent the median-value in each group.
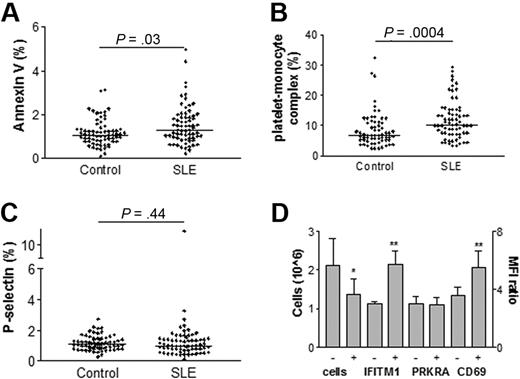
Increased activation of platelets in SLE patients
To investigate if the increased expression of the 4 type I IFN-regulated PRKRA, IFITM1, CD58, and CD69 identified above had any effect on platelet activation, the expressions of several platelet activation markers were analyzed. Annexin V binding and amounts of platelet-monocyte complexes were found to be increased in SLE patients (P = .03 and P = .0004, respectively, Figure 2). However, platelets from SLE patients did not show a significantly increased expression of P-selectin (P = .44, Figure 2). There was a weak but statistically significant correlation between PRKRA and P-selectin (P = .02, r = 0.30), monocyte-platelet complexes (P = .04, r = 0.26) and CD69 (P = .02, r = 0.29) suggesting an association between the type I IFN-response and platelet activation. Patients using drugs containing acetylsalicylic acid, hydroxychloroquine or immune suppressive drugs (azathioprine, mycophenolatmotefil, rituximab, methotrexate, cyclophosphamide or cyclosporine A) did not differ in any of the investigated platelet markers compared with SLE patients without those drugs. There were no strong indications that treatment with nonsteroidal anti-inflammatory drugs (NSAIDs) would have an impact on our results. There was, however, a decreased CD69 (P = .011) and P-selectin (P = .043) expression, but no other activation markers or type I IFN-regulated proteins were altered in these NSAID treated patients (data not shown). Thus, we demonstrated an increased activation of platelets from SLE patients associated with the type I IFN-response.
Increased platelet activation in SLE patients and stimulation of MEG-01 cells. Platelet binding of annexin V (A), platelet-monocyte complexes (B), and expression of P-selectin (C) were measured by flow cytometry in healthy controls and SLE patients. The values are expressed as percentage of cells being positive compared with an antibody isotype control. The lines represent the median-value in each group. MEG-01 cells were cultured in the presence or absence of IFNα and type I IFN-regulated proteins were measured by flow cytometry (D). The results are expressed as the mean fluorescence index (MFI) ratio or as the cell concentration with 1 SD. The statistical significance is noted above each set of data with P values or signs where * indicates P < .05 and **, P < .01.
Increased platelet activation in SLE patients and stimulation of MEG-01 cells. Platelet binding of annexin V (A), platelet-monocyte complexes (B), and expression of P-selectin (C) were measured by flow cytometry in healthy controls and SLE patients. The values are expressed as percentage of cells being positive compared with an antibody isotype control. The lines represent the median-value in each group. MEG-01 cells were cultured in the presence or absence of IFNα and type I IFN-regulated proteins were measured by flow cytometry (D). The results are expressed as the mean fluorescence index (MFI) ratio or as the cell concentration with 1 SD. The statistical significance is noted above each set of data with P values or signs where * indicates P < .05 and **, P < .01.
Increased serum concentrations of IFNα and immune complexes in SLE patients
Because a type I IFN signature was found and ICs can induce a type I IFN response,22 the serum levels of IFNα and ICs were measured. SLE patients had increased levels of serum IFNα compared with healthy controls (mean SLE 1.29 ± 6.14 pg/mL and mean control 0.02 ± 0.06 pg/mL, P = .01), as well as increased levels of ICs compared with healthy controls (median SLE 0.11 [0.11-0.97] mg/mL and median control 0.03 [0.03-0.67] mg/mL, P = .0005). SLE patients with increased levels of serum IFNα had increased levels of ICs when measured by Western blot (P = .003, data not shown). Furthermore, a weak correlation (P = .004, r = 0.35) was found between ICs and serum levels of IFNα suggesting that ICs might be involved in the regulation of serum IFNα. In summary, we could demonstrate that the SLE patients had increased serum concentrations of ICs and IFNα.
Increased type I IFN-regulated protein expression in MEG-01 cells after IFNα treatment
To investigate if IFNα could induce the expression of the type I IFN-regulated proteins identified in the SLE platelet samples, a megakaryocytic cell line (MEG-01) was used. When stimulated with IFNα, MEG-01 cells increased their expression of IFITM1 and CD69 (P = .004 and P = .002, respectively) and we could also demonstrate a decreased number of cells (P = .02, Figure 2D). However, the PRKRA protein expression was not increased upon IFNα stimulation and CD58 was not tested in this assay (Figure 2D). In line with this observation, an inverse correlation between PRKRA protein expression and platelet count (P = .007, r = −0.33) was found, confirming the antiproliferative effect of IFNα in vivo. In conclusion, we could demonstrate that IFNα influenced the megakaryocytic type I IFN-regulated protein expression suggesting an interaction of IFNα with megakaryocytes in the bone marrow of SLE patients.
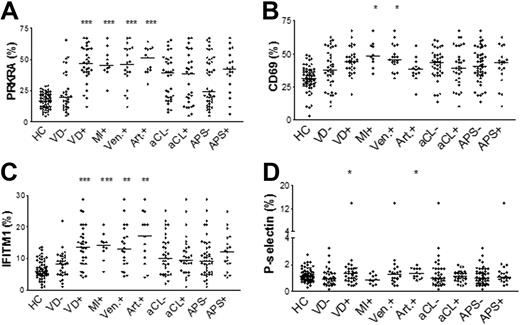
Type I IFN-regulated proteins are associated with vascular disease in SLE
Markers of platelet activation and levels of type I IFN-regulated proteins were statistically correlated. It was therefore of interest to investigate if these platelet activation markers and type I IFN-regulated protein expressions were associated with VD in SLE. Patients with a history of any VD event displayed increased levels of PRKRA, IFITM1 and P-selectin (P < .0001, P = .0002 and P = .045, respectively, Figure 3). SLE patients with a previous history of MI had increased protein levels of CD69, PRKRA and IFITM1 (P = .04, P = .0004, and P = .0004, respectively). SLE patients with a history of venous thrombosis had increased protein levels of PRKRA, IFITM1 and CD69 (P = .0001, P = .003, and P = .03, respectively) and patients with a previous history of arterial thrombosis had increased levels of PRKRA, IFITM1 and P-selectin (P < .0001, P = .003 and P = .03, respectively, Figure 3). Presence of anticardiolipin (aCL) antibodies ever during disease or the APS were not associated with any of the investigated platelet markers. Patients treated with Warfarin had increased levels of IFITM1 (P = .02), PRKRA (P = .009) and platelet-monocyte complexes (P = .011). As expected, in this group of patients all had a previous episode of VD, which most likely explains these findings. Altogether, these results suggest that SLE patients with a previous history of VD have more activated platelets with increased expression of type I IFN-regulated proteins than SLE patients without a previous history of VD.
Increased expression of type I IFN-regulated proteins in patients with a history of VD. Patients with vascular disease (VD+), myocardial infarction (MI), venous thrombosis (Ven.+), and arterial thrombosis (Art.+) are compared with patients without VD (VD−). Patients with a history of aCL antibodies (aCL+) are compared with patients without a history of aCL antibodies (aCL−) and patients with antiphospholipid antibody syndrome (APS+) are compared with patients without APS (APS−). Healthy controls (HC) are also included. The lines represent the median-values. The statistical significance is noted above each set of data where * indicates P < .05; **, P < .01; and ***, P < .001.
Increased expression of type I IFN-regulated proteins in patients with a history of VD. Patients with vascular disease (VD+), myocardial infarction (MI), venous thrombosis (Ven.+), and arterial thrombosis (Art.+) are compared with patients without VD (VD−). Patients with a history of aCL antibodies (aCL+) are compared with patients without a history of aCL antibodies (aCL−) and patients with antiphospholipid antibody syndrome (APS+) are compared with patients without APS (APS−). Healthy controls (HC) are also included. The lines represent the median-values. The statistical significance is noted above each set of data where * indicates P < .05; **, P < .01; and ***, P < .001.
Discussion
The underlying mechanisms of the increased risk of VD in SLE are unclear, and the role of platelets has not previously been thoroughly examined. In this study we present a strong association between type I IFN-regulated proteins in platelets and VD in SLE and we hypothesise that interferogenic immune complexes (ICs) promote IFNα production by plasmacytoid dendritic cells that may influence gene expression in megakaryocytes resulting in increased levels of type I IFN-regulated proteins. We found that platelets derived from these megakaryocytes had increased expression of the type I IFN-regulated genes and proteins as demonstrated by microarray, real-time PCR and flow cytometry. Furthermore, SLE patients with a history of VD had clearly higher levels of platelet type I IFN-regulated proteins as well as more activated platelets compared with patients without a history of VD.
The type I IFN signature has been described previously in PBMCs from SLE patients.3 The use of microarray analysis on platelets is new with only a few publications due to difficulties in obtaining high purity platelets.19,23,24 Here we could, for the first time, demonstrate up-regulation of several type I IFN-regulated genes in platelets from SLE patients and also confirm increased expression on platelets of the proteins encoded by the type I IFN-regulated genes. Not all of the investigated genes were significantly increased as estimated by real-time PCR. This was most likely due to the limited number of samples in cohort I. However, the corresponding protein expression was significantly increased in the larger cohort II. CD69 was found to be up-regulated in platelets from SLE patients, especially in patients with previous episodes of MI. CD69 is a constitutively expressed protein on platelets and the platelet CD69 mRNA level has previously been identified as a strong discriminator of acute ST-segment elevation MI by microarray analysis.25 Up-regulation of CD69 expression by IFNα has been described in neutrophils,26 and we found that megakaryocytes, cultured in the presence of IFNα, increased the expression of CD69 and other type I IFN-regulated proteins. We hypothesise that IFNα stimulates megakaryocytes in the bone marrow to express these proteins. Importantly, there was a strong association between PRKRA expression and history of VD, independently of aPL antibodies. Prospective studies on the importance of PRKRA and CD69 expression as biomarkers for VD in SLE are clearly warranted. It is, however, important to note that the median time between VD event and blood sampling was 8 years. It is possible that the IFN-regulated platelet proteins are constitutively over-expressed in these patients. Ongoing studies are investigating the use of these markers in a larger cohort of SLE patients, patients with other autoimmune diseases, as well as patients with VD without underlying inflammatory disease.
The underlying biologic mechanism of the association between the type I IFN gene and protein profile is not clear, and it was not the aim of this study to investigate this. Two of the type I IFN-regulated proteins, PRKRA and IFITM1, are both involved in the IFN-mediated antiviral defense. IFITM1 has been described to decrease cell growth27 and inhibit influenza A viral replication.28 PRKRA acts indirectly by forming heterodimers with the interferon-inducible RNA-dependent protein kinase PKR.29 Upon dimerization, PKR is activated and the eukaryotic translation initiation factor 2 is phosphorylated, leading to decreased protein synthesis and increased apoptosis.30-32 PRKRA is also required for the accumulation of mature miRNA and is an important part of the RNA-induced silencing complex that processes pre-miRNA to functional miRNA.33 Whether these properties of PRKRA and IFITM1 could influence activation of platelets leading to VD in SLE remains to be examined.
Inflammation in general has been suggested to play an important role in the development of VD and the presence of pro-inflammatory cytokines increases platelet aggregation in patients with acute coronary syndrome.34 There are a number of findings indicating an important role of IFNα, a key cytokine in SLE pathogenesis, in the development of VD. High IFNα serum levels in SLE have been associated with decreased numbers of endothelial progenitor cells and impaired ability of endothelial progenitor cells to differentiate into mature cells, which could promote atherosclerosis.35,36 Furthermore, a risk allele has been found in the type I IFN-regulated gene STAT4 that was found to be associated with ischemic cerebrovascular events in SLE.16 A STAT4 risk allele has also recently been associated with the primary APS.37 In summary, genetic variants within the type I IFN system (STAT4) associated with VD and IFNα mediated endothelial dysfunction all contribute to the pathogenesis of VD. The correlation found between type I IFN-regulated proteins and platelet activation markers demonstrated in this paper adds a novel mechanism for development of VD in SLE.
Because we postulate in this paper that IFNα could be involved in VD and platelet activation, it would be relevant to study patients with chronic hepatitis C who are commonly treated with recombinant IFNα. No increase in platelet activation markers are seen in these patients. Instead platelet activation is reported to decrease during IFNα therapy,38 probably due to a diminished viral load. The lack of VD and platelet activation in the IFNα treated patients might be due to the much shorter time period with IFNα exposure in hepatitis compared with the life-long exposure in SLE, but also to the fact that development of VD in SLE is a complex process with many different mechanisms involved.
Platelet activation status was principally measured in 3 ways; phosphatidylserine and P-selectin surface expression and platelet-monocyte complex formation, and increased platelet activation could be demonstrated by 2 of the methods. This was in accordance with others who have demonstrated increased phosphatidylserine8 and platelet-monocyte complexes9 in patients with SLE. Despite the increased activation state of circulating platelets, we did not see any increase in platelet P-selectin expression. Measurement of circulating platelet-monocyte complexes, however, is a more sensitive marker of platelet activation than surface expression of P-selectin because of the rapid loss of surface expression of the latter.39 It is also possible that P-selectin might be down-regulated due to the increased levels of IFNα.40
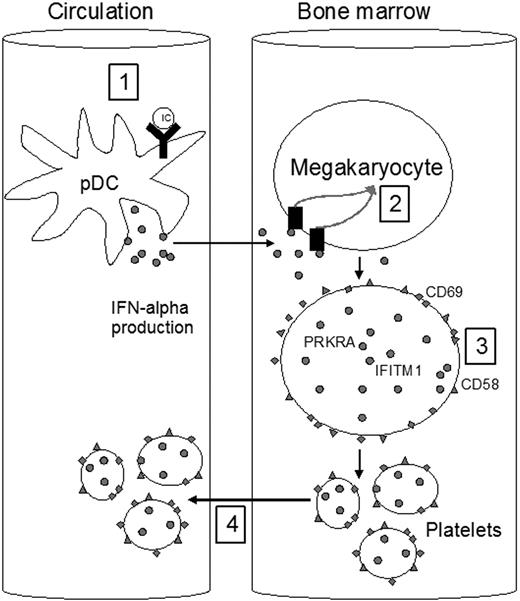
In conclusion, we have, for the first time, found a type I IFN-signature in platelets from SLE patients. This signature was clearly associated with a history of VD. As illustrated in Figure 4, our data support that IFNα, derived from interferogenic IC-stimulated plasmacytoid dendritic cells, up-regulates the megakaryocytic type I IFN-regulated genes and proteins and affects the activation of platelets. Platelets with the type I IFN-signature seem to be more activated and may potentially be more prone to take part in thrombotic events and contribute to the development of VD. We suggest that platelets with the type I IFN signature might constitute a novel marker of VD in SLE. Further studies are required to clarify the mechanisms involved and prospective studies are needed to investigate its usefulness as a predictive marker of VD.
Type I IFN-profile in platelets. A schematic model describing interactions between immune complexes (ICs), interferonα (IFNα), and cells supported by the experiments reported here. RNA and DNA containing ICs induce IFNα production from plasmacytoid dendritic cells (pDCs) in the circulation (1). IFNα induces increased expression of type I IFN-regulated genes and proteins in the megakaryocytes in the bone marrow (2-3). The platelets are released from the megakaryocytes and enter the circulation (4).
Type I IFN-profile in platelets. A schematic model describing interactions between immune complexes (ICs), interferonα (IFNα), and cells supported by the experiments reported here. RNA and DNA containing ICs induce IFNα production from plasmacytoid dendritic cells (pDCs) in the circulation (1). IFNα induces increased expression of type I IFN-regulated genes and proteins in the megakaryocytes in the bone marrow (2-3). The platelets are released from the megakaryocytes and enter the circulation (4).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The study was supported by grants from the Swedish Research Council (2008-2201), the Medical Faculty at Lund University, Alfred Österlund's Foundation, The Crafoord Foundation, Greta and Johan Kock's Foundation, King Gustaf V's 80th Birthday Foundation, Lund University Hospital, the Swedish Rheumatism Association, Swedish Society of Medicine, and the Foundation of the National Board of Health and Welfare.
Authorship
Contribution: All authors participated in the design of the study and read and approved the paper; C.L. performed real-time PCR and all flow cytometry, performed the statistical analyses, and wrote the paper; S.A. performed the microarray and gene cluster analysis and real-time PCR and edited the paper; M.A. performed the immune complex detection by enzyme-linked immunosorbent assay and Western blot and edited the paper; B.G., A.J., L.T., G.S., and D.E., edited the paper; and A.A.B. supervised the project and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Lood, Department of Laboratory Medicine, Section of Microbiology, Immunology and Glycobiology, Lund University, Sölvegatan 23, SE-223 62 Lund, Sweden; e-mail: christian.lood@med.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal