Abstract
Trib1, Trib2, and Trib3 are mammalian homologs of Tribbles, an evolutionarily conserved Drosophila protein family that mediates protein degradation. Tribbles proteins function as adapters to recruit E3 ubiquitin ligases and enhance ubiquitylation of the target protein to promote its degradation. Increased Trib1 and Trib2 mRNA expression occurs in human myeloid leukemia and induces acute myeloid leukemia in mice, whereas Trib3 has not been associated with leukemia. Given the high degree of structural conservation among Tribbles family members, we directly compared the 3 mammalian Tribbles in hematopoietic cells by reconstituting mice with hematopoietic stem cells retrovirally expressing these proteins. All mice receiving Trib1 or Trib2 transduced hematopoietic stem cells developed acute myeloid leukemia, whereas Trib3 mice did not. Our previous data indicated that Trib2-mediated degradation of the transcription factor, CCAAT/enhancer-binding protein-alpha (C/EBPα), is important for leukemogenesis. Similar to Trib2, Trib1 induced C/EBPα degradation and inhibited its function. In contrast, Trib3 failed to inactivate or promote efficient degradation of C/EBPα. These data reveal that the 3 Tribbles homologs differ in their ability to promote degradation of C/EBPα, which account for their differential ability to induce leukemia.
Introduction
Tribbles encode an evolutionarily conserved protein family that influences proliferation, motility, metabolism, and oncogenic transformation. Originally identified in Drosophila, Tribbles homologs are characterized by a central serine/threonine kinase-like domain (KD) and a C-terminal binding site for E3 ubiquitin ligases. Although the KD is the defining feature of the Tribbles family, these proteins are considered to be catalytically inactive because they lack conserved residues from the characteristic adenosine triphosphate binding site and catalytic core motif within the KD.1-4 Accordingly, Tribbles probably function as adapter or scaffold proteins. Although the 3 mammalian Tribbles proteins (Trib1, Trib2, and Trib3, collectively referred to as “Trib1-3”) are highly homologous in the KD and the C-terminal E3 ubiquitin ligase binding site, they show limited similarity in the N- and C-terminal domains (supplemental Figure 1; available on the Blood Web site; see the Supplemental Materials link at the top of the online article). It is not known whether the expansion of the Tribbles family from 1 to 3 members in higher eukaryotes has resulted in functional differences among the homologs or enables differential tissue expression. In support of the latter, Trib1 demonstrates the highest level of expression in bone marrow (BM), peripheral blood leukocytes (PB), thyroid gland, and pancreas; Trib2 is highly expressed in human thymus and PB; whereas Trib3 is most highly expressed in human liver, with lower expression in hematopoietic compartments, such as BM, PB, spleen, and thymus.5,6
In several contexts, Tribbles proteins facilitate ubiquitin-dependent degradation of their target proteins. Drosophila tribbles mediates degradation of the cdc25 homolog string, resulting in a protracted G2/M transition during gastrulation and morphogenesis.1,2,4 In addition, Drosophila tribbles regulates oogenesis by inducing ubiquitylation and proteasomal degradation of slbo, the Drosophila homolog of CCAAT/enhancer-binding protein (C/EBP) transcription factors.3 Similar to Drosophila tribbles, mammalian Tribbles family members also stimulate protein degradation. Trib3 stimulates degradation of acetyl CoA carboxylase (ACC), an enzyme important in fatty acid synthesis, by recruiting the E3 ubiquitin ligase, COP1.7 The presence of Trib3 enhances the ability of COP1 to bind and ubiquitylate ACC, ultimately leading to ACC degradation. Trib2 promotes degradation of the transcription factors C/EBPα and C/EBPβ.8,9 In addition to their roles in protein degradation, Tribbles may exert additional functions; for example, Trib3 binds and inhibits AKT, whereas Trib1 and Trib3 modulate mitogen-activated protein kinase (MAPK) activity.10-17
Recent studies demonstrate that high levels of Trib1 and Trib2 expression are associated with human hematopoietic malignancies. Trib1 is elevated in acute myeloid leukemia (AML) and myelodysplastic syndrome patient samples with gene amplifications, and Trib2 is elevated in a subset of AML patient samples.8,18-20 These findings are supported by murine experiments in which retroviral expression of either Trib1 or Trib2 in BM progenitors caused AML.8,21 For Trib2, degradation of C/EBPα appears to play an important role in the pathogenesis of murine AML.8 Significantly, Trib2-associated human AMLs were found in a cohort of AMLs characterized by decreased C/EBPα function. It is not known whether Trib1-induced AML uses a similar mechanism.
Because Trib1, Trib2, and Trib3 share greater than 45% sequence homology, we investigated the ability of all 3 mammalian family members to induce AML in mice and to promote degradation of C/EBPα. Here we show that both Trib1 and Trib2, but not Trib3, induce AML and promote efficient degradation of C/EBPα. The tight correlation between Trib-mediated degradation of C/EBPα and the ability of the different family members to induce leukemia in mice indicates that the leukemogenic potential of Tribbles proteins is dependent on their ability to efficiently degrade C/EBPα. Furthermore, these data suggest that therapies that inhibit C/EBPα degradation may be effective in Tribbles-induced AML.
Methods
Constructs and retroviruses
All Trib2-related constructs were cloned as described previously.8 Murine Trib1 and Trib3 cDNAs were flag-tagged or myc-tagged and subcloned into pcDNA3.1, MigR1, or MSCV-IRES-NGFR vectors. Swap domain mutants were cloned into a version of the MigR1 retroviral vector that contains a myc-tag using polymerase chain reaction-driven overlap extension.22 The integrity of Trib1-3 and the swap mutants were confirmed by sequencing. Retroviral supernatants were generated by transient transfection of 293T cells and titered with 3T3 cells as previously described.23
BM transplantation
BM transplantations were performed as described previously.8,24 Briefly, BM cells were harvested from 8- to 10-week-old C57BL/6 mice (Taconic Farms) 5 days after intravenous 5-fluorouracil administration. BM cells were retrovirally transduced by spinoculation with retroviral supernatants in the presence of interleukin-3 (IL-3), IL-6, stem cell factor, and WEHI-conditioned medium. Equivalent retroviral titers were used for Trib1-3 and MigR1 retroviral supernatants as described.24,25 Cells (0.5 × 106) were injected intravenously into lethally irradiated B6 recipients. Mice were followed with bimonthly tail vein bleeds. Mice with leukemia were killed after development of severe cachexia and/or lethargy or blasts in the PB. All experiments were performed in accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals26 under an animal protocol approved by the University of Pennsylvania Animal Care and Use Committee.
Methycellulose culture
BM cells were enriched for progenitors and transduced as described for BM transplantations. On day 3 after harvest, retrovirally transduced cells were plated in triplicate in methylcellulose media (Methocult M3231; StemCell Technologies) supplemented with cytokines (10 ng/mL granulocyte-macrophage colony-stimulating factor [GM-CSF], 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL stem cell factor, PeproTech and BD Biosciences) at 1 × 104 cells per plate. Cells from primary plates (colonies scored at 7-9 days) were transferred to secondary (colonies scored at 7 days) and tertiary (colonies scored at 7 days) plates containing the indicated cytokines.
Quantitative transcript analysis
RNA was purified using the QIAGEN RNeasy Mini Kit. cDNA was synthesized with the Superscript II kit (Invitrogen). Real-time quantitative reverse-transcribed polymerase chain reaction was carried out on an ABI 7900. Transcripts were amplified with Sybr Green PCR Master Mix (ABI). Primers included the following: Trib1 forward 5′-AATCGCCGACTACCTGCTGCTAC-3′, Trib1 reverse 5′-GACGGCGGAAACAATCTGCTTGAA-3′, Trib2 forward, 5′-AGCCCGACTGTTCTACCAGA-3′, Trib2 reverse, 5′-AGCGTCTTCCAAACTCTCCA-3′, Trib3 forward 5′-CAC CTC CCG CCT CAG ACT T-3′, and Trib3 reverse 5′-GTA GGC AGC CGG GCA TAA-3′.
Cell lines and differentiation assay
The 32D cells were maintained in Iscove modified Dulbecco medium (Invitrogen), with 10% fetal calf serum, 10% WEHI-3B supernatant, 100 U/mL penicillin, 100 μg/mL streptomycin, and 12μM l-glutamine. At 48 hours after transduction, 32D cells (5 × 104) were plated in 10 ng/mL IL-3 (PeproTech) or 25 ng/mL granulocyte colony-stimulating factor (G-CSF) (PeproTech) and assessed for granulocytic differentiation by fluorescence-activated cell sorter analysis (CD11b, eBioscience) 2 to 5 days after induction of differentiation with G-CSF.
The 293T cells and HeLa cells were maintained in Dulbecco modified Eagle medium with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 12μM l-glutamine. The 293T or HeLa cells were transfected with 6 to 12 μg of indicated plasmid or pcDNA3 control plasmid using calcium chloride. Cells were harvested for protein expression 48 hours after transfection.
Western blotting
To detect C/EBPα, cells were treated with or without 10μM MG132 (Calbiochem) for 1 hour (32D cells) or 2 hours (HeLa cells), washed in phosphate-buffered saline, lysed directly in 2× sodium dodecyl sulfate sample buffer, and detected with anti-C/EBPα (sc-61, Santa Cruz Biotechnology). Whole cell lysates were prepared with modified Radio Immunoprecipitation Assay (RIPA) buffer (1% NP-40, 50mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 150mM NaCl, 1mM ethylenediaminetetraacetic acid, 1mM phenylmethylsulfonyl fluoride, 1mM NaF, 1mM Na3VO4 supplemented with protease inhibitors; Sigma-Aldrich). Antibodies used were anti-HA (12CA5, Abcam), anti-flag (M2; Sigma-Aldrich), anti-ACC (rabbit polyclonal; Cell Signaling Technology), and anti-myc (9E10).
Results
Trib1 and Trib2, but not Trib3, induce AML in mice
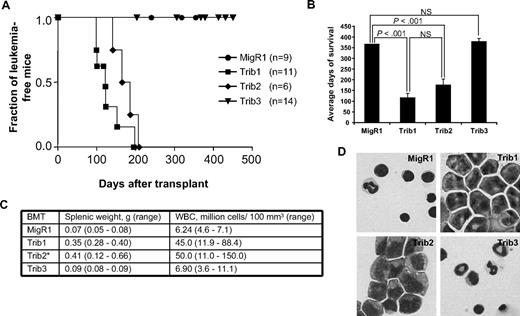
Mammalian Trib1, Trib2, and Trib3 demonstrate considerable sequence similarity, particularly in the KD and the C-terminal binding site for the E3 ubiquitin ligase COP1 (supplemental Figure 1). To gain insight into the functional significance of this homology, we compared the propensity of Trib1, Trib2, and Trib3 to induce leukemia in the murine BM transplantation model. After verifying that proteins of the expected molecular weight were expressed at similar levels (supplemental Figure 2A), we used normalized retroviral supernatants expressing Trib1, Trib2, or Trib3 to transduce 5-fluorouracil-treated BM progenitors that were subsequently transferred to lethally irradiated syngeneic C57BL/6 mice. Consistent with previous data, 100% of Trib1 and Trib2 mice developed AML (Figure 1A).8,21 Although Trib1 mice developed leukemia with slightly faster kinetics than Trib2 mice, this difference was not statistically significant (116 ± 28 days after transplantation for Trib1 mice and 174 ± 21 days after transplantation for Trib2 mice, Figure 1A-B). Trib1 and Trib2 mice exhibited splenomegaly, PB leukocytosis, and increased blasts in the BM (Figure 1C-D). Flow cytometric analyses revealed that Trib1 leukemic cells were largely green fluorescent protein–positive (GFP+) and expressed intermediate levels of the myeloid markers CD11b and Gr1, as well as F4/80 and c-kit (supplemental Figure 2), a phenotype similar to Trib2-induced AML.8 Both Trib1- and Trib2-induced AML could be serially transplanted to sublethally irradiated syngeneic recipient mice (data not shown). Trib3 mice, like MigR1 controls, did not develop leukemia or significant pathology (Figure 1) despite maintaining GFP+ cells and harboring an intact provirus (supplemental Figure 3). Thus, despite significant structural similarity to Trib1 and Trib2, Trib3 did not induce AML in mice receiving BM transplantations.
Trib1 and Trib2, but not Trib3, induce AML. (A) Kaplan-Meier survival curve of mice receiving BM transplant with Trib1 (n = 11), Trib2 (n = 6), Trib3 (n = 14), or control MigR1 (n = 9) transduced cells. Results are derived from 2 independent experiments. (B) Average survival for indicated BM transplantation. P values determined by Student t test are indicated. NS indicates no significant difference. The difference between Trib1 and Trib2 yielded P > .1 (Student t test). (C) Average splenic weight and white blood cell count of MigR1, Trib1, Trib2, and Trib3 mice. *The splenic weights and white blood cell counts for the Trib2 mice are published.8 (D) Wright-Giemsa stain of BM cytospin from mice receiving the indicated BM transplantation.
Trib1 and Trib2, but not Trib3, induce AML. (A) Kaplan-Meier survival curve of mice receiving BM transplant with Trib1 (n = 11), Trib2 (n = 6), Trib3 (n = 14), or control MigR1 (n = 9) transduced cells. Results are derived from 2 independent experiments. (B) Average survival for indicated BM transplantation. P values determined by Student t test are indicated. NS indicates no significant difference. The difference between Trib1 and Trib2 yielded P > .1 (Student t test). (C) Average splenic weight and white blood cell count of MigR1, Trib1, Trib2, and Trib3 mice. *The splenic weights and white blood cell counts for the Trib2 mice are published.8 (D) Wright-Giemsa stain of BM cytospin from mice receiving the indicated BM transplantation.
Trib1 and Trib2, but not Trib3, convey serial plating potential
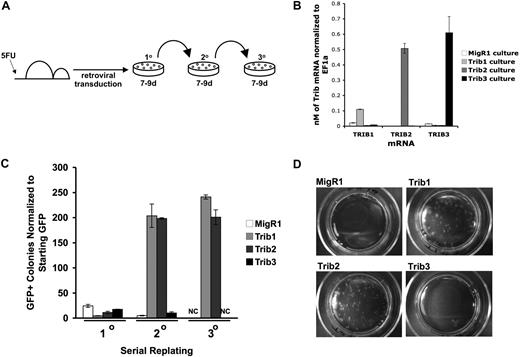
We next sought to compare the ability of Trib1, Trib2, and Trib3 to promote self-renewal, which might require a lower threshold activity than leukemic transformation in vivo. To assay the in vitro self-renewal potential of cells expressing Tribbles homologs, we transduced BM progenitors and serially plated the cells in methylcellulose (Figure 2A). After the first plating, mRNA expression of Trib3 in Trib3-transduced cells was higher than Trib1 mRNA and comparable with Trib2 mRNA in the respectively transduced cells (Figure 2B). Similar to the vector control MigR1, Trib1-, Trib2-, and Trib3-transduced BM induced colony growth in the initial plating; however, only Trib1- and Trib2-transduced BM cells gave rise to an increased number of colonies in the secondary plating (Figure 2C). By the tertiary plating, only Trib1- and Trib2-transduced BM cells continued to support colony growth (Figure 2C-D). These data indicate that Trib1 and Trib2, but not Trib3, are capable of promoting self-renewal in hematopoietic progenitors. The inability of Trib3 to serially plate was not the result of a lack of expression as Western blotting revealed that the flag-tagged Trib3 protein was expressed in these cells (data not shown).
Trib1 and Trib2, but not Trib3, confer serial plating activity in methylcellulose cultures. (A) Serial colony assays were performed as shown using primary BM cells transduced with retroviruses. (B) Primary methylcellulose cultures were sorted for GFP and used for RNA isolation. Absolute quantification of Trib1, Trib2, or Trib3 mRNA normalized to EF1α from sorted BM cells after 8 days of culture in methylcellulose. Values are mean (± SD) for triplicate samples. Data are representative of 2 independent experiments. (C) The colony numbers were counted during 3 rounds of serial culture and normalized to starting GFP. Values are mean (± SD) for triplicate samples. NC indicates no colonies. Data are representative of 3 independent experiments. (D) Plates from the third serial culture are shown.
Trib1 and Trib2, but not Trib3, confer serial plating activity in methylcellulose cultures. (A) Serial colony assays were performed as shown using primary BM cells transduced with retroviruses. (B) Primary methylcellulose cultures were sorted for GFP and used for RNA isolation. Absolute quantification of Trib1, Trib2, or Trib3 mRNA normalized to EF1α from sorted BM cells after 8 days of culture in methylcellulose. Values are mean (± SD) for triplicate samples. Data are representative of 2 independent experiments. (C) The colony numbers were counted during 3 rounds of serial culture and normalized to starting GFP. Values are mean (± SD) for triplicate samples. NC indicates no colonies. Data are representative of 3 independent experiments. (D) Plates from the third serial culture are shown.
Trib1 and Trib2, but not Trib3, induce efficient degradation of C/EBPα
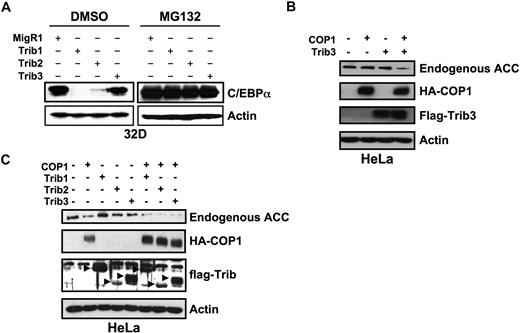
To investigate the mechanistic basis for the differential ability of Trib1-3 to induce colony growth and AML, we compared the ability of Trib1-3 to degrade C/EBPα. We previously demonstrated that Trib2 binds and stimulates proteasome-dependent C/EBPα degradation and that Trib2-induced C/EBPα degradation played an important role in leukemogenesis.8 We thus hypothesized that Trib1, but not Trib3, would promote C/EBPα degradation. We assessed Trib-induced degradation of endogenous C/EBPα by transducing the 32D myeloid cell line with retroviral supernatants expressing Trib1-3. Consistent with our hypothesis, Trib1 and Trib2 induced complete degradation of C/EBPα, but Trib3 did not (Figure 3A). Degradation of C/EBPα by Trib1 and Trib2 was rescued by the addition of the proteasome inhibitor MG132, indicating that C/EBPα degradation is proteasome-dependent (Figure 3A). These data further strengthen the association between Trib-induced C/EBPα degradation and leukemogenesis.
Trib1 and Trib2, but not Trib3, induce efficient degradation of C/EBPα. (A) Western blot for C/EBPα in sorted 32D cells transduced with MigR1, Trib1, Trib2, or Trib3 and treated with either the proteasome inhibitor MG132 or the DMSO control. Endogenous C/EBPα expression was determined with an anti-C/EBPα antibody. β-Actin served as the protein loading control. Data are representative of 3 independent experiments. (B) Western blot for endogenous ACC in HeLa cells transfected with Trib3 and the E3 ubiquitin ligase COP1. (C) Western blot for endogenous ACC in HeLa cells transfected with the indicated Tribbles constructs and COP1. In panels B and C, ACC was detected with an anti-ACC antibody, COP1 was detected with an anti-HA antibody, Tribs were detected with an anti-FLAG antibody, and β-actin was the protein loading control. Data are representative of 2 independent experiments.
Trib1 and Trib2, but not Trib3, induce efficient degradation of C/EBPα. (A) Western blot for C/EBPα in sorted 32D cells transduced with MigR1, Trib1, Trib2, or Trib3 and treated with either the proteasome inhibitor MG132 or the DMSO control. Endogenous C/EBPα expression was determined with an anti-C/EBPα antibody. β-Actin served as the protein loading control. Data are representative of 3 independent experiments. (B) Western blot for endogenous ACC in HeLa cells transfected with Trib3 and the E3 ubiquitin ligase COP1. (C) Western blot for endogenous ACC in HeLa cells transfected with the indicated Tribbles constructs and COP1. In panels B and C, ACC was detected with an anti-ACC antibody, COP1 was detected with an anti-HA antibody, Tribs were detected with an anti-FLAG antibody, and β-actin was the protein loading control. Data are representative of 2 independent experiments.
Because the inability of Trib3 to induce efficient degradation of C/EBPα may be attributed to a reduced binding affinity for C/EBPα, we investigated the capacity of the 3 Tribbles proteins to directly bind C/EBPα in vitro (supplemental Figure 4). For these studies, Trib1-3 proteins were prepared as glutathione S-transferase (GST) fusion proteins and used to recover radiolabeled C/EBPα produced by in vitro transcription and translation. Compared with the GST fusion protein of the ankyrin repeat domain of Notch1 as a negative control, all 3 GST-Tribbles proteins were able to recover C/EBPα, but the amount of C/EBPα captured by Trib3 trended lower than either Trib1 or Trib2, especially at the highest Tribbles input. A more quantitative assessment of C/EBPα binding by the 3 Tribbles proteins was precluded by the aggregation-prone nature of the recombinant Tribbles molecules, which was most pronounced for GST-Trib3 (P.H.D., M.E.V., O.S., L.X., K. Toscano, S.U., S.C.B., W.S.P., unpublished observations, May 2009). Nevertheless, when taken together with the observation that Trib1 and Trib2 promote efficient degradation of C/EBPα whereas Trib3 does not, these binding data are consistent with the idea that Trib3 exhibits a lower binding affinity for C/EBPα, which may account for the different degradation patterns.
Previously, Trib3 was shown to regulate ACC by recruiting the E3 ligase, COP1, to ACC and enhancing ACC degradation via the ubiquitin-proteasome pathway.7 To demonstrate that our Trib3 construct is functional, we transfected HeLa cells with Trib3 and COP1. HeLa cells were used for these studies as ACC expression is low in 32D cells and Trib3 degraded endogenous ACC in HeLa cells.7 Expression of Trib3 and COP1 induced degradation of endogenous ACC (Figure 3B). Expression of Trib1 or Trib2 also induced ACC degradation (Figure 3C); however, in this experiment, expression of COP1 alone seemed to induce mild ACC degradation. These data suggest that some substrates, such as ACC, can be targeted by all 3 Tribbles proteins; whereas others, such as C/EBPα, are preferentially targeted by specific Tribbles family members. To investigate the possibility that Trib3-induced protein degradation was cell-type specific, we tested the ability of Trib3 to degrade C/EBPα in HeLa cells where Trib3 degraded ACC. Whereas coexpression of Trib2 and C/EBPα resulted in proteasome-dependent C/EBPα degradation, Trib3 did not degrade C/EBPα, either in the presence or absence of exogenous COP1 (supplemental Figure 5). Trib2 was able to degrade C/EBPα in the presence or absence of exogenous COP1, whereas ACC degradation was greater in the presence of exogenous COP1. This suggests that C/EBPα degradation is mediated by endogenous COP1 or another protein with similar activity and that this level of activity is insufficient to degrade ACC.
Trib1 and Trib2 inhibit myeloid differentiation
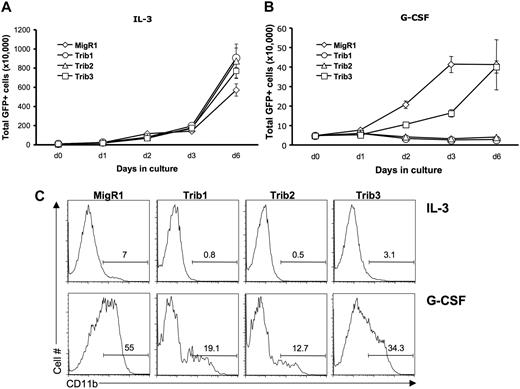
Our previous data demonstrated that Trib2-mediated C/EBPα degradation inhibited granulocytic differentiation.8 To determine the effect of Trib1 and Trib3 on granulocytic differentiation, we transduced 32D cells with Trib1-3. The 32D cells proliferate exponentially in the presence of IL-3 but can be induced to undergo granulocytic differentiation in the presence of G-CSF.27 This differentiation is dependent on C/EBPα.28,29 On G-CSF treatment, 32D cells undergo minimal proliferation and differentiate, which can be measured by expression of the myeloid surface marker CD11b. In the presence of IL-3, Trib1-3 expression did not significantly alter proliferation (Figure 4A). As expected, Trib2-transduced 32D cells failed to differentiate and died when stimulated with G-CSF (Figure 4B). Trib1, like Trib2, induced death of 32D cells in G-CSF (Figure 4B) and inhibited differentiation as measured by CD11b expression (Figure 4C). In contrast, Trib3-transduced 32D cells survived and differentiated (Figure 4B-C). The Trib3-transduced 32D cells displayed slower growth kinetics and decreased CD11b expression compared with the MigR1 retroviral vector, suggesting that Trib3 may have mild inhibitory effects on C/EBPα activity in this assay. Together, these data show that both Trib1 and Trib2 efficiently block C/EBPα-dependent 32D cell differentiation, whereas Trib3 does not.
Trib1 and Trib2, but not Trib3, inhibit myeloid differentiation. The 32D cells were transduced with MigR1, Trib1, Trib2, or Trib3 and plated in IL-3 (A) or G-CSF (B) 48 hours after transduction. The number of GFP-expressing cells were determined at the indicated time points. Values are mean (± SD) for triplicate samples. (C) Flow cytometric analysis of CD11b expression, which indicates differentiation, is shown at day 3 after IL-3 or G-CSF treatment. Data are representative of 3 independent experiments.
Trib1 and Trib2, but not Trib3, inhibit myeloid differentiation. The 32D cells were transduced with MigR1, Trib1, Trib2, or Trib3 and plated in IL-3 (A) or G-CSF (B) 48 hours after transduction. The number of GFP-expressing cells were determined at the indicated time points. Values are mean (± SD) for triplicate samples. (C) Flow cytometric analysis of CD11b expression, which indicates differentiation, is shown at day 3 after IL-3 or G-CSF treatment. Data are representative of 3 independent experiments.
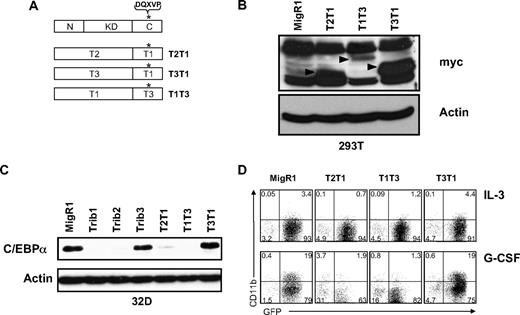
The Trib3 C-terminus is capable of degrading C/EBPα
Trib1-3 contain a conserved COP1 binding site in the C-terminus, and recruitment of COP1 through this site results in Trib3-mediated ACC degradation.7 Because all 3 Tribbles family members were able to recruit COP1 in a coimmunoprecipitation assay (supplemental Figure 6), we made C-terminal swap mutants of Trib1 and Trib3 (Figure 5A) to reveal whether the C-terminus of Trib3, which contains the COP1 binding site, can effectively recruit COP1 to C/EBPα and induce its degradation. We also made the control construct T2T1, which contains the N-terminus and kinase-like domain of Trib2 and the C-terminus of Trib1. After verifying expression of the mutants using an antibody to detect the myc-tag (Figure 5B), we assessed the ability of the domain swap mutants to induce C/EBPα degradation (Figure 5C). As expected, the control swap mutant T2T1 induced C/EBPα degradation. In addition, T1T3 induced C/EBPα degradation, showing that the Trib3 C-terminus can substitute for the C-terminus of Trib1 and effectively recruit COP1. Conversely, the T3T1 mutant did not promote C/EBPα degradation, suggesting that the Trib3 N-terminus and kinase-like domain are incapable of inducing C/EBPα degradation, even in the presence of the functional Trib1 C-terminus.
The Trib3 C-terminus can functionally replace the Trib1 C-terminus. (A) Schematic of C-terminal domain swap mutants. *The COP1 binding site DQXVP. (B) Western blot for protein expression of indicated domain swap mutants in transfected 293T cells. The expressed proteins were detected with an anti-myc antibody (9E10) and are indicated with arrows. β-Actin was the protein loading control. (C) Western blot for C/EBPα in sorted 32D cells transduced with the indicated retrovirus. Endogenous C/EBPα expression was detected with an anti-C/EBPα antibody. β-Actin served as the protein loading control. (D) Flow cytometric analysis for CD11b in 32D cells transduced with indicated retrovirus, then plated in IL-3 or G-CSF for 3 days. Data are representative of 3 independent experiments.
The Trib3 C-terminus can functionally replace the Trib1 C-terminus. (A) Schematic of C-terminal domain swap mutants. *The COP1 binding site DQXVP. (B) Western blot for protein expression of indicated domain swap mutants in transfected 293T cells. The expressed proteins were detected with an anti-myc antibody (9E10) and are indicated with arrows. β-Actin was the protein loading control. (C) Western blot for C/EBPα in sorted 32D cells transduced with the indicated retrovirus. Endogenous C/EBPα expression was detected with an anti-C/EBPα antibody. β-Actin served as the protein loading control. (D) Flow cytometric analysis for CD11b in 32D cells transduced with indicated retrovirus, then plated in IL-3 or G-CSF for 3 days. Data are representative of 3 independent experiments.
We also investigated the ability of the Tribbles swap mutants to induce 32D cell differentiation (Figure 5D). Consistent with the ability to stimulate degradation of C/EBPα, both T2T1 and T1T3 inhibited 32D cell differentiation in G-CSF, whereas T3T1 did not. Together, our results suggest that Trib1 and Trib2 operate distinctly from Trib3 to stimulate C/EBPα degradation and induce AML in mice.
Discussion
In the present study, we demonstrate that Trib1 and Trib2 induce AML in a murine retroviral BM transplantation model, but that Trib3 is incapable of causing AML in this model. Our data provide the first evidence that Trib3 behaves in a manner distinct from Trib1 and Trib2 in vivo and highlight important physiologic and molecular effects that may account for these differences. We previously showed that C/EBPα is a Trib2 target and Trib2-mediated degradation of C/EBPα is an important aspect of Trib2-mediated leukemogenesis. We now show that Trib1 also induces degradation of C/EBPα. In contrast, Trib3 failed to promote efficient degradation of C/EBPα. Not only does this finding reveal differential substrate use by the Tribbles family members, it also strengthens the association between Tribbles-induced leukemogenesis and C/EBPα degradation.
Our studies also show that Trib1 and Trib2, but not Trib3, inhibit G-CSF-induced 32D cell differentiation. In this assay, differentiation requires C/EBPα-dependent signaling. This process is abrogated in the presence of Trib1- and Trib2-induced C/EBPα degradation. Trib3 did not induce 32D cell differentiation as efficiently as the control vector, suggesting that Trib3 may have a mild effect on C/EBPα degradation. Nevertheless, this effect was insufficient to either promote self-renewal of BM progenitors or induce leukemia.
To begin to dissect the functional difference between Trib3 and Trib1/Trib2, we focused on the C-terminal domain that contains the COP1 binding site. Recruitment of COP1 is required for Trib3-induced ACC degradation7 and Trib2-induced C/EBPα degradation (K.K. and W.S.P., submitted for publication). We show that all 3 Tribbles proteins interact with COP1. In functional experiments, we demonstrated that replacing the Trib1 C-terminus with Trib3 C-terminal sequences or replacing Trib2 C-terminal sequences with the Trib1 counterpart retained the capacity for these chimeric proteins to stimulate C/EBPα degradation. In contrast, replacement of Trib3 C-terminal sequences with those from Trib1 failed to induce C/EBPα degradation. These data suggest that the failure of Trib3 to effectively induce C/EBPα degradation and leukemia is probably not related to COP1 recruitment. We also investigated whether the N-terminus of Trib3 inhibited its ability to induce efficient degradation of C/EBPα; however, deletion of the Trib3 N-terminus (amino acids 1-65) failed to enhance the ability of Trib3 to mediate C/EBPα degradation (data not shown). Together, these data suggest that Trib3 is incapable of promoting efficient degradation of C/EBPα and inducing AML because of functional differences from Trib1 and Trib2 that map to the KD rather than the N- or C-termini.
Although the Tribbles homologs vary in their ability to promote C/EBPα degradation, each protein is capable of stimulating degradation of ACC in HeLa cells. The biochemical and structural basis for this difference remains to be understood. It is possible that the affinity for C/EBPα plays an important role as Trib3 appears to bind less readily to C/EBPα. Nevertheless, additional studies are required to fully understand why Trib1-3 vary in their ability to induce degradation of C/EBPα.
In addition to inducing C/EBPα degradation, both Trib1 and Trib2 inhibit C/EBPβ as well. In adipocytes, Trib2 degrades C/EBPβ9 and lipopolysaccharide-stimulated macrophages from Trib1 knockout mice display enhanced C/EBPβ expression and functional activity.30 These findings, in conjunction with our results, suggest that mammalian Trib1 and Trib2 function to inhibit C/EBP proteins in a manner similar to slbo inactivation by Drosophila tribbles.3
A previous study of Trib1-induced murine AML suggested that an important component of Trib1-induced leukemia was increased MAPK signaling.21 In conditions where Trib1 and Trib2 promoted C/EBPα degradation in 32D cells, we did not observe increased Erk 1/2 phosphorylation in these cells by intracellular flow cytometry (supplemental Figure 7). Furthermore, neither Trib1 nor Trib2 increased the level of Erk 1/2 phosphorylation over the vector control or Trib3 (supplemental Figure 7). Thus, C/EBPα degradation demonstrates a stronger correlation with Tribbles-induced leukemogenesis than MAPK activation. Nevertheless, these data do not rule out a role for MAPK signaling in the pathogenesis of Tribbles-induced AML.
In conclusion, our studies have identified distinct functions for mammalian Tribs. Both Trib1 and Trib2 induce efficient degradation of C/EBPα and induce AML in our murine model. In contrast, Trib3 failed to facilitate degradation of C/EBPα or induce leukemia. These studies strengthen the correlation between Trib-mediated C/EBPα degradation and leukemia. Understanding the mechanistic basis for differential substrate use by different Tribs will probably reveal new strategies for inhibiting Trib-induced C/EBPα degradation in leukemia and other tumors characterized by C/EBPα degradation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the laboratory of W.S.P. for important contributions to these studies and the University of Pennsylvania Stemmler ASU, University of Pennsylvania Abramson Cancer Center Flow Cytometry, and Abramson Cancer Center cores.
This work was supported by the National Institutes of Health (grant CA093615; S.C.B., W.S.P.; grant T32CA009140; P.H.D., M.Z.-Z.; and Penn Prep Award R25GM071745; M.E.V.) and the Leukemia & Lymphoma Society (Special Fellow Award; K.K.).
National Institutes of Health
Authorship
Contribution: P.H.D., K.K., S.U., M.E.V., L.X., M.Z.-Z., and O.S. performed experiments; C.R. provided reagents; P.H.D. and S.U. made the figures; and P.H.D., S.U., S.C.B., and W.S.P. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of K.K. is Department of Biochemistry, BioSciences Institute, University College Cork, Cork, Ireland.
Correspondence: Warren S. Pear, Department of Pathology and Laboratory Medicine, Abramson Family Cancer Research Institute, University of Pennsylvania, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: wpear@mail.med.upenn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal