Abstract
We have previously shown that the plant-derived compound parthenolide (PTL) can impair the survival and leukemogenic activity of primary human acute myeloid leukemia (AML) stem cells. However, despite the activity of this agent, PTL also induces cellular protective responses that likely function to reduce its overall cytotoxicity. Thus, we sought to identify pharmacologic agents that enhance the antileukemic potential of PTL. Toward this goal, we used the gene expression signature of PTL to identify compounds that inhibit cytoprotective responses by performing chemical genomic screening of the Connectivity Map database. This screen identified compounds acting along the phosphatidylinositol 3-kinase and mammalian target of rapamycin pathways. Compared with single agent treatment, exposure of AML cells to the combination of PTL and phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitors significantly decreased viability of AML cells and reduced tumor burden in vitro and in murine xenotransplantation models. Taken together, our data show that rational drug combinations can be identified using chemical genomic screening strategies and that inhibition of cytoprotective functions can enhance the eradication of primary human AML cells.
Introduction
The biologic heterogeneity within primary human tumors has become an increasingly prevalent consideration for the development of new therapies.1,2 This heterogeneity is evident for acute myeloid leukemia (AML) in which the data suggest that a subpopulation of the bulk AML, the AML stem cells (AML-SCs), are essential for initiating and perpetuating disease as well as resistance to standard chemotherapy, likely providing a residual cell population that promotes relapse.3-7 Toward improving clinical outcomes, we and others have proposed and devised therapies that effectively circumvent the resistance of AML-SCs in preclinical studies.5-8 Indeed, previous studies demonstrated that agents capable of simultaneously inhibiting nuclear factor-κB (NF-κB) and inducing oxidative stress were effective for ablation of AML-SCs. Both of these functions are performed by the single agent, parthenolide (PTL). However, PTL is a suboptimal pharmaceutical, and concentrations of PTL required to ablate AML-SCs are not likely to be achievable in patients.8 This limitation has prompted efforts to discover pharmacologically superior analogs such as dimethyl-amino-parthenolide (DMAPT), which is currently in clinical trials.9 In addition, in silico gene expression–based screens using the PTL transcriptional signature as a template have identified compounds such as celastrol10 and 4-hydroxy-2-nonenal11-15 that demonstrate PTL-like mechanistic and functional properties.16
Here, we hypothesized that chemical genomic approaches can predict compounds that are likely to enhance the efficacy of PTL or DMAPT. To test this hypothesis, we performed chemical genomic screening against the recently expanded Connectivity Map chemogenomic database build02 (CMap2), which now includes 6100 chemical perturbations of cancer cells and represents the most substantial repository of chemogenomic data, allowing for exploration of relationships between compounds on the basis of transcriptional profiles (reviewed in Lamb et al17 ). Several recent studies indicate the ability of chemical genomics to recapitulate and/or modulate chemical and biologic processes that are relevant to therapy through comparison of gene expression patterns.10,16,18-20 Since the PTL transcriptional signature is enriched for cytoprotective pathways that likely decrease the potency of PTL, we used the Connectivity Map database to identify compounds that impair this cytoprotection and enhance the efficacy of PTL. We discovered compounds acting along the PI3K and mTOR pathways.
Methods
Compounds, primary tissue, and cell culture isolation of CD34+ AML total RNA
Primary cryopreserved AML specimens were thawed and cultured in serum-free medium21 for 1 hour followed by treatment with either PTL (BIOMOL), wortmannin or LY-294002 (EMD Biosciences), temsirolimus (LC Laboratories) or DMAPT (prepared as described previously).22 Normal bone marrow from volunteer donors was treated in the same fashion. The TEX cell line was a kind gift from Dr John Dick.23 Studies were performed with approval from the University of Rochester Institutional Review Board, guidelines of the National Institutes of Health, and principles of the Declaration of Helsinki.
Parthenolide gene expression signature
The gene expression signature for CD34+ primary AML upon PTL exposure was described previously.16 Briefly, cells were cultured for 1 hour ex vivo and exposed to PTL for 6 hours. Total RNA for 12 pairs of PTL-treated and untreated primary patient samples was harvested and submitted to the University of Rochester Functional Genomics Core Facility for hybridization on Affymetrix HG-U133 + 2.0 GeneChips. Data were normalized using the robust multi-array average (RMA) procedure.24 Genes were selected using probewise paired t tests. Multiple test correction was performed using the Benjamini-Hochberg procedure. Pathway analysis was performed using Ingenuity Pathway Analysis Software Version 4.0. Data are deposited in the Gene Expression Omnibus, accession number GSE7538.
Gene expression-based chemogenomic screens
The genes with the 75 most extreme test statistics were chosen for the up-regulated and down-regulated genes, yielding a 150-gene signature. Next, we interrogated the Connectivity Map (build02) using the described Connectivity Score statistic, which combines signed Kolmogorov-Smirnov test statistics, and determined specificity using the publicly available Web-based interface available at http://www.broadinstitute.org/cmap.17 Robustness of phosphatidylinositol 3-kinase/mammalian target of rapamycin (PI3K/mTOR) inhibitors as the most extreme class of agent to antagonize the PTL signature was confirmed through repeated interrogation of the CMap2 dataset across a range of false discovery rate cutoffs using R/BioConductor.25 The Connectivity Map dataset is made available by the Broad Institute at http://www.broadinstitute.org/cmap. The genes used to perform the Connectivity Map analysis are included in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometric assays
To determine viability, cells were stained using annexin V–fluorescein isothiocyanate (FITC; BD Biosciences) and 7-aminoactinomycin (7-AAD, Molecular Probes-Invitrogen) to detect phosphatidylserine exposition and cell permeability, respectively. Cells were additionally stained with antibodies against phenotypic markers CD34, CD38, and CD123 (BD Biosciences) to identify phenotypically defined subpopulations. At least 50 000 events were recorded per condition on an LSR II flow cytometer (BD Biosciences). Data analysis was conducted using FlowJo 8.2 software for Mac OS X (TreeStar). Cells that were negative for annexin V and 7-AAD were scored as viable. To determine oxidative state of AML cells, staining was performed with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA), monobromobimane (mBBr), and 7-AAD (Molecular Probes-Invitrogen).
Colony-forming unit and NOD/SCID xenotransplantation assays
MethoCult (StemCell Technologies) was used to assess the colony-forming ability of both AML and normal hematopoietic progenitors, after treatment with the indicated drugs. Growth conditions and scoring were performed as described.26 For xenograft studies, nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice were sublethally irradiated with 2.7 Gy (270 Rads) using a RadSource X-Ray irradiator before transplantation. Equal numbers of primary AML cells (2 million cells per mouse) were then injected via the tail vein in a final volume of 0.2 mL of phosphate-buffered saline (PBS) with 0.5% fetal bovine serum (FBS). Treatment with DMAPT and temsirolimus was started 3 weeks after transplantation. DMAPT was administered via intraperitoneal (IP) injection at a dose of 100 mg/kg 3× daily. Temsirolimus was administered via intraperitoneal injection 3×/wk at 5 mg/kg. The treatment regimen was maintained for 3 weeks. Animals were then killed and the mouse BM analyzed by flow cytometry. Tumor burden was ascertained by determining the percentage of human CD45 cells in the mouse bone marrow (BM). In addition, cells were stained with antibodies specific to human CD34, CD38 and mouse CD45 antibodies (BD Biosciences). For the secondary injections, human CD34+ cells were isolated from the BM of the untreated and the DMAPT + temsirolimus–treated groups using an antibody-coupled magnetic bead separation kit (Miltenyi Biotec) according to the manufacturer's protocol. Equal numbers of human CD34+ cells (800 000 cells per mouse) were injected into sublethally irradiated NOD/SCID mice. Mice were killed 8 weeks after secondary transplantation and the BM analyzed by flow cytometry to ascertain the tumor burden. Peripheral blood counts were determined using a Heska HemaTrue analyzer.
Immunoblots and quantitative real-time polymerase chain reaction
Primary AML cells were treated for 6 hours in serum-free medium with PTL,27 temsirolimus, or both at the indicated doses. At 6 hours after treatment, one fraction of the treated cells was isolated for RNA preparation according to the Trizol protocol or for protein preparation as described previously.28 Protein lysates were subjected to immunoblot analysis with antibodies to phospho-p65 (S536), phospho-p70S6 kinase (T421/S424), phospho-Akt (S473), phospho-4E-BP1 (T37/46), total Akt, total 4E-BP1 (Cell Signaling Technology), total p70S6K (Santa Cruz Biotechnology), heme oxygenase I (HO-1, Stressgen), and actin (Sigma-Aldrich). RNA was submitted for analysis using TaqMan assays per manufacturer's recommended protocols (Applied Biosystems).
Confocal microscopy
Cell fixation was performed using methanol pre-chilled to −20°C. Upon permeabilization with blocking buffer (10% fetal bovine serum and 0.1% Tween 20 in 1× phosphate-buffered saline [pH 7.4]), staining was performed with anti-Nrf2 (C-20) antibody (Santa Cruz Biotechnology) in blocking buffer for 2 hours at room temperature. Cells were then washed and stained with goat-anti–rabbit Alexa488 secondary antibody. Slides were mounted using Fluoromount-G 234 (SouthernBiotech). Fluorescence was observed using a 100× objective (1.4 numeric aperture) on a Leica SP1 inverted scanning confocal microscope. Image acquisition was performed using Leica confocal software (Leica TCS SP5).
Synergy analysis
Assessment of synergy between compounds was performed according to the Chou-Talalay median-effects method29 as implemented in the CalcuSyn 2.0 software package (Biosoft). Isobologram analysis was also perfomed using CalcuSyn. Shifts in LD50 were determined using GraphPad Prism Version 5.0 software (GraphPad Software).
Statistical analysis
Statistical significance for cell viability and colony-forming assays was performed using one-way analysis of variance followed by the Tukey multiple comparison test to determine the significance of particular comparisons, except where otherwise indicated. Two-way analysis of variance was used to determine significance in the NOD/SCID xenotransplant assays followed by Tukey posttests. Analyses and graphs were performed using GraphPad Prism software.
Results
Search for cytoprotective pathways activated when AML cells are challenged with parthenolide or DMAPT
From our analysis of PTL, we noted that many of the molecular responses evoked by drug treatment were likely to represent cellular cytoprotective responses (supplemental Figure 1). Thus, we reasoned that screening for compounds that are known to inhibit cytoprotective pathways and counteract the transcriptional responses to PTL would serve as potential synergizing agents. For example, if the PTL signature were opposite the signature of an mTOR inhibitor, it would suggest that PTL is potentially an mTOR activator. To this end, we screened the 6100 gene expression profiles (“instances”) of drug treatments in the CMap2 database using a 150-gene PTL response signature.16 The resulting compounds were prioritized according to the CMap2 “connectivity score” metrics.30 A positive connectivity score reflects a compound with a gene expression profile most similar to PTL, whereas a negative connectivity score reflects a compound with a gene expression profile most opposite to PTL. From the 6100 drug treatment instances, the instance achieving the greatest positive connectivity score with respect to our PTL response signature derived in primary AML was PTL treatment of HL60 cells, suggesting the accuracy of this screen (not shown). However, among compounds achieving negative connectivity scores, significant enrichment was noted for compounds acting along the PI3K and mTOR pathway (P < 10−5). These compounds included wortmannin, LY-294002, and sirolimus (Figure 1A). Inhibitors of the PI3K/mTOR pathways demonstrated an overall tendency to achieve negative connectivity scores (Figure 1B). To determine whether the finding of PI3K/mTOR inhibitors was not limited to the particular 150-gene PTL response signature used to screen the CMap2 database, we tested a range of PTL gene expression signatures over a range of statistical cutoffs varying from 0.1 to 0.0001. PI3K and mTOR inhibitors were reliable findings, consisting of between 40% and 85% of the top 20 compounds (Figure 1C) resulting from our screen when considering wortmannin, LY-294002, and sirolimus (median percentage = 60%). In contrast, upon querying CMap2 with 100 randomly generated gene signatures, the median and mode for the number of PI3K/mTOR inhibitor hits was zero among the top 20 results (range, 0-3). Thus, the finding of PI3K and mTOR inhibitors was not localized to a specific statistical cutoff, reflecting the increased likelihood that PTL activates the PI3K and mTOR pathways. Moreover, the connection to PI3K and mTOR inhibitors was highly specific, produced by less than 2.1% of queries conducted with 312 gene expression signatures in the Broad Institute molecular signature database (MSigDB). Finally, we determined whether the observation of “negatively connected” PI3K and mTOR inhibitors was a general response to cytotoxic agents. Neither daunorubicin, etoposide, nor paclitaxel demonstrated this effect (supplemental Figure 3).
PI3K/mTOR inhibitor gene expression signature opposes the cytoprotective signature of PTL. (A) Rank and description of PI3K or mTOR inhibitors (wortmannin, LY-294002, and sirolimus [rapamycin]) resulting from interrogation of the CMap2 database. Results are ranked according to extremeness of the negative connectivity score. The connectivity score statistic is indicated in the “Score” column. (B) Bar plot indicating the distribution of PI3K or mTOR inhibitors (wortmannin, LY-294002, or sirolimus) along the 6100 search results ordered according to similarity to PTL. Each horizontal black line indicates an instance of either wortmannin, LY-294002, or sirolimus. The green region indicates instances achieving positive connectivity scores. The gray region indicates instances with no relationship to the PTL response signature. The red region indicates instances of compounds with negative connectivity scores. The gray and green regions indicate instances of the PI3K and mTOR inhibitors class that are either unrelated to PTL or similar to PTL, respectively. The enrichment for the PI3K/mTOR inhibitor compound class is significant (P < 10−5). Results in the red region are most relevant to this screen. (C) The frequency of wortmannin, LY-294002, and sirolimus instances among the top 20 negatively connected hits arising from screens with varying PTL response signatures differing by false discovery rate cutoff. The horizontal axis indicates the false discovery rate (FDR) cutoff; the vertical axis represents the frequency of these instances.
PI3K/mTOR inhibitor gene expression signature opposes the cytoprotective signature of PTL. (A) Rank and description of PI3K or mTOR inhibitors (wortmannin, LY-294002, and sirolimus [rapamycin]) resulting from interrogation of the CMap2 database. Results are ranked according to extremeness of the negative connectivity score. The connectivity score statistic is indicated in the “Score” column. (B) Bar plot indicating the distribution of PI3K or mTOR inhibitors (wortmannin, LY-294002, or sirolimus) along the 6100 search results ordered according to similarity to PTL. Each horizontal black line indicates an instance of either wortmannin, LY-294002, or sirolimus. The green region indicates instances achieving positive connectivity scores. The gray region indicates instances with no relationship to the PTL response signature. The red region indicates instances of compounds with negative connectivity scores. The gray and green regions indicate instances of the PI3K and mTOR inhibitors class that are either unrelated to PTL or similar to PTL, respectively. The enrichment for the PI3K/mTOR inhibitor compound class is significant (P < 10−5). Results in the red region are most relevant to this screen. (C) The frequency of wortmannin, LY-294002, and sirolimus instances among the top 20 negatively connected hits arising from screens with varying PTL response signatures differing by false discovery rate cutoff. The horizontal axis indicates the false discovery rate (FDR) cutoff; the vertical axis represents the frequency of these instances.
Given the typical role of PI3K signaling in growth and survival, and previous reports implicating these pathways in the survival of AML,31,32 we reasoned that activation of this pathway by PTL could represent a protective mechanism evoked by the cellular insult of PTL treatment. If true, then addition of PI3K and/or mTOR pathway inhibitors should enhance the cytotoxicity of PTL.
Activation of the PI3K/mTOR pathway in response to PTL
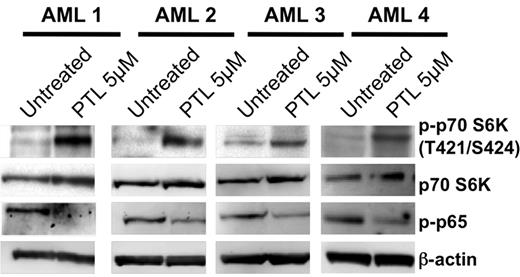
To directly examine whether PTL activated the PI3K pathway, we examined the status of phospho-p70S6K (Thr-421/Ser-424), a direct target of mTOR33 that is phosphorylated as a common response to PI3K and mTOR activation. Treatment with 5μM PTL induced phosphorylation of p70S6K in all patient samples examined (Figure 2), whereas levels of total were p70S6K unaffected. The mTOR target, 4EBP,34 demonstrated substantial baseline phosphorylation (Thr-37/46) that was very modestly to unappreciably increased upon PTL exposure (supplemental Figure 6), possibly reflecting regulatory differences in mTOR substrates. However, the activation of PI3K/mTOR pathway was further supported by the PTL-induced phosphorylation of its immediate downstream target, Akt/PKB (supplemental Figure 5).35 PTL treatment also resulted in the inhibition of NF-κB, as shown by the loss of phosphorylation on the NF-κB p65 subunit (Figure 2). Notably, the activation occurred in connection with the ability of PTL to inhibit NF-κB. Thus, the results indicate activation of the PI3K/mTOR axis and suggest that this activation is connected to PTL activity as evidenced by increased phosphorylation of p70S6K in association with decreased phospho-p65.
Activation of the PI3K/mTOR pathway in response to PTL. Representative immunoblot analyses for 3 primary AML specimens treated with 5μM PTL for 6 hours. Blots were probed as indicated with antibodies against either phospho-p70 S6 kinase (T421/S424), total p70 S6 kinase, phospho-p65 (S536), or β-actin.
Activation of the PI3K/mTOR pathway in response to PTL. Representative immunoblot analyses for 3 primary AML specimens treated with 5μM PTL for 6 hours. Blots were probed as indicated with antibodies against either phospho-p70 S6 kinase (T421/S424), total p70 S6 kinase, phospho-p65 (S536), or β-actin.
PI3K and mTOR inhibitors demonstrate synergism with PTL at the bulk, progenitor, and stem cell level
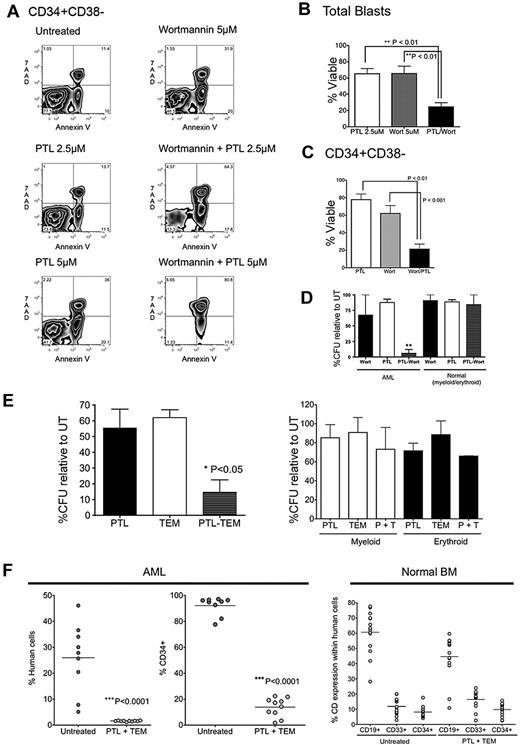
We next determined whether PI3K and mTOR inhibitors could enhance the efficacy of PTL. Primary AML cells were exposed to a subeffective dose of PTL and either the PI3K inhibitor, wortmannin, or the mTOR inhibitor, rapamycin (sirolimus). Cell survival in different AML subpopulations was assessed at 24 hours after treatment. Treatment with PTL alone and wortmannin alone were moderately toxic to phenotypically described AML-SCs, demonstrating a modest effect on overall viability relative to untreated controls (Figure 3A-C). However, the combination of PTL and wortmannin demonstrated greater toxicity against AML-SCs than treatment with either PTL alone (P < .01) or wortmannin alone (P < .001). Next, we tested the effects of the mTOR inhibitor, rapamycin, and the PI3K inhibitor, LY-294002, with similar results (supplemental Figure 4). Combination indices (CIs) to determine synergy for the drug combinations (ie, CI < 1) were calculated according to Chou-Talalay method,29 which indicated that the combinations were synergistic (CI = 0.61 for PTL and wortmannin; CI = 0.33 for PTL and rapamycin).
Treatment with PI3K/mTOR inhibitors synergizes with PTL's antileukemia activity. Primary AML samples were treated for 18 hours with either 5μM wortmannin, 2.5μM PTL, or 5μM PTL, alone or in combination. (A) Representative example of flow cytometric analysis of CD34+CD38− populations (AML6). Dot plots show 7AAD (y-axis) vs annexin V (x-axis). (B) Percentage of viable cells normalized to untreated control for total blasts (n = 5; AML2, AML6, AML8, AML10, AML11) and (C) CD34+CD38− populations (n = 5; AML2, AML6, AML8, AML10, AML11). Error bars represent SEM. (D) AML vs normal cells were treated with 5μM wortmannin alone, 2.5μM PTL alone, or in combination for 18 hours in suspension culture, followed by plating in methylcellulose culture. The percentage of colony-forming units (CFUs) was normalized to untreated control (n = 3; AML4, AML6, AML8). (E) Percentage of colonies relative to untreated control after treatment for 18 hours with either 2.5 μg/mL of temsirolimus (TEM), 5μM PTL, either alone or in combination. Primary AML specimens are shown in the left panel and primary normal marrow specimens are shown in the right panel (n = 3; AML4, AML6, AML8). (F) Percentage of engraftment of AML1 achieved in NOD/SCID mice receiving AML (left panel) or normal BM (right panel) cells after 18 hours of culture with or without 2.5 μg/mL TEM and 5μM PTL. Each symbol represents a single animal analyzed at 6 to 8 weeks after transplantation. Mean engraftment is indicated by the horizontal bars. Normal CD34+ (untreated vs PTL + TEM), P = .280. Normal CD33+ (untreated vs PTL + TEM), P = .084; normal CD19+ (untreated vs PTL + TEM), P = .009. “% human cells” indicates the percentage of total bone marrow cells positive for labeling with hCD45 antibody. “%CD34+” is gated on hCD45+ cells.
Treatment with PI3K/mTOR inhibitors synergizes with PTL's antileukemia activity. Primary AML samples were treated for 18 hours with either 5μM wortmannin, 2.5μM PTL, or 5μM PTL, alone or in combination. (A) Representative example of flow cytometric analysis of CD34+CD38− populations (AML6). Dot plots show 7AAD (y-axis) vs annexin V (x-axis). (B) Percentage of viable cells normalized to untreated control for total blasts (n = 5; AML2, AML6, AML8, AML10, AML11) and (C) CD34+CD38− populations (n = 5; AML2, AML6, AML8, AML10, AML11). Error bars represent SEM. (D) AML vs normal cells were treated with 5μM wortmannin alone, 2.5μM PTL alone, or in combination for 18 hours in suspension culture, followed by plating in methylcellulose culture. The percentage of colony-forming units (CFUs) was normalized to untreated control (n = 3; AML4, AML6, AML8). (E) Percentage of colonies relative to untreated control after treatment for 18 hours with either 2.5 μg/mL of temsirolimus (TEM), 5μM PTL, either alone or in combination. Primary AML specimens are shown in the left panel and primary normal marrow specimens are shown in the right panel (n = 3; AML4, AML6, AML8). (F) Percentage of engraftment of AML1 achieved in NOD/SCID mice receiving AML (left panel) or normal BM (right panel) cells after 18 hours of culture with or without 2.5 μg/mL TEM and 5μM PTL. Each symbol represents a single animal analyzed at 6 to 8 weeks after transplantation. Mean engraftment is indicated by the horizontal bars. Normal CD34+ (untreated vs PTL + TEM), P = .280. Normal CD33+ (untreated vs PTL + TEM), P = .084; normal CD19+ (untreated vs PTL + TEM), P = .009. “% human cells” indicates the percentage of total bone marrow cells positive for labeling with hCD45 antibody. “%CD34+” is gated on hCD45+ cells.
We next determined whether the effects of inhibition of these pathways are discernable in functional assays. To this end, we tested the ability of PTL to functionally impair the formation of AML colonies, but not erythroid and myeloid colonies, alone or in combination with the rapamycin analog, temsirolimus (TEM; Figure 3E left panel). Indeed, PTL and TEM each demonstrated a moderate ability to impair the formation of AML colonies (52% and 62% colonies, respectively, relative to untreated controls). However, PTL + TEM demonstrated a substantial improvement over either PTL alone (P < .05) or TEM alone (P < .05), significantly reducing the formation of AML colonies to 11% relative to untreated controls. Importantly, PTL, TEM, and PTL + TEM did not significantly impair formation of normal erythroid and myeloid colonies (Figure 3E right panel).
To determine the ability of PTL + TEM to impair AML-SC function, primary AML cells or BM cells obtained from healthy donors were treated with the combination of drugs ex vivo for 24 hours and injected into sublethally irradiated NOD/SCID mice. At 6 weeks posttransplantation, BM was harvested and examined for the presence of human cells. We observed that the combination resulted in a significant decrease of AML cells to engraft in NOD/SCID mice (Figure 3F left panel); in contrast, normal BM cell engraftment was only modestly affected (Figure 3F right panel). As with PTL and rapamycin, the effects of PTL and TEM were synergistic (CI = 0.48). Detailed isobologram analysis for the interaction between PTL and TEM is shown in supplemental Figure 7.
Inhibition of PI3K/mTOR impairs antioxidant responses to PTL
Pathway analysis of the PTL response signature indicated involvement of the NF-E2–related factor 2 (Nrf2) pathway (supplemental Figures 1-2), a transcription factor involved in activating the expression of detoxifying and antioxidant enzymes.36 Nrf2 is reportedly translocated to nucleus via PI3K activation.37 Previous studies indicate that the oxidizing ability of PTL is essential for its ability to ablate AML-SCs.26 Accordingly, exposure of CD34+ AML cells to PTL for 6 hours resulted in stabilization of nuclear-localized Nrf2 and expression of Nrf2 target genes.16 Given the role of Nrf2 in promoting the transcription of cytoprotective genes, including antioxidant genes, we examined whether PI3K and mTOR inhibitors could prevent activation of Nrf2 and/or its downstream targets.
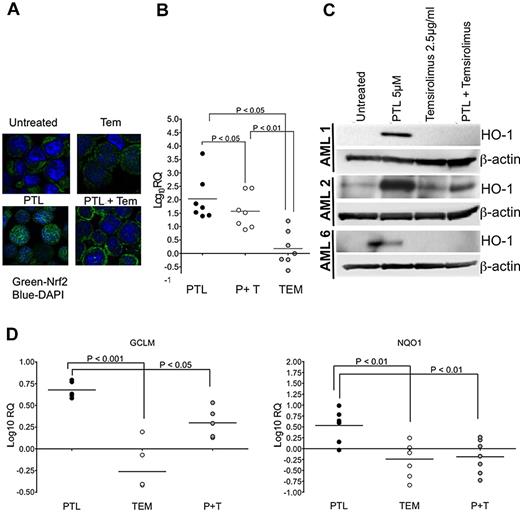
To test whether PTL-mediated translocation of Nrf2 to the nucleus involved the PI3K/mTOR pathway, we ascertained the localization of Nrf2 in AML cells. PTL exposure for 6 hours resulted in the nuclear accumulation of Nrf2 (Figure 4A bottom left) relative to untreated controls (Figure 4A top left). TEM alone resulted in no change in the cellular distribution of Nrf2 (Figure 4A top right) relative to untreated control. In contrast to PTL, exposure of cells to the combination of PTL and TEM prevented accumulation of Nrf2 in the nucleus, where its distribution is apparently cytoplasmic (Figure 4A bottom right). Because Nrf2 demonstrates a cytoplasmic distribution with the combination of PTL + TEM, but a nuclear distribution with PTL alone, we expected that the antioxidant responses controlled by Nrf2 in response to PTL would diminish with the combination of PTL and TEM. To test this prediction, we determined the effect of the interaction between PTL and TEM on a well-known target of Nrf2, heme oxygenase 1 (HO-1),38 at the transcriptional and translational level at 6 hours posttreatment. Using quantitative real-time PCR (qPCR) on primary CD34+ AML cells, we observed that PTL + TEM generally impaired the ability of PTL to induce HO-1 expression in a significant manner (P < .05; Figure 4B). The effects of TEM in terms of suppression of HO-1 expression were also apparent at the translational level as determined by immunoblot analysis of total HO-1 protein in primary CD34+ AML specimens (Figure 4C). To further confirm the suppressed activity of Nrf2, we tested additional Nrf2 targets using qPCR, including glutamate-cysteine ligase regulatory subunit (GCLM)39 and NAD(P)H:quinone oxidoreductase (NQO1).40 Indeed, the PTL-mediated induction of GCLM (Figure 4D left panel) was significantly suppressed with the PTL + TEM combination (P < .001). Similar effects were observed with NQO1 (P < .01). Based on these findings, we next sought to determine whether the observable impairment of antioxidant gene expression would demonstrate the expected functional consequence of increased reactive oxygen species (ROS).
Molecular response to PTL and temsirolimus. AML samples were treated with parthenolide (PTL), alone or in combination with 2.5 μg/mL of temsirolimus (TEM). (A) Confocal micrograph for TEX cells after treatment for 6 hours with either 2.5 μg/mL TEM, 5μM PTL, either alone or in combination. Cells were stained with anti-Nrf2 antibody (green) and nuclear dye DAPI (4,6 diamidino-2-phenylindole, blue). (B) qPCR analyses of heme oxygenase I in CD34+ AML specimens after 6 hours of treatment with PTL, TEM, or PTL + TEM (P + T). Data are shown relative to untreated controls for AML1-AML7. Fold changes are represented on a log-10 scale (log10RQ). (C) Immunoblot analyses for the indicated treatments at the 6-hour timepoint. Blots were probed with antibodies against either heme oxygenase 1 (HO-1) or β-actin. (D) qPCR analyses of GCLM (left panel; AML1, AML2, AML4, AML5, AML6) and NQO1 (right panel; AML1-AML7) in CD34+ AML specimens after 6 hours of treatment with PTL, TEM, or PTL + TEM (P + T). Data are shown relative to untreated controls. Fold changes are represented on a log-10 scale (log10RQ).
Molecular response to PTL and temsirolimus. AML samples were treated with parthenolide (PTL), alone or in combination with 2.5 μg/mL of temsirolimus (TEM). (A) Confocal micrograph for TEX cells after treatment for 6 hours with either 2.5 μg/mL TEM, 5μM PTL, either alone or in combination. Cells were stained with anti-Nrf2 antibody (green) and nuclear dye DAPI (4,6 diamidino-2-phenylindole, blue). (B) qPCR analyses of heme oxygenase I in CD34+ AML specimens after 6 hours of treatment with PTL, TEM, or PTL + TEM (P + T). Data are shown relative to untreated controls for AML1-AML7. Fold changes are represented on a log-10 scale (log10RQ). (C) Immunoblot analyses for the indicated treatments at the 6-hour timepoint. Blots were probed with antibodies against either heme oxygenase 1 (HO-1) or β-actin. (D) qPCR analyses of GCLM (left panel; AML1, AML2, AML4, AML5, AML6) and NQO1 (right panel; AML1-AML7) in CD34+ AML specimens after 6 hours of treatment with PTL, TEM, or PTL + TEM (P + T). Data are shown relative to untreated controls. Fold changes are represented on a log-10 scale (log10RQ).
Inhibition of PI3K/mTOR increases oxidative stress induced by PTL
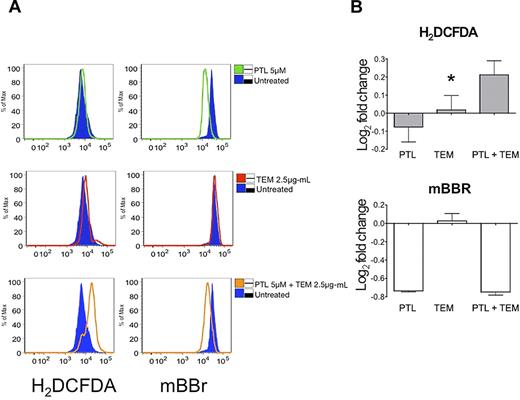
Induction of oxidative stress (along with NF-κB inhibition) has previously been described as one of the hallmarks of agents that selectively impair primary AML cells.26 Thus, we investigated whether the combination of TEM with PTL acted to increase overall oxidative burden. As shown in Figure 5, PTL alone is a potent free thiol scavenger as indicated by loss of fluorescence labeling with the thiol reactive probe mBBr (Figure 5A top left panel; Figure 5B bottom panel). This activity of PTL has previously been described as a primary mechanism by which the compound induces oxidative stress.26,41 Interestingly, at sublethal doses of PTL used in this study, there is not an overt increase in cellular reactive oxygen species (ROS), as detected by the well-characterized ROS probe, H2DCF-DA (Figure 5A top left panel; Figure 5B top panel). Moreover, TEM alone fails to appreciably modulate either free thiol species or ROS (Figure 5A middle panels; Figure 5B). In contrast, the combination of PTL + TEM induces both a loss of free thiols and an increase in ROS, indicative of a greater overall oxidative insult with the combined regimen (Figure 5A, bottom panels; Figure 5B). Thus, TEM blocks the PTL-mediated activation of Nrf2 and its antioxidant target genes, resulting in increased oxidative stress.
The combination of PTL and TEM increases the overall oxidative insult to AML cells. Cells were treated with PTL, TEM, or PTL + TEM at the concentrations indicated alongside untreated controls. (A) Histogram overlay indicating relative oxidative changes in AML1 as measured by H2DCF-DA (left column) and mBBr (right column). Untreated controls are indicated by the shaded blue histogram. Drug treatments are indicated by colored open histograms top row: untreated vs PTL, Middle row: untreated vs TEM, Bottom row: untreated vs PTL + TEM (B) Bar chart indicates the base-2 log fold change of mean fluorescence intensity measured by H2DCF-DA (top left panel) or mBBr (top right panel). *P < .05 (n = 3; AML1).
The combination of PTL and TEM increases the overall oxidative insult to AML cells. Cells were treated with PTL, TEM, or PTL + TEM at the concentrations indicated alongside untreated controls. (A) Histogram overlay indicating relative oxidative changes in AML1 as measured by H2DCF-DA (left column) and mBBr (right column). Untreated controls are indicated by the shaded blue histogram. Drug treatments are indicated by colored open histograms top row: untreated vs PTL, Middle row: untreated vs TEM, Bottom row: untreated vs PTL + TEM (B) Bar chart indicates the base-2 log fold change of mean fluorescence intensity measured by H2DCF-DA (top left panel) or mBBr (top right panel). *P < .05 (n = 3; AML1).
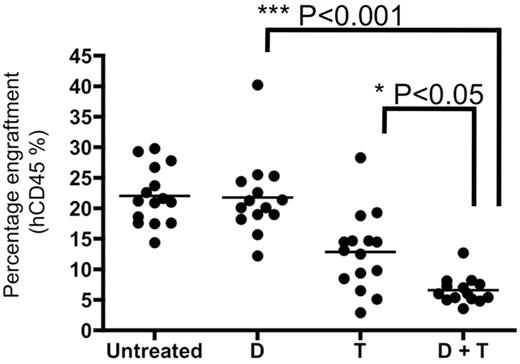
The combination of DMAPT and temsirolimus impairs engraftment of AML-SCs in NOD/SCID mice and reduces tumor burden in established AML xenotransplants
Based on the biologic activity of PTL in combination with PI3K and mTOR inhibition, we sought to determine the efficacy of this strategy for reducing tumor burden in established human-mouse xenografts using clinical agents. The effects of the interaction between TEM and DMAPT, the clinical analog of PTL,22,42 were tested at doses that are only minimally active for each drug when used as single agents.9,43 NOD/SCID mice were sublethally irradiated and injected with 2 million AML cells per animal. At 2-3 weeks after engraftment, we initiated a treatment schedule consisting of DMAPT, TEM, or DMAPT + TEM for 3 weeks, after which tumor burden as a function of percent bone marrow engraftment was determined (Figure 6). DMAPT at the doses used had no significant effect on tumor burden relative to untreated animals, and TEM was able to produce a modest effect on tumor burden (∼ 50% of untreated). However, DMAPT + TEM decreased tumor burden to ∼ 25% of untreated controls and DMAPT alone (P < .05). When the remaining tumor was injected into sublethally irradiated secondary recipient NOD/SCID mice, we observed substantial impairment of tumor growth (supplemental Figure 8). Finally, we also examined nonspecific toxicity of the DMAPT and TEM drug combination in naive NOD/SCID mice. The data indicate no significant impairment of normal hematopoiesis, and no observable changes in weight or general health (supplemental Figure 9).
In vivo treatment with PTL and TEM combinations results in decrease of tumor burden. Primary AML cells (AML1) were injected into sublethally irradiated mice. Three weeks after transplant, mice were either left untreated or treated with DMAPT (D) or TEM (T), alone or in combination for 3 weeks. DMAPT was administered at a dose of 100 mg/kg 3× daily. TEM was administered 3 times a week at 5 mg/kg. Mice were euthanatized and bone marrow was analyzed for percentage of human cells using anti-human CD45 antibody.
In vivo treatment with PTL and TEM combinations results in decrease of tumor burden. Primary AML cells (AML1) were injected into sublethally irradiated mice. Three weeks after transplant, mice were either left untreated or treated with DMAPT (D) or TEM (T), alone or in combination for 3 weeks. DMAPT was administered at a dose of 100 mg/kg 3× daily. TEM was administered 3 times a week at 5 mg/kg. Mice were euthanatized and bone marrow was analyzed for percentage of human cells using anti-human CD45 antibody.
Discussion
In the present study we describe a chemogenomic strategy that rapidly selected agents that enhance the cytotoxicity of PTL. This effort successfully identified PI3K/mTOR-pathway inhibitors as compounds that effectively synergize with PTL to induce selective eradication of primary human AML cells, including AML-SCs. Furthermore, the effects against bulk, stem, and progenitor AML cells were evident both in vitro with patient specimens and in vivo using xenotransplantation models of AML.
Based on these findings, we propose that chemogenomic approaches are applicable to the identification of unknown synergy between 2 or more candidate agents. The results described herein underscore the capabilities of chemogenomic profiling efforts such as the CMap, where the overall pattern of transcriptional changes are ascertained for a collection of compounds.17,30 Unlike simple live/dead screens, the transcriptional data associated with compounds are reusable for the discovery of novel relationships between compounds. As proteomic and epigenomic data become available in the chemical space, the precision with which drug discovery can be performed in this manner is likely to improve dramatically.
It is notable that the relationship between PTL and PI3K/mTOR inhibitors discovered by the CMap was anticorrelative, that is, PI3K and mTOR inhibitors tended to exert opposite effects on gene expression changes associated with PTL exposure. A reasonable and intuitive initial interpretation of this result is that PI3K/mTOR inhibitors should antagonize the effects of PTL. However, our study demonstrates synergism between PTL and PI3K/mTOR inhibitors. Synergism is likely observed in this particular case because the PTL signature at the time point we selected largely represents genes that serve cytoprotective functions that diminish PTL function. Indeed, “negatively connected” drugs such as mTOR inhibitors suppressed cytoprotection as suggested by the ability of TEM to impair Nrf2 function upon PTL challenge. We propose that the ability to predict synergy/antagonism with the CMap is strongly dependent on an understanding of the function of the input genes.
In addition to yielding a promising compound combination, the CMap2-based screen provided surprising insight into the biology of AML at the bulk, stem, and progenitor level. Indeed, the primary target for eradicating AML-SCs in many studies has been NF-κB. This study indicates that exposure of primary AML cells to PTL up-regulates PI3K/mTOR signaling. Although the underlying mechanism of this induction remains to be elucidated, we propose that it may represent a secondary protective response that is activated upon loss of the survival benefit conferred by NF-κB activity. Thus, going forward, detection of secondary survival responses associated with different therapeutics may enable both improved drug efficacy and greater success in translating compounds to clinic.
Another notable aspect of this work is the results obtained using TEM in combination with DMAPT for treatment of established xenografts (Figure 6). Treatment of animals with DMAPT alone did not appreciably affect tumor burden at the dose and schedule used for this study. Nonetheless, DMAPT was able to synergize with TEM and cause a significant decrease in tumor burden with the DMAPT + TEM combination. This observation is relevant to the drug development process because it suggests that compounds failing to demonstrate overt antitumor effects as single agents may still exert effects that are of significant value when combined with other agents. Thus, we suggest that comprehensive transcriptional profiling of patients undergoing treatment with experimental agents may be critical in predicting the overall value of a candidate agent. Indeed, experimental agents that fail to elicit an overt antitumor response may still be modulating pathways highly relevant to therapy. In the absence of objective tumor responses, such activities will likely be missed unless some form of comprehensive analysis such as gene expression profiling is used during the early phases of testing new drugs.
We also observed that another component of the cytoprotective response associated with PTL is Nrf2 activation. Inhibition of either PI3K or mTOR abrogated the ability of PTL to cause nuclear accumulation of Nrf2. Accordingly, failure to activate Nrf2 further resulted in prevention or impairment of the activation of Nrf2-regulated genes. Previous reports have indicated the ability of PI3K to directly activate Nrf2 (supplemental Figure 2).37 However, in addition to PI3K, we note here that mTOR activity appears to be an additional requirement for Nrf2 activation in primary AML specimens, a mechanism for pursuit in future studies.
Taken together, our findings demonstrate that chemogenomic strategies are not only relevant to the identification of cancer drugs, but also that such methods may be of significant benefit in designing potential multiagent therapeutic regimens. Moreover, by focusing on properties conserved in all AML cell populations (ie, stem, progenitor, and blast), we propose that regimens most likely to maximize eradication of intrinsically heterogeneous tumor populations can be developed using high-dimensional drug response data such as that available at CMap2.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work is dedicated in memory of Mr Arthur Kroll, whose generous support and passion for research has been instrumental in developing new approaches to leukemia therapy. We also gratefully acknowledge Martin Carroll, Ari Melnick, and William Matthews for critical reading of the manuscript.
This work was supported by grants from the Leukemia & Lymphoma Society (LLS TRP 6099-06), the New York State Stem Cell Foundation (NYSTEM C024964), and the Department of Defense (W81XWH-07-1-0601). D.C.H. is supported in part by the Raymond and Beverly Sackler Center for Biomedical and Biophysical Sciences.
Authorship
Contribution: D.C.H. designed research, performed experiments, developed methods, wrote software, analyzed data, and wrote the paper; S.S. designed research, performed experiments, and wrote the paper; M.M., R.M.R., M.B., C.A.C. and L.W. performed experiments; P.A.C. performed experiments and analyzed data; M.L.G. designed research, performed experiments, analyzed data, and wrote the paper; and C.T.J. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: C.T.J. and P.A.C. have a financial interest in Leuchemix Inc. The remaining authors declare no competing financial interests.
Correspondence: Duane C. Hassane, Department of Pathology and Laboratory Medicine, Institute for Computational Biomedicine, Weill Cornell Medical College, 1305 York Ave, Box 140, New York, NY 10021; e-mail: dhassane@med.cornell.edu.
References
Author notes
D.C.H. and S.S. contributed equally to this work.
M.L.G. and C.T.J. contributed equally to this work.

![Figure 1. PI3K/mTOR inhibitor gene expression signature opposes the cytoprotective signature of PTL. (A) Rank and description of PI3K or mTOR inhibitors (wortmannin, LY-294002, and sirolimus [rapamycin]) resulting from interrogation of the CMap2 database. Results are ranked according to extremeness of the negative connectivity score. The connectivity score statistic is indicated in the “Score” column. (B) Bar plot indicating the distribution of PI3K or mTOR inhibitors (wortmannin, LY-294002, or sirolimus) along the 6100 search results ordered according to similarity to PTL. Each horizontal black line indicates an instance of either wortmannin, LY-294002, or sirolimus. The green region indicates instances achieving positive connectivity scores. The gray region indicates instances with no relationship to the PTL response signature. The red region indicates instances of compounds with negative connectivity scores. The gray and green regions indicate instances of the PI3K and mTOR inhibitors class that are either unrelated to PTL or similar to PTL, respectively. The enrichment for the PI3K/mTOR inhibitor compound class is significant (P < 10−5). Results in the red region are most relevant to this screen. (C) The frequency of wortmannin, LY-294002, and sirolimus instances among the top 20 negatively connected hits arising from screens with varying PTL response signatures differing by false discovery rate cutoff. The horizontal axis indicates the false discovery rate (FDR) cutoff; the vertical axis represents the frequency of these instances.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/26/10.1182_blood-2010-04-278044/5/m_zh89991063100001.jpeg?Expires=1769216117&Signature=cxpQ694Xf5YvmYVTW9g0MT2NKOtAHA76l~CDR4pom3xWA5Gzhizf8Kj5a1lY1WkNqTvh2jlLhS5-FF63TpGm4sFtPWJBHzOcNueFH9Z~Cnc4t9xOZ~0JMBgXnf28tI121GAFxMF9LjH0G3ujd0FxmmVT3oDFPTMky4x4GFWaCTqw6BiIFGMdCEdvyDngyPFvbsUIUM-uqBbvvZp-cxekNbMXtWvy8CVKacIMR5zEQuSRlUAFqwUBtRG43axn2IbGb9MnsAO3-nEbVGO5vwnE3VAuVz4sk~uTDh~mKSK3r41zerKAEO~16CmJgU3ORDqeNsrZJsceeTXQzX5PL3FQzw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal