Abstract
Regulatory B cells control inflammation and autoimmunity in mice, including the recently identified IL-10–competent B10 cell subset that represents 1% to 3% of spleen B cells. In this study, a comparable IL-10–competent B10 cell subset was characterized in human blood. B10 cells were functionally identified by their ability to express cytoplasmic IL-10 after 5 hours of ex vivo stimulation, whereas progenitor B10 (B10pro) cells required 48 hours of in vitro stimulation before they acquired the ability to express IL-10. B10 and B10pro cells represented 0.6% and approximately 5% of blood B cells, respectively. Ex vivo B10 and B10pro cells were predominantly found within the CD24hiCD27+ B-cell subpopulation that was able to negatively regulate monocyte cytokine production through IL-10–dependent pathways during in vitro functional assays. Blood B10 cells were present in 91 patients with rheumatoid arthritis, systemic lupus erythematosus, primary Sjögren syndrome, autoimmune vesiculobullous skin disease, or multiple sclerosis, and were expanded in some cases as occurs in mice with autoimmune disease. Mean B10 + B10pro-cell frequencies were also significantly higher in patients with autoimmune disease compared with healthy controls. The characterization of human B10 cells will facilitate their identification and the study of their regulatory activities during human disease.
Introduction
B cells are generally considered to positively regulate immune responses by producing antigen-specific antibody and helping to induce optimal CD4+ T-cell activation.1 However, B cells and specific B-cell subsets can also negatively regulate immune responses in mice.2-6 The absence or loss of these regulatory B cells exacerbates disease symptoms in contact hypersensitivity, experimental autoimmune encephalomyelitis, chronic colitis, collagen-induced arthritis, and lupus-like models of autoimmunity.7-15 In many of these cases, B cells regulate inflammation, asthma, and T cell–mediated autoimmunity through the production of interleukin-10 (IL-10).8-10,12-16 Both human and mouse IL-10 exhibit numerous pleiotrophic activities in vitro and in vivo, including suppression of both Th1 and Th2 polarization and inhibition of antigen presentation and proinflammatory cytokine production by dendritic cells, monocytes, and macrophages.17
In mice, a subset of IL-10–competent regulatory B cells can be functionally identified by their ability to express cytoplasmic IL-10 after 5 hours of in vitro stimulation with lipopolysaccharide (LPS), phorbol myristate acetate (PMA), and ionomycin, with monensin included in the cultures to block IL-10 secretion.12,13 These IL-10–competent B cells have been labeled as B10 cells to identify them as the predominant, if not exclusive, source of B-cell IL-10 production and to distinguish them from other regulatory B-cell subsets that may also exist.5 For example, inducible IL-12–producing B cells regulate intestinal inflammation.18 B10 cells are found within the spleens of naive wild-type mice at frequencies of 1% to 3%, where they predominantly represent a subset of the phenotypically unique CD1dhiCD5+CD19hi B-cell subpopulation that shares overlapping cell surface markers with multiple phenotypically defined B-cell subsets.11-14,19,20 Additional B cells within the CD1dhiCD5+ B-cell subpopulation acquire the ability to function like B10 cells during 48 hours of in vitro stimulation with LPS or agonistic CD40 monoclonal antibody (mAb).5 These B10 progenitor (B10pro) cells are then able to express cytoplasmic IL-10 after stimulation with PMA, ionomycin, and monensin for 5 hours.21 B10 cells also require diverse B-cell antigen receptors for their development,21 and their regulatory functions are Ag-restricted in vivo.12,13 Spleen B10-cell numbers increase significantly in diabetes- and lupus-prone mice,14,21 and the adoptive transfer of antigen-primed CD1dhiCD5+ B cells reduces inflammation during contact hypersensitivity and autoimmune disease.12,13,22
The identification and characterization of an IL-10–producing B-cell subset in mice raise the issue of whether B cells with these functional properties exist in humans. Studies of B-cell IL-10 production in humans have yielded diverse results that are currently difficult to unify into a coherent model.23-28 It is also unknown whether human B10 cells share overlapping physiologic triggers with mouse B10 cells that lead to IL-10 production and their expansion in vitro.12,13,21 Therefore, the purpose of the current study was to enumerate and characterize the IL-10 competent B10 and B10pro cell subsets in humans.
Methods
Cells
Heparinized blood was obtained from healthy donors (age, 14-73 years) or from patients. Patients with rheumatoid arthritis met the American College of Rheumatology 1987 revised classification criteria.29 Patients with systemic lupus erythematosus satisfied the 1982 classification criteria.30 Patients with primary Sjögren syndrome fulfilled the American-European consensus group criteria.31 Patients with autoimmune vesiculobullous skin disease, including bullous pemphigoid, pemphigus foliaceus, pemphigus vulgaris, and dermatitis herpetiformis, had typical clinical and histologic findings, with diagnostic findings on direct immunofluorescence of perilesional skin or oral mucosa.32,33 Patients with multiple sclerosis fulfilled the 2005 McDonald criteria for relapsing remitting or primary progressive multiple sclerosis34 or secondary progressive multiple sclerosis as defined using the Lublin and Reingold criteria.35 Tissues were obtained anonymously from persons without identifiable hematologic disorders, with the purified B cells immediately cryopreserved (> 90% cell viability). Cryopreserved cord blood samples were obtained from the Duke University Stem Cell Laboratory and the Carolinas Cord Blood Bank.
Blood mononuclear cells were isolated by centrifugation over a discontinuous Lymphoprep (Axis-Shield PoC As) gradient. Cell numbers were quantified by hemocytometer, with relative lymphocyte percentages among viable cells (based on scatter properties) determined by flow cytometry. In some experiments, B cells were enriched using RosetteSep (StemCell Technologies) following the manufacturer's protocols. CD19-mAb-coated microbeads (Miltenyi Biotec) were used to purify blood B cells by positive selection following the manufacturer's instructions. When necessary, the cells were enriched a second time using a fresh magnetic-activated cell sorting column to obtain more than 99% purities.
Written informed consent was obtained in all cases according to the Declaration of Helsinki. All protocols were approved by the respective ethics boards: Institutional Review Board of Duke University Medical Center, the Human Protection Committee of Dana-Farber Cancer Institute/Harvard Medical School, University of Rochester Institutional Review Board, and the Institutional Review Board of St Luke's-Roosevelt Hospital Center Institute for Health Sciences.
Antibodies, immunofluorescence analysis, and cell sorting
Antihuman mAbs included: IgD (IA6–2) from BD Biosciences; CD21 (BU33), CD22 (RFB4), CD23 (D.6) from Ancell; IgM (MHM-88), CD1d (51.1), CD5 (UCHT2), CD19 (HIB19), CD24 (ML5), CD25 (BC96), CD27 (O323), CD38 (HIT2), CD40 (HB14), CD48 (BJ40), and CD148 (A3), tumor necrosis factor-α (TNF-α; MAb11), functional grade CD3 (HIT3a), and phycoerythrin-conjugated or unconjugated anti–IL-10 (JES3–19F1, JES3–9D7) mAbs from BioLegend. Antihuman IgM Ab was from Jackson ImmunoResearch Laboratories.
Single-cell suspensions were stained on ice using predetermined optimal concentrations of each antibody for 20 to 60 minutes and fixed as described.36 Cells with the light scatter properties of lymphocytes were analyzed by 2–6 color immunofluorescence staining and FACScan, FACSCalibur, or FACSCanto II flow cytometers (BD Biosciences). Dead cells were excluded from the analysis based on their forward- and side-light scatter properties and the use of LIVE/DEAD Fixable Violet Dead Cell Stain Kits (Invitrogen). All histograms are shown on a 4-decade logarithmic scale, with gates shown to indicate background isotype-matched control mAb staining set with less than 2% of the cells being positive. Blood CD24hiCD27+ and CD24lowCD27− B cells were isolated using a FACSVantage SE flow cytometer (BD Biosciences) with 90% to 95% purities.
Analysis of IL-10 production
Intracellular IL-10 analysis by flow cytometry was as described.12 Briefly, cells were resuspended (2 × 106 cells/mL) in medium (RPMI 1640 media containing 10% fetal calf serum, 200 μg/mL penicillin, 200 U/mL streptomycin, and 4mM L-glutamine [all from Invitrogen]) and stimulated with LPS (10 μg/mL, Escherichia coli serotype 0111: B4; Sigma-Aldrich), CpG (ODN 2006, 10 μg/mL; Invivogen), or other Toll-like receptor (TLR) agonists (TLR1, Pam3CSK4, 1 μg/mL; TLR2, heat-killed Listeria monocytogenes, 108 cells/mL; TLR3, polyriboinosinic acid/polyribocytidylic acid, 10 μg/mL; TLR5, S typhimurium flagellin, 1 μg/mL; TLR6, Pam2CGDPKHPKSF, 1 μg/mL; TLR7, imiquimod, 1 μg/mL; TLR8, ssRNA40, 1 μg/mL; Invivogen), CD40L (1 μg/mL; R&D Systems), antihuman CD40 mAb (1 μg/mL; BioLegend), PMA (50 ng/mL; Sigma-Aldrich), ionomycin (1 μg/mL; Sigma-Aldrich), brefeldin A (BFA, 1 × solution/mL; BioLegend), monensin (2mM; eBioscience), and antiIgM antibody (10 μg/mL) as indicated in 48-well flat-bottom plates before staining and flow cytometry analysis. For analysis of cell proliferation, lymphocytes were stained with carboxyfluorescein succinimidyl ester (CFSE) Vybrant CFDA SE fluorescent dye (5 μM; Invitrogen) according to the manufacturer's instructions. For IL-10 detection, Fc receptors were blocked using FcγR-binding inhibitor (eBioscience). Stained cells were fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer's instructions and stained with anti–IL-10 mAb.
Secreted IL-10 was quantified by enzyme-linked immunosorbent assay (ELISA). Purified B cells (4 × 105) were cultured in 0.2 mL of complete medium in a 96-well flat-bottom tissue culture plates. Culture supernatant fluid IL-10 concentrations for triplicate samples were quantified using IL-10 OptEIA ELISA kits (BD Biosciences) following the manufacturer's protocols.
IL10 transcript expression
In some experiments, IL-10-secreting blood B cells were identified after 4 hours of in vitro stimulation using an IL-10 secretion detection kit (Miltenyi Biotec) with subsequent staining for CD19 expression before cell sorting into IL-10+CD19+ and IL-10−CD19+ populations. RNA extraction, cDNA generation, and real-time PCR analysis were as described.13 IL-10 primers were: sense 5′-CTTCGAGATC TCCGAGATGC CTTC-3′, antisense 5′-ATTCTTCACC TGCTCCACGG CCTT-3′. GAPDH primers were: sense 5′-GCCACCCAGA AGACTGTGGA TGGC-3′ and antisense 5′-CATGTAGGCC ATGAGGTCCA CCAC-3′.
In vitro functional assays
Blood CD24hiCD27+CD19+ and CD24lowCD27−CD19+ B cells from healthy controls were purified by cell sorting and stimulated with CpG (10 μg/mL) plus CD40L (1 μg/mL) for 24 hours. Naive CD4+CD25− T cells were purified by magnetic-activated cell sorting (Miltenyi Biotec) and cultured in replicate wells either alone (1 × 106/mL) or with CD40/CpG-stimulated B cells (1 × 106/mL) in tissue culture plates coated with CD3 mAb (0.5 μg/mL) for 72 hours with either BFA or phorbol ester and ionomycin plus BFA (PIB) added during the final 5 hours. The cells were stained for cell surface CD4 and cytoplasmic TNF-α expression before analysis by flow cytometry. Alternatively, CD40/CpG-stimulated sorted B-cell populations (1 × 106/mL) were cultured for 20 hours with equal numbers of blood monocytes that had been purified by plastic adherence or magnetic-activated cell sorting, with anti–IL-10 mAb (10 μg/mL) added to some cultures. The cultured cells were then washed, stimulated with LPS (1 μg/mL) for 4 hours in the presence of BFA, and stained for cell surface CD14 and cytoplasmic TNF-α or interferon-γ expression before analysis by flow cytometry.
Statistical analysis
All data are shown as mean ± SEM. Significant differences between sample means were determined using the Student t test.
Results
Enumeration of human IL-10-producing B cells
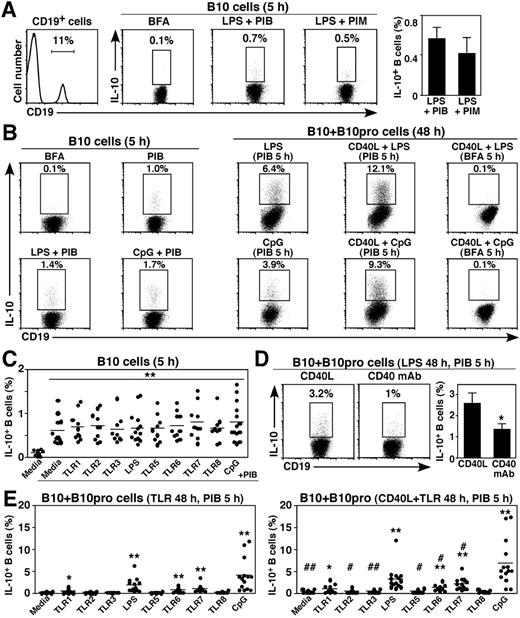
Mouse B10-cell frequencies have been described, with background cytoplasmic IL-10 staining of less than or equal to 0.2% commonly observed with B cells from IL-10−/− mice.12,13,21 A similar strategy was therefore optimized to enumerate human blood IL-10–competent B cells. B cells spontaneously expressing IL-10 were below the threshold for reliable quantification by immunofluorescence staining, even when the cells were cultured with BFA to stop Golgi transport (Figure 1A). However, a distinct subset of cytoplasmic IL-10+ B cells was observed at low 0.25% to 2% frequencies after ex vivo stimulation using PIB (Figure 1A-B). Stimulation with PIB for 5 hours induced 0.8% ± 0.1% of blood B cells on average to express IL-10 (n = 14, 1.9 ± 0.3 × 10−3 B10 cells/mL, Figure 1B-C). B-cell stimulation with TLR agonists did not substantially alter mean B10-cell numbers, although adding either CpG oligonucleotides (CpG, TLR9 agonist) or LPS to the PIB cultures increased IL-10+ B-cell frequencies in some persons (Figure 1A-C). Stimulation beyond 5 hours or using 10-fold higher PMA or ionomycin concentrations resulted in extensive B-cell death, which complicated B10-cell enumeration. Background IL-10 mAb staining was also reduced by the exclusion of cell doublets and dead cells from the analysis. BFA was also used to block IL-10 secretion rather than monensin because it optimized human B-cell cytoplasmic IL-10 expression (Figure 1A). B cells cultured in BFA served as negative controls because they gave results similar to isotype control mAb staining. Thus, blood B10 cells were rare but readily quantified in healthy humans.
Enumeration of human blood IL-10-competent B10 and B10pro cells. (A) Visualizing IL-10+ B cells. Purified blood mononuclear cells were cultured with BFA, LPS plus PMA, ionomycin and BFA (PIB), or LPS plus PMA, ionomycin, and monensin (PIM) for 5 hours and stained for cell viability, cell surface CD19 expression, and cytoplasmic IL-10. Representative cytoplasmic IL-10 staining by viable, single CD19+ B cells is shown in the flow cytometry dot-plots, with percentages indicating cytoplasmic IL-10+ B-cell frequencies within the indicated gates. Blood mononuclear cells that were cultured with BFA alone before immunofluorescence staining served as negative controls, with background staining similar to that obtained using isotype-matched control mAbs. Bar graphs represent mean (± SEM) B10-cell frequencies from 3 persons. (B) Representative IL-10 production by B cells from a person with relatively high B10-cell frequencies. B10 cells were identified after in vitro stimulation for 5 hours as in panel A. Alternatively, IL-10+ B-cell frequencies were determined after in vitro B10pro cell maturation by stimulation with LPS, CD40L + LPS, CpG, or CD40L + CpG, with PIB added during the final 5 hours of 48-hour cultures. As negative controls for IL-10 staining, only BFA was added to some cultures during the final 5 hours. Percentages indicate the frequencies of cytoplasmic IL-10+ B cells within the indicated gates among total CD19+ B cells. (C) B10-cell frequencies in persons after with TLR agonist stimulation as in panels A and B. Dots represent results from single persons after 5-hour culture with BFA alone, PIB, or the indicated TLR agonist + PIB. Horizontal bars represent means. (D) CD40L induced optimal B10 + B10pro cell maturation during 48 hours in vitro cultures with either recombinant CD40L or CD40 mAb, plus LPS for 48 hours, with PIB added during the final 5 hours. Bar graphs represent means (± SEM) from 5 persons. Similar results were obtained in 2 independent experiments. (E) Representative B10 + B10pro-cell frequencies after in vitro maturation and stimulation. Blood mononuclear cells were cultured for 48 hours with media alone or media containing CD40L, along with the indicated TLR agonists, with PIB added during the last 5 hours of culture. Significant differences between cultures with or without CD40L are indicated: #P < .05; ##P < .01. (C-E) Significant differences between means of controls and individual stimuli are indicated: *P < .05; **P < .01.
Enumeration of human blood IL-10-competent B10 and B10pro cells. (A) Visualizing IL-10+ B cells. Purified blood mononuclear cells were cultured with BFA, LPS plus PMA, ionomycin and BFA (PIB), or LPS plus PMA, ionomycin, and monensin (PIM) for 5 hours and stained for cell viability, cell surface CD19 expression, and cytoplasmic IL-10. Representative cytoplasmic IL-10 staining by viable, single CD19+ B cells is shown in the flow cytometry dot-plots, with percentages indicating cytoplasmic IL-10+ B-cell frequencies within the indicated gates. Blood mononuclear cells that were cultured with BFA alone before immunofluorescence staining served as negative controls, with background staining similar to that obtained using isotype-matched control mAbs. Bar graphs represent mean (± SEM) B10-cell frequencies from 3 persons. (B) Representative IL-10 production by B cells from a person with relatively high B10-cell frequencies. B10 cells were identified after in vitro stimulation for 5 hours as in panel A. Alternatively, IL-10+ B-cell frequencies were determined after in vitro B10pro cell maturation by stimulation with LPS, CD40L + LPS, CpG, or CD40L + CpG, with PIB added during the final 5 hours of 48-hour cultures. As negative controls for IL-10 staining, only BFA was added to some cultures during the final 5 hours. Percentages indicate the frequencies of cytoplasmic IL-10+ B cells within the indicated gates among total CD19+ B cells. (C) B10-cell frequencies in persons after with TLR agonist stimulation as in panels A and B. Dots represent results from single persons after 5-hour culture with BFA alone, PIB, or the indicated TLR agonist + PIB. Horizontal bars represent means. (D) CD40L induced optimal B10 + B10pro cell maturation during 48 hours in vitro cultures with either recombinant CD40L or CD40 mAb, plus LPS for 48 hours, with PIB added during the final 5 hours. Bar graphs represent means (± SEM) from 5 persons. Similar results were obtained in 2 independent experiments. (E) Representative B10 + B10pro-cell frequencies after in vitro maturation and stimulation. Blood mononuclear cells were cultured for 48 hours with media alone or media containing CD40L, along with the indicated TLR agonists, with PIB added during the last 5 hours of culture. Significant differences between cultures with or without CD40L are indicated: #P < .05; ##P < .01. (C-E) Significant differences between means of controls and individual stimuli are indicated: *P < .05; **P < .01.
Human B10pro cell identification
In mice, B10pro cell maturation into IL-10-competent B10 cells is induced by 48 hours of stimulation with either LPS or agonistic CD40 mAb.21 Human blood B10pro cells capable of maturing into IL-10–competent cells after in vitro culture were also identified. The total frequency of B10 and B10pro cells (B10 + B10pro) is quantified in this assay, as the B cells that acquire IL-10 competence in vitro (eg, matured B10pro cells) cannot be differentiated from preexisting B10 cells that inherently express cytoplasmic IL-10 after 5 hours of PIB stimulation. Culturing human B cells in media alone for 48 hours resulted in approximately 0.2% of B cells expressing cytoplasmic IL-10 after PIB stimulation during the last 5 hours of culture (Figure 1B). Adding LPS, CpG, or recombinant CD40 ligand (CD40L, CD154) alone, together, or in combination with BFA to the 48-hour cultures did not increase IL-10-producing B10-cell frequencies. However, after PIB stimulation during the last 5 hours of culture, B10 + B10pro-cell frequencies increased to 0.6% ± 0.1%, 1.9% ± 0.4%, 0.8% ± 0.1%, 1.2% ± 0.2%, and 4.1% ± 1.0% after 48 hours of TLR1 agonist, LPS, TLR6 agonist, TLR7 agonist, or CpG stimulation, respectively (Figure 1E left panel). Compared with media or most TLR agonists alone, the addition of CD40L to the cultures significantly enhanced mean B10 + B10pro-cell frequencies (Figure 1E right panel). CD40L induced higher IL-10+ B-cell frequencies than agonistic CD40 mAb (Figure 1D). Thus, dual CD40 and TLR stimulation induced the highest frequencies of B10pro cells to become IL-10–competent B10 cells, with the highest frequencies (7.0% ± 1.4%) and numbers of IL-10+ cells (1.6 ± 0.3 × 104 cells/mL, n = 14) induced after 48 hours of CD40L plus CpG stimulation. B10-cell percentages relative to all B cells decreased with age among 60 healthy persons, with a significant inverse correlation between B10 + B10pro-cell frequencies and age (P < .05, data not shown). Thus, B10pro cell maturation in vitro optimized IL-10–competent B-cell enumeration.
B10-cell numbers in newborn blood and adult tissues
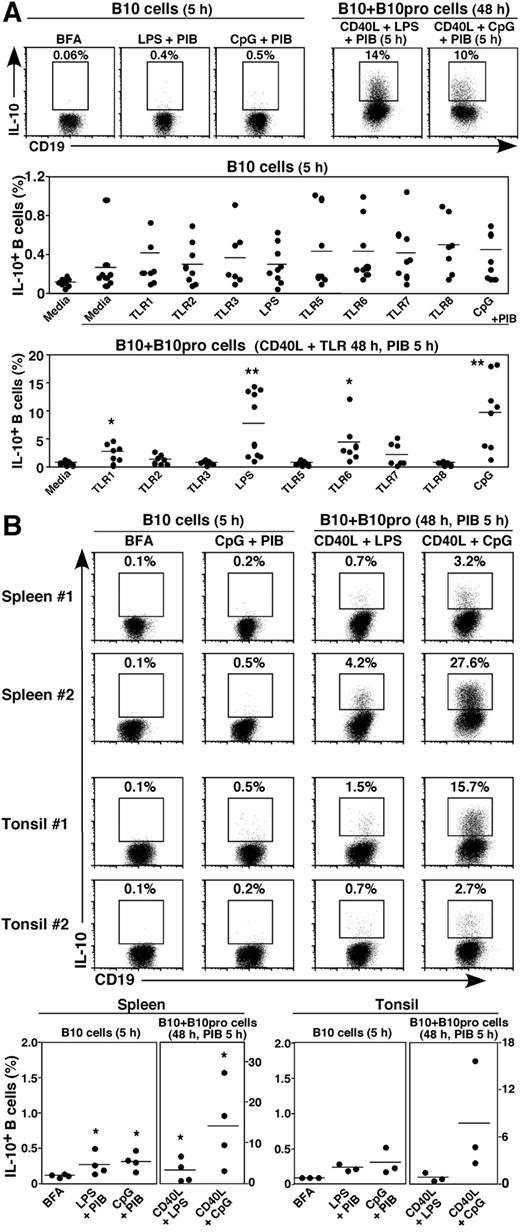
Newborn blood contained both B10 and B10pro cells. Mean B10-cell frequencies in cord blood after 5 hours of CpG + PIB stimulation were 0.45% ± 0.14% (Figure 2A). B10 + B10pro-cell frequencies were similar or higher in cord blood relative to adult blood after culture with CD40L and TLR agonists: TLR1 (2.6% ± 0.6%), LPS (7.6% ± 1.8%), TLR6 (4.2% ± 1.4%), or TLR9 (CpG, 9.6 ± 2.3%) agonists with PIB added during the final 5 hours of culture. B10 cells were also found within spleens (0.31% ± 0.06%, n = 4, CpG + PIB) and tonsils (0.31% ± 0.11%, n = 3, CpG + PIB) of adults without known disease (Figure 2B). Thus, newborn and adult blood and tissues contain quantifiable numbers of B10 and B10pro cells.
Human B10 and B10pro cells in (A) cord blood and (B) spleen and tonsil. B10 cells and B10 + B10pro cells were identified after in vitro stimulation for 5 hours and 48 hours, respectively, as in Figure 1. Representative results are shown along with graphs indicating IL-10+ B-cell frequencies within persons. Cells cultured with BFA alone served as negative controls for background IL-10 staining. Significant differences between means of BFA controls and individual stimuli are indicated: *P < .05; **P < .01.
Human B10 and B10pro cells in (A) cord blood and (B) spleen and tonsil. B10 cells and B10 + B10pro cells were identified after in vitro stimulation for 5 hours and 48 hours, respectively, as in Figure 1. Representative results are shown along with graphs indicating IL-10+ B-cell frequencies within persons. Cells cultured with BFA alone served as negative controls for background IL-10 staining. Significant differences between means of BFA controls and individual stimuli are indicated: *P < .05; **P < .01.
Regulation of B10 cell IL-10 production and secretion in vitro
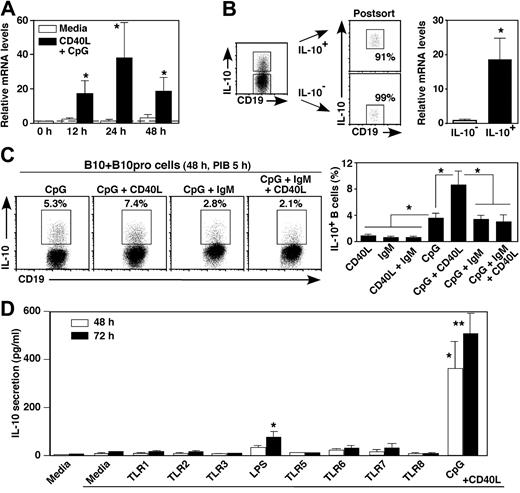
The time course of blood B-cell IL-10 induction was assessed in vitro by quantifying IL-10 transcripts. By 12, 24, and 48 hours, B-cell stimulation with CD40L plus CpG induced 6.8-, 24-, and 5.9-fold higher IL10 transcript levels, respectively, than was observed for unstimulated B cells (P < .05; Figure 3A). Blood B cells that were actively secreting IL-10 after in vitro stimulation expressed IL10 transcripts at 19-fold higher levels than were detected in IL-10− B cells (Figure 3B). Thus, IL10 gene transcription parallels the induction of B10-cell cytoplasmic IL-10 expression.
Human blood B-cell stimulation induces IL-10 transcription and secretion in vitro. (A) Time course of IL10 transcript induction. Purified CD19+ B cells were cultured with media alone or CD40L + CpG for the times indicated, with IL10 transcripts quantified by real-time reverse-transcribed polymerase chain reaction analysis. Bar graphs indicate mean relative IL10 transcript (± SEM) levels in 6 persons. (B) B cells secreting IL-10 express IL10 transcripts. Purified blood B cells were cultured with PMA and ionomycin for 4 hours before CD19 staining and secreted IL-10 capture (left panel). Cell surface IL-10+ and IL-10− B cells were isolated using the indicated gates and subsequently reassessed for IL-10 secretion (right panels) before relative IL10 transcript levels were quantified by real-time reverse-transcribed polymerase chain reaction analysis. Mean fold differences (± SEM) for IL10 transcript levels from 3 different persons are shown, with transcript levels normalized so that the relative mean IL-10− B-cell value is 1.0. (C) Cell surface signals that regulate cytoplasmic IL-10 expression. Blood B cells were cultured with CpG, CD40L, and antiIgM Ab (IgM) as indicated for 48 hours with PIB added during the final 5 hours of culture. Representative frequencies of IL-10-producing cells are shown, with bar graphs representing mean (± SEM) percentages in 5 persons. (D) TLR agonists that induce IL-10 secretion. Purified CD19+ B cells were cultured with media alone, CD40L, or with TLR agonists and CD40L as indicated for 48 or 72 hours. IL-10 secreted into the culture supernatant fluid was quantified by ELISA. Bar graphs represent mean IL-10 (± SEM) concentrations from more than or equal to 4 different persons. (A-D) Similar results were obtained in 2 independent experiments. Significant differences between means of cells cultured in media alone and stimulated cultures are indicated: *P < .05; **P < .01.
Human blood B-cell stimulation induces IL-10 transcription and secretion in vitro. (A) Time course of IL10 transcript induction. Purified CD19+ B cells were cultured with media alone or CD40L + CpG for the times indicated, with IL10 transcripts quantified by real-time reverse-transcribed polymerase chain reaction analysis. Bar graphs indicate mean relative IL10 transcript (± SEM) levels in 6 persons. (B) B cells secreting IL-10 express IL10 transcripts. Purified blood B cells were cultured with PMA and ionomycin for 4 hours before CD19 staining and secreted IL-10 capture (left panel). Cell surface IL-10+ and IL-10− B cells were isolated using the indicated gates and subsequently reassessed for IL-10 secretion (right panels) before relative IL10 transcript levels were quantified by real-time reverse-transcribed polymerase chain reaction analysis. Mean fold differences (± SEM) for IL10 transcript levels from 3 different persons are shown, with transcript levels normalized so that the relative mean IL-10− B-cell value is 1.0. (C) Cell surface signals that regulate cytoplasmic IL-10 expression. Blood B cells were cultured with CpG, CD40L, and antiIgM Ab (IgM) as indicated for 48 hours with PIB added during the final 5 hours of culture. Representative frequencies of IL-10-producing cells are shown, with bar graphs representing mean (± SEM) percentages in 5 persons. (D) TLR agonists that induce IL-10 secretion. Purified CD19+ B cells were cultured with media alone, CD40L, or with TLR agonists and CD40L as indicated for 48 or 72 hours. IL-10 secreted into the culture supernatant fluid was quantified by ELISA. Bar graphs represent mean IL-10 (± SEM) concentrations from more than or equal to 4 different persons. (A-D) Similar results were obtained in 2 independent experiments. Significant differences between means of cells cultured in media alone and stimulated cultures are indicated: *P < .05; **P < .01.
The response of human B10 + B10pro cells to CD40L, CpG, and antigen receptor-generated signals was also examined. Compared with CD40L alone, CpG induced the highest levels of B10pro cell maturation into IL-10–competent B10 cells, which was further increased when both CD40L and CpG were added to the cultures (Figure 3C). By contrast, IgM ligation using mitogenic antibody did not induce cytoplasmic IL-10 expression but actually inhibited the B10pro cell maturation effects of CpG + CD40L stimulation. In vitro IgM signals also inhibit mouse B10pro cell maturation and cytoplasmic IL-10 induction.21 Among TLR agonists, LPS and CpG were also the most potent stimuli for inducing IL-10 secretion by human blood B cells (Figure 3D). The addition of CD40L to the cultures was not required but enhanced LPS- and CpG-induced IL-10 secretion. Thus, similar signals induce human and mouse B10pro and B10 cells to mature and express cytoplasmic IL-10 in vitro.
Phenotypic characterization of IL-10–competent B cells
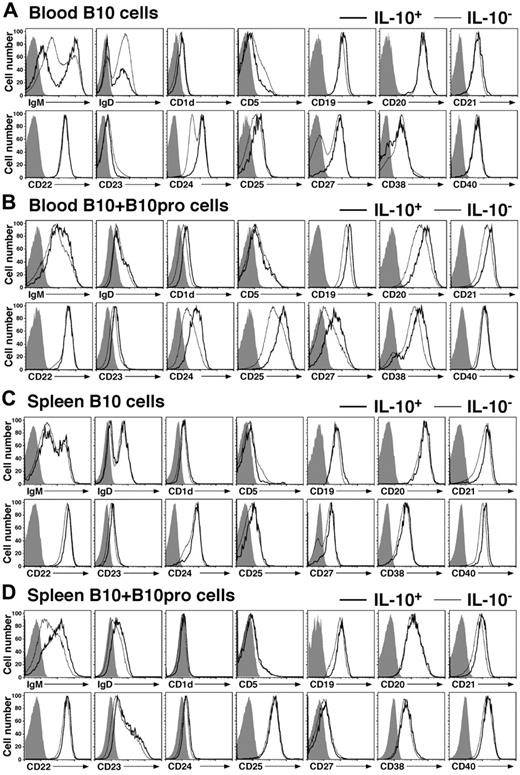
Whether human B10 cells represent a unique or known B-cell subset was determined by analyzing their cell surface phenotype. Cell surface IgM, IgD, CD1d, CD5, CD10, CD19, CD21, CD22, CD23, CD24, CD25, CD27, CD38, and CD40 densities did not change when B cells were stimulated with PIB, LPS + PIB, or CpG + PIB for 5 hours and/or permeabilized. Furthermore, the transport of newly synthesized proteins to the cell surface is inhibited by the addition of BFA to the cultures. Therefore, these markers were used to phenotype freshly isolated B10 cells. Half of blood B10 cells expressed high IgM levels and low IgD levels (Figure 4A). Both CD24 and CD27 expression was high on the majority of B10 cells, whereas IL-10− B cells expressed either high or low density CD24 and CD27. CD19 and CD25 expression was also higher on B10 cells than IL-10− B cells. Otherwise, the remaining cell surface markers were absent or expressed similarly by both B10 cells and IL-10− B cells. The same results were obtained after PIB, LPS + PIB, or CpG + PIB stimulation (data not shown). Thereby, freshly isolated blood IL-10+ B10 cells were predominantly found within the CD24hi or CD27+ B-cell subpopulations.
Phenotypes of human blood and tissue B10 cells ex vivo and B10 + B10pro cells after in vitro culture. (A) Representative cell surface phenotype of blood B cells cultured with LPS + PIB for 5 hours. (B) Representative cell surface phenotype of blood B10 + B10pro cells after stimulation with CD40L + LPS for 48 hours with PIB added during the final 5 hours of culture. (C) Representative cell surface phenotype of spleen B10 cells cultured with CpG + PIB for 5 hours. (D) Representative cell surface phenotype of spleen B10 + B10pro cells after stimulation with CD40L + CpG for 48 hours with PIB added during the final 5 hours of culture. (A-D) Cultured cells were stained for viability and cell surface molecule expression, permeabilized, stained with anti–IL-10 mAb, and analyzed by flow cytometry. Representative cell surface molecule expression by IL-10+ (thick line) and IL10− (thin line) CD19+ B cells from 3 persons. Shaded histograms represent isotype-matched control mAb staining.
Phenotypes of human blood and tissue B10 cells ex vivo and B10 + B10pro cells after in vitro culture. (A) Representative cell surface phenotype of blood B cells cultured with LPS + PIB for 5 hours. (B) Representative cell surface phenotype of blood B10 + B10pro cells after stimulation with CD40L + LPS for 48 hours with PIB added during the final 5 hours of culture. (C) Representative cell surface phenotype of spleen B10 cells cultured with CpG + PIB for 5 hours. (D) Representative cell surface phenotype of spleen B10 + B10pro cells after stimulation with CD40L + CpG for 48 hours with PIB added during the final 5 hours of culture. (A-D) Cultured cells were stained for viability and cell surface molecule expression, permeabilized, stained with anti–IL-10 mAb, and analyzed by flow cytometry. Representative cell surface molecule expression by IL-10+ (thick line) and IL10− (thin line) CD19+ B cells from 3 persons. Shaded histograms represent isotype-matched control mAb staining.
The phenotype of blood B10 + B10pro cells was also assessed after 48 hours of culture with CD40L + CpG and 5 hours of LPS + PIB stimulation. Prolonged B-cell stimulation induced significant changes in the cell surface phenotype of both IL-10+ and IL-10− B cells, with most B cells induced to express CD25 and CD38 (Figure 4B). Nonetheless, B10 + B10pro cells on average expressed higher densities of CD1d, CD19, CD20, CD21, CD23, CD24, CD25, CD27, and CD38 compared with IL-10− B cells, consistent with an activated phenotype. Spleen B10 cells were also predominantly CD27+, although the expression of most cell surface molecules was similar if not identical for B10 cells and IL-10− B cells (Figure 4C). Spleen B10 + B10pro cells and IL-10− B cells also had similar phenotypes after 48 hours of stimulation in vitro (Figure 4D). Thereby, blood B10 cells represented a subset of the CD24hiCD27+ subpopulation, whereas IL-10 expression remained the best marker for categorizing B10 cells and B10 + B10pro cells.
Blood B10 cells are enriched within the CD24hiCD27+ B-cell subpopulation
In addition to being predominantly CD24hi and CD27+, most B10 cells also expressed additional cell surface markers of activation (CD48hi) and memory (CD148hi) that were not affected by the 5-hour culture conditions used to induce B-cell cytoplasmic IL-10 expression (Figure 5A-B). Even when the spectrum of blood donors was compared, B10 cells were always predominantly CD24hi and CD27+ (Figure 5C). Cell surface IgD, CD27, and CD38 expression profiles have also been used frequently to define human B-cell subsets.37,38 However, when blood B10 cells were analyzed based on their CD38 versus IgD, or CD27 versus IgD expression profiles, IL-10+ B10 cells from representative blood donors did not fall into distinct subpopulations (Figure 5C).
Ex vivo blood B10 and B10pro cells share cell surface markers with memory B cells. (A) Blood B10 cells predominantly exhibit a CD24hiCD27+CD48hiCD148hi phenotype. Purified blood B cells were cultured with CpG + PIB for 5 hours before immunofluorescence staining for viability, cell surface molecule expression, and cytoplasmic IL-10. Cell surface CD24, CD27, CD38, CD48, and CD148 expression by IL-10+ (thick line) and IL-10− (thin line) CD19+ cells was assessed by flow cytometry. (B) Cytoplasmic IL-10 induction does not affect the cell surface phenotype of B cells. CD19+ blood B cells were cultured with media on ice (thin line) or with CpG + PIB (thick line) for 5 hours before immunofluorescence staining and flow cytometry analysis as in panel A. (A-B) Shaded histograms represent isotype-matched control mAb staining. Results represent those obtained for 3 persons. (C) Distributions of B10 cells within B-cell subsets defined by CD24, CD27, IgD/CD38, and IgD/CD27 expression. Purified blood B cells were cultured with LPS + PIB for 5 hours before immunofluorescence staining and flow cytometry analysis as in panel A. The horizontal and vertical lines on each contour plot are shown for reference, with the lower left quadrants delineating the IgD−CD38− and IgD−CD27− subsets determined by control mAb staining. Results represent those obtained for 5 persons. (D) The ex vivo CD24hiCD27+ B-cell subset includes the majority of B10 cells. Purified B cells were cultured with LPS + PIB for 5 hours before immunofluorescence staining for cell surface CD19, CD24, and CD27 expression and cytoplasmic IL-10 expression, with subsequent flow cytometry analysis. (E) B10pro cells derive from the CD24hiCD27+ B-cell subset. Purified blood B cells were sorted into the CD24hiCD27+ and CD24lowCD27− B-cell subsets, as indicated by the gates shown with purities more than 90% when reanalyzed by flow cytometry. The purified B cells were cultured with CD40L plus either LPS or CpG for 48 hours, with PIB added during the final 5 hours of culture before the relative percentages of IL-10+ B cells within the indicated gates was determined. Similar results were obtained in 2 independent experiments. (F) Ex vivo CD24hiCD27+ B cells are the predominant source of secreted IL-10. Purified blood B cells were sorted into the CD24hiCD27+ and CD24lowCD27− B-cell subsets as in panel E and cultured with the indicated stimuli for 72 hours. IL-10 secreted into the culture supernatant fluid was quantified by ELISA. Bar graphs represent mean IL-10 (± SEM) concentrations from triplicate ELISA determinations. Significant differences between means from CD24hiCD27+ and CD24lowCD27− B cells are indicated: **P < .01. Differences between means from cells in media or with stimuli are indicated: ##P < .01. (G) B10-cell proliferation in vitro. Blood mononuclear cells were labeled with CFSE and cultured with CD40L and CPG (top panels) or CD40L and LPS (top panels) for 48 to 96 hours, with PIB added for the last 5 hours of culture. Histograms (right) represent CFSE expression by the IL-10+ (thick line) or IL-10− (thin line) B-cell subsets. Results are representative of 2 independent experiments.
Ex vivo blood B10 and B10pro cells share cell surface markers with memory B cells. (A) Blood B10 cells predominantly exhibit a CD24hiCD27+CD48hiCD148hi phenotype. Purified blood B cells were cultured with CpG + PIB for 5 hours before immunofluorescence staining for viability, cell surface molecule expression, and cytoplasmic IL-10. Cell surface CD24, CD27, CD38, CD48, and CD148 expression by IL-10+ (thick line) and IL-10− (thin line) CD19+ cells was assessed by flow cytometry. (B) Cytoplasmic IL-10 induction does not affect the cell surface phenotype of B cells. CD19+ blood B cells were cultured with media on ice (thin line) or with CpG + PIB (thick line) for 5 hours before immunofluorescence staining and flow cytometry analysis as in panel A. (A-B) Shaded histograms represent isotype-matched control mAb staining. Results represent those obtained for 3 persons. (C) Distributions of B10 cells within B-cell subsets defined by CD24, CD27, IgD/CD38, and IgD/CD27 expression. Purified blood B cells were cultured with LPS + PIB for 5 hours before immunofluorescence staining and flow cytometry analysis as in panel A. The horizontal and vertical lines on each contour plot are shown for reference, with the lower left quadrants delineating the IgD−CD38− and IgD−CD27− subsets determined by control mAb staining. Results represent those obtained for 5 persons. (D) The ex vivo CD24hiCD27+ B-cell subset includes the majority of B10 cells. Purified B cells were cultured with LPS + PIB for 5 hours before immunofluorescence staining for cell surface CD19, CD24, and CD27 expression and cytoplasmic IL-10 expression, with subsequent flow cytometry analysis. (E) B10pro cells derive from the CD24hiCD27+ B-cell subset. Purified blood B cells were sorted into the CD24hiCD27+ and CD24lowCD27− B-cell subsets, as indicated by the gates shown with purities more than 90% when reanalyzed by flow cytometry. The purified B cells were cultured with CD40L plus either LPS or CpG for 48 hours, with PIB added during the final 5 hours of culture before the relative percentages of IL-10+ B cells within the indicated gates was determined. Similar results were obtained in 2 independent experiments. (F) Ex vivo CD24hiCD27+ B cells are the predominant source of secreted IL-10. Purified blood B cells were sorted into the CD24hiCD27+ and CD24lowCD27− B-cell subsets as in panel E and cultured with the indicated stimuli for 72 hours. IL-10 secreted into the culture supernatant fluid was quantified by ELISA. Bar graphs represent mean IL-10 (± SEM) concentrations from triplicate ELISA determinations. Significant differences between means from CD24hiCD27+ and CD24lowCD27− B cells are indicated: **P < .01. Differences between means from cells in media or with stimuli are indicated: ##P < .01. (G) B10-cell proliferation in vitro. Blood mononuclear cells were labeled with CFSE and cultured with CD40L and CPG (top panels) or CD40L and LPS (top panels) for 48 to 96 hours, with PIB added for the last 5 hours of culture. Histograms (right) represent CFSE expression by the IL-10+ (thick line) or IL-10− (thin line) B-cell subsets. Results are representative of 2 independent experiments.
Because the CD24hiCD27+ B-cell subpopulation represented 24% ± 5% (n = 7) of blood B cells on average, it was assessed whether CD24 and CD27 could be used as overlapping markers to enrich for B10 cells. When purified CD24hiCD27+ and CD24lowCD27− B cells were cultured individually with LPS + PIB for 5 hours to induce IL-10 expression, B10-cell frequencies were more than 10-fold higher within the CD24hiCD27+ subpopulation compared with CD24lowCD27− B cells (Figure 5D). To determine whether ex vivo B10pro cells also localize within the CD24hiCD27+ subpopulation, purified CD24hiCD27+ and CD24lowCD27− B cells were cultured individually to induce B10pro cell maturation. Again, the frequency of IL-10+ B10 + B10pro cells was more than 10-fold higher within the CD24hiCD27+ subpopulation compared with CD24lowCD27− cells (Figure 5E). The capacity of freshly isolated CD24hiCD27+ and CD24lowCD27− B cells to secrete IL-10 was also assessed. Again, IL-10 was predominantly produced by the CD24hiCD27+ B-cell subpopulation in response to CD40L, LPS, LPS + CD40L, CpG, or CpG + CD40L stimulation for 72 hours (Figure 5F). Thus, blood B10 and B10pro cells predominantly represented a small subset of cells within the CD24hiCD27+ B-cell subpopulation.
Because B10 and B10pro cells express markers for activated and memory B cells, their ability to proliferate in response to mitogens was assessed. Purified blood B cells were labeled with CFSE before in vitro culture. There was little, if any, IL-10+ or IL-10− B-cell proliferation during 48-hour cultures regardless of whether the cells were stimulated with LPS or CpG. However, IL-10+ B cells proliferated significantly in response to CpG at 96 hours, whereas IL-10− B-cell proliferation was modest (Figure 5G). Thus, blood B10 and B10 + B10pro cells preferentially proliferated in response to mitogen stimulation, consistent with their CD24hiCD27+ phenotype.
B10 cells regulate innate immunity
Resting or activated mouse B10 cells do not significantly influence mitogen-driven T-cell activation or proliferation in vitro.22 However, activated B10 cells do inhibit TNF-α production by antigen-specific CD4+ T cells during antigen-driven in vitro assays. In vitro T-cell–B-cell coculture systems were therefore assessed to determine whether human B10 cells regulate mitogen-driven T-cell function. First, purified blood CD24hiCD27+ B cells or CD24lowCD27− B cells were stimulated with CD40L and CpG for 24 hours, washed extensively, and added to cultures of purified CD4+ T cells that were stimulated with plate-bound CD3 mAb for 72 hours. In these assays, both CD24hiCD27+ B cells and CD24lowCD27− B cells inhibited CD4+ T-cell expression of TNF-α equally (Figure 6A). Unstimulated B cells did not affect CD4+ T-cell expression of TNF-α in these assays, and the addition of anti–IL-10 mAb to cultures containing activated CD24hiCD27+ or CD24lowCD27− B cells did not rescue TNF-α expression by CD4+ T cells (data not shown). Thus, activated human B cells can inhibit mitogen-induced TNF-α production by CD4+ T cells through IL-10-independent pathways that are not unique to CD24hiCD27+ B cells.
B10-cell regulation of innate immunity. (A) B10 cell effects on mitogen-stimulated T-cell cytokine production. Purified blood CD24hiCD27+ or CD24lowCD27− B cells were stimulated with CD40L plus CpG for 24 hours, isolated, and then cultured with CD3 mAb-stimulated CD4+ T cells for 72 hours. After PMA plus ionomycin stimulation, CD4+ T-cell TNF-α expression was assessed by flow cytometry (heavy lines). CD4+ T cells cultured alone are shown as positive controls (thin lines). Background cell staining using unstimulated T cells is shown (shaded lines). (B) B10 cells regulate monocyte cytokine production. Purified blood CD24hiCD27+ or CD24lowCD27− B cells were stimulated with CD40L plus CpG for 24 hours and were cultured with blood monocytes for 20 hours before cytoplasmic TNF-α expression by CD14+ monocytes was assessed after 4 hours of LPS stimulation (heavy lines). Anti–IL-10 mAb was added to some cultures as indicated (dashed lines). Monocytes cultured alone are shown as positive controls (thin lines), with background cell staining using unstimulated monocytes shown (shaded lines). (A-B) Results represent those obtained in more than or equal to 2 independent experiments.
B10-cell regulation of innate immunity. (A) B10 cell effects on mitogen-stimulated T-cell cytokine production. Purified blood CD24hiCD27+ or CD24lowCD27− B cells were stimulated with CD40L plus CpG for 24 hours, isolated, and then cultured with CD3 mAb-stimulated CD4+ T cells for 72 hours. After PMA plus ionomycin stimulation, CD4+ T-cell TNF-α expression was assessed by flow cytometry (heavy lines). CD4+ T cells cultured alone are shown as positive controls (thin lines). Background cell staining using unstimulated T cells is shown (shaded lines). (B) B10 cells regulate monocyte cytokine production. Purified blood CD24hiCD27+ or CD24lowCD27− B cells were stimulated with CD40L plus CpG for 24 hours and were cultured with blood monocytes for 20 hours before cytoplasmic TNF-α expression by CD14+ monocytes was assessed after 4 hours of LPS stimulation (heavy lines). Anti–IL-10 mAb was added to some cultures as indicated (dashed lines). Monocytes cultured alone are shown as positive controls (thin lines), with background cell staining using unstimulated monocytes shown (shaded lines). (A-B) Results represent those obtained in more than or equal to 2 independent experiments.
Even though mouse B10-cell regulation of T-cell function may require antigen-specific in vitro assays that are not amenable to human studies, mouse monocyte cytokine production is directly regulated by B10 cells through IL-10-dependent pathways that are not antigen-specific (M.H. et al, manuscript in preparation). The ability of human B10 cells to regulate innate monocyte responses was therefore assessed. Remarkably, TNF-α production was significantly reduced when monocytes were cultured with activated blood CD24hiCD27+ B cells, and this effect was blocked by anti–IL-10 mAb (Figure 6B). The addition of anti–IL-10 mAb to monocytes cultured alone did not affect TNF-α production (data not shown), whereas CD24lowCD27− B cells variably affected monocyte TNF-α production through IL-10–independent pathways. Thus, IL-10 production by CD24hiCD27+ B cells regulates monocyte TNF-α production.
B10 cells in patients with autoimmune disease
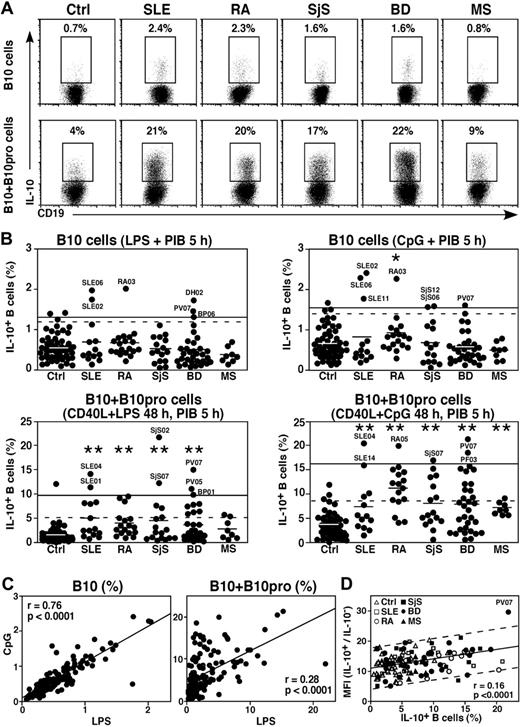
To determine whether blood B10-cell numbers are altered during inflammation and autoimmune disease, B10 and B10pro cells were examined in 91 patients with systemic lupus erythematosus, rheumatoid arthritis, primary Sjögren syndrome, autoimmune vesiculobullous skin disease, or multiple sclerosis (Figure 7A-B). Most patients were undergoing active treatment with immunomodulatory agents and/or low doses of prednisone (Table 1). B10-cell frequencies were significantly higher in one pemphigus vulgaris patient (PV07) and 1 dermatitis herpetiformis patient (DH02) not undergoing immunosuppressive therapy. Two lupus patients (SLE02, SLE06) and 1 rheumatoid arthritis patient (RA03) also had significantly higher B10-cell frequencies, but retrospective evaluation of their disease status, autoantibody profile, and treatment regimen did not indicate why these persons had higher B10-cell frequencies. No patients expressed significantly lower B10-cell frequencies than age-matched controls. Mean B10-cell frequencies were significantly higher for rheumatoid arthritis patients after culture with CpG but not for other patient groups (Figure 7B), even though B10-cell frequencies increased similarly in most cases after either LPS or CpG stimulation (Figure 7C left panel).
Blood B10-cell frequencies in patients with autoimmune disease. (A) Representative B-cell cytoplasmic IL-10 expression by control (Ctrl) persons, and lupus (SLE), rheumatoid arthritis (RA), Sjögren syndrome (SjS), blistering skin disease (BD), and multiple sclerosis (MS) patients with relatively high B10-cell frequencies after in vitro CpG + PIB stimulation for 5 hours. B10 + B10pro cell maturation was induced by 48-hour CD40L + CpG stimulation, with PIB added during the final 5 hours of culture. Percentages indicate IL-10+ B-cell frequencies among CD19+ B cells. (B) IL-10+ B-cell frequencies as in panel A, with each dot representing single persons. Horizontal bars represent group means. The solid horizontal lines represent means plus 2 SD (95% confidence interval) for controls; and dashed lines, means plus 2 SD for all values. The patients are described in Table 1. Significant differences between means of patient groups and healthy controls are indicated: *P < .05; **P < .01. (C) Relative frequencies of B10 cells and B10 + B10pro cells identified for control persons and patients with autoimmune disease as in panel B compared after CpG or LPS stimulation, with each dot representing a person. (D) Relationship between cytoplasmic IL-10 expression levels and B10 + B10pro-cell frequencies in control persons and patients after stimulation with CD40L + CpG, with PIB added during the final 5 hours of 48-hour cultures. Linear mean fluorescence intensities (MFI) for IL-10+ and IL-10− B cells were determined using the gates indicated in panel A with the values shown representing a ratio of IL-10+ to IL-10− MFIs. A linear regression line (± 95% prediction bands, dashed lines) is shown for reference.
Blood B10-cell frequencies in patients with autoimmune disease. (A) Representative B-cell cytoplasmic IL-10 expression by control (Ctrl) persons, and lupus (SLE), rheumatoid arthritis (RA), Sjögren syndrome (SjS), blistering skin disease (BD), and multiple sclerosis (MS) patients with relatively high B10-cell frequencies after in vitro CpG + PIB stimulation for 5 hours. B10 + B10pro cell maturation was induced by 48-hour CD40L + CpG stimulation, with PIB added during the final 5 hours of culture. Percentages indicate IL-10+ B-cell frequencies among CD19+ B cells. (B) IL-10+ B-cell frequencies as in panel A, with each dot representing single persons. Horizontal bars represent group means. The solid horizontal lines represent means plus 2 SD (95% confidence interval) for controls; and dashed lines, means plus 2 SD for all values. The patients are described in Table 1. Significant differences between means of patient groups and healthy controls are indicated: *P < .05; **P < .01. (C) Relative frequencies of B10 cells and B10 + B10pro cells identified for control persons and patients with autoimmune disease as in panel B compared after CpG or LPS stimulation, with each dot representing a person. (D) Relationship between cytoplasmic IL-10 expression levels and B10 + B10pro-cell frequencies in control persons and patients after stimulation with CD40L + CpG, with PIB added during the final 5 hours of 48-hour cultures. Linear mean fluorescence intensities (MFI) for IL-10+ and IL-10− B cells were determined using the gates indicated in panel A with the values shown representing a ratio of IL-10+ to IL-10− MFIs. A linear regression line (± 95% prediction bands, dashed lines) is shown for reference.
Patient characteristics
| Diagnosis no. . | Sex . | Age, y . | Disease duration, y . | Autoantibody/clinical status . | Immunosuppressive therapy . |
|---|---|---|---|---|---|
| RA01 | M | 54 | 11 | RF = 600 IU/mL; anti-CCP = 68.5 U/mL | MTX, ADA, Pred 5 mg/day |
| RA02 | F | 44 | 4 | RF = 352 IU/mL; anti-CCP>100 U/mL | MTX |
| RA03 | F | 54 | 14 | RF = 146 IU/mL | MTX, LEF |
| RA04 | F | 85 | 18 | RF = negative | MTX, IFX |
| RA05 | M | 69 | 19 | RF = 208 IU/mL; anti-CCP>100 U/mL | MTX, Pred 5 mg/day |
| RA06 | F | 71 | 8 | RF = negative | MTX |
| RA07 | F | 67 | 13 | RF = 333 IU/mL | ETN |
| RA08 | F | 58 | 25 | RF = 53 IU/mL | MTX, ETN, Pred 3 mg/day |
| RA09 | F | 68 | 7 | RF = 339 IU/mL | MTX, Pred 3 mg/day |
| RA10 | M | 75 | 13 | RF = 420 IU/mL; anti-CCP = 18.9 U/mL | MTX, LEF, Pred 10 mg/day |
| RA11 | F | 73 | 7 | RF and anti-CCP = negative | MTX, LEF |
| RA12 | M | 61 | 27 | RF = 107 IU/mL | MTX, LEF |
| RA13 | F | 66 | 6 | RF = positive | ADA |
| RA14 | F | 84 | 11 | RF = 275 IU/mL; anti-CCP = 28 U/mL | ETN |
| RA15 | F | 52 | 3 | RF = negative; anti-CCP>100 U/mL | ETN |
| RA16 | F | 76 | 18 | RF = positive | MTX |
| RA17 | M | 63 | 11 | RF = negative; anti-CCP>100 U/mL | MTX, SSZ, Pred 1 mg/day |
| RA18 | F | 43 | 15 | RF = 23 IU/mL | MTX, IFX |
| RA19 | F | 62 | 30 | RF = 148 IU/mL; anti-CCP>100 U/mL | LEF; Pred 5 mg/day |
| SLE01 | F | 65 | 11 | ANA = 1:2560; IgG CL and anti-dsDNA = positive | HCQ, LEF, Pred 5 mg/day |
| SLE02 | M | 31 | 3 | ANA = 1:640; anti-RNP, anti-Sm, and anti-Ro = positive | HCQ, Pred 3 mg/day |
| SLE03 | M | 63 | 32 | ANA = 1:640; anti-dsDNA and IgG anti-CL = positive | HCQ, Pred 5 mg/day |
| SLE04 | F | 37 | 5 | ANA = 1:2560; anti-Ro = positive | None |
| SLE05 | F | 43 | 15 | ANA = 1:160; RF = 36 IU/mL | HCQ, MMF |
| SLE06 | F | 46 | 8 | ANA = positive; anti-Ro = positive | MMF 2 g/day, Pred 10 mg/day |
| SLE07 | M | 31 | 23 | ANA = 1:160; anti-dsDNA and IgG anti-CL = positive | HCQ |
| SLE08 | F | 47 | 10 | ANA = 1:2560; anti-dsDNA, anti-Ro and anti-La = positive; RF = 103 IU/mL | None |
| SLE09 | F | 37 | 16 | ANA = positive; anti-dsDNA, anti-IgM and IgG CL = positive | HCQ; Pred 10 mg/day |
| SLE10 | F | 48 | 7 | ANA = 1:2560; anti-dsDNA, anti-Ro and anti-La = positive | Pred 5 mg/day |
| SLE11 | F | 37 | 25 | ANA = 1:640 | HCQ |
| SLE12 | F | 49 | 6 | ANA = 1:640; anti-dsDNA and anti-Ro = positive | HCQ |
| SLE13 | F | 48 | 20 | ANA = 1:640; anti-dsDNA and anti-Ro = positive | Pred 5 mg/day |
| SLE14 | F | 58 | 13 | ANA = positive; anti-dsDNA, anti-Ro and anti-La = positive | MMF, HCQ, Pred 10 mg/day |
| SjS01 | F | 52 | 1 | ANA = 1:2560; RF = 354 IU/mL; anti-Ro and anti-La = positive | None |
| SjS02 | F | 65 | 15 | ANA = 1:2560; RF = 22 IU/mL; anti-Ro = positive | HCQ |
| SjS03 | F | 57 | 37 | ANA = 1:160; anti-Ro = positive | MMF, Pred 40 mg/day |
| SjS04 | F | 67 | 22 | ANA = 1:2560; RF 110 IU/mL, anti-Ro and anti-La = positive | HCQ |
| SjS05 | F | 60 | 9 | ANA = 1:2560; RF = 126 IU/mL, anti-Ro = positive | HCQ |
| SjS06 | F | 58 | 21 | ANA = 1:2560; anti-Ro and anti-La = positive | None |
| SjS07 | F | 41 | 13 | ANA = 1:2560; anti-Ro and anti-La = positive | HCQ |
| SjS08 | F | 59 | 8 | ANA = 1:2560; RF = 508 IU/mL; anti-Ro = positive | None |
| SjS09 | F | 42 | 4 | ANA = 1:2560; RF = 110 IU/mL; anti-Ro and anti-La = positive | HCQ |
| SjS10 | F | 58 | 5 | ANA = 1:2560; anti-Ro and anti-La = positive | HCQ, Pred 3 mg/day |
| SjS11 | M | 66 | 5 | ANA = 1:2560; anti-Ro and anti-La = positive | None |
| SjS12 | F | 76 | 13 | ANA = 1:2560; anti-Ro = positive; RF = 32 IU/mL | None |
| SjS13 | M | 51 | 1 | ANA = 1:160 | HCQ |
| SjS14 | F | 68 | 28 | ANA = 1:2560; anti-Ro = positive | HCQ |
| SjS15 | F | 66 | 12 | ANA = 1:2560 | None |
| SjS16 | F | 64 | 8 | ANA = 1:2560; anti-Ro = positive | None |
| SjS17 | F | 78 | 13 | ANA = positive; anti-Ro = positive | HCQ |
| BP01 | M | 72 | 0.3 | Anti-BP180 = 84 U/mL; anti-BP230 = 115 U/mL, no clinical disease | Pred 60 mg/day |
| BP02 | M | 54 | 1.2 | Anti-BP180 = 72 U/mL; anti-BP230 = negative, no clinical disease | MMF, Pred 12 mg/day |
| BP03 | F | 56 | 2 | Anti-BP180 = 51 U/mL; anti-BP230 = negative, no clinical disease | Pred 20 mg/day |
| BP04 | M | 75 | 4.3 | Anti-BP180 = 45 U/mL; anti-BP230 = 3, mild disease | None |
| BP05 | F | 66 | 1.8 | Anti-BP180 = 96 U/mL; anti-BP230 = 131, severe disease | None |
| BP06 | M | 77 | 0.5 | Anti-BP180 = 5 U/mL; anti-BP230 = 95, mild disease | None |
| BP07 | F | 67 | 17 | Anti-BP180 = 46 U/mL; anti-BP230 = negative, mild disease | RTX (20 months earlier) |
| BP08 | F | 54 | 2 | Anti-BP180 = 22 U/mL; anti-BP230 = 7, no clinical disease | RTX (13 months earlier), Pred 15 mg/day |
| BP09 | F | 62 | 5.4 | Anti-BP180 = 30 U/mL; anti-BP230 = 3, mild disease | RTX (52 months earlier) |
| BP10 | M | 73 | 0.25 | Anti-BP180 = 222 U/mL; anti-BP230 = 7, no clinical disease | Pred 50 mg/day |
| PF01 | M | 54 | 8.6 | Anti-DSG1 = 134 U/mL; anti-DSG3 = negative, mild disease activity | AZA |
| PF02 | M | 55 | 9.8 | Anti-DSG1 = negative; anti-DSG3 = negative, mild disease activity | RTX (30 months earlier) |
| PF03 | M | 46 | 6.6 | Anti-DSG1 = 416; anti-DSG3 = negative, moderate disease activity | None |
| PF04 | M | 50 | 5.3 | Anti-DSG1 = 1906; anti-DSG3 = negative, moderate disease activity | MMF |
| PF05 | M | 72 | 2.8 | Anti-DSG1 = 118; anti-DSG3 = negative, no clinical disease | Dapsone 100 mg/day |
| PF06 | F | 47 | 0.4 | Anti-DSG1 = 211; anti-DSG3 = negative, moderate clinical disease activity | Pred 40 mg/day |
| PV02 | M | 43 | 3 | Anti-DSG1 = negative; anti-DSG3 = 213 U/mL, moderate oral disease activity | MMF, Pred 20 mg/day |
| PV03 | M | 73 | 3.3 | Anti-DSG1 = negative; anti-DSG3 = 948 U/mL, mild oral disease | Pred 12 mg/day |
| PV04 | F | 55 | 4.7 | Anti-DSG1 = negative; anti-DSG3 = 406 U/mL, moderate oral disease activity | RTX (15 months earlier) |
| PV05 | M | 59 | 8.3 | Anti-DSG1 = negative; anti-DSG3 = 50 U/mL, no disease activity | AZA |
| PV06 | F | 48 | 8.6 | Anti-DSG1 = negative; anti-DSG3 = 25 U/mL, no disease activity | None |
| PV07 | M | 45 | 0.25 | Anti-DSG1 = 968; anti-DSG3 = 735, severe disease involving 20% of the skin | MMF; Pred 80 mg/day |
| PV08 | M | 84 | 0.2 | Anti-DSG1 = 75; anti-DSG3 = 146 U/mL, moderate disease activity | None |
| PV09 | M | 64 | 9.8 | Anti-DSG1 = 14; anti-DSG3 = 115 U/mL, mild disease activity | Pred 20 mg/day; IFX |
| PV10 | M | 59 | 3.8 | Anti-DSG1 = 1; anti-DSG3 = 49 U/mL, no disease activity | AZA |
| PV11 | M | 55 | 6 | Anti-DSG1 = 34; anti-DSG3 = 35 U/mL, mild oral disease activity | Pred 20 mg/day; MMF |
| PV12 | F | 58 | 9.5 | Anti-DSG1 = 1; anti-DSG3 = 43 U/mL, mild oral disease activity | AZA |
| PV13 | M | 75 | 5 | Anti-DSG1 = 1; anti-DSG3 = 42 U/mL, mild disease | RTX (6 months earlier), Pred 1 mg/day |
| PV14 | M | 70 | 8.4 | Anti-DSG1 = 110; anti-DSG3 = 113 U/mL, no clinical disease | AZA |
| PV15 | F | 37 | 0.3 | Anti-DSG1 = 104; anti-DSG3 = 810 U/mL, moderate disease activity | Pred 40 mg/day |
| PV16 | M | 78 | 15 | Anti-DSG1 = 297; anti-DSG3 = 447 U/mL, moderate disease activity | None |
| DH1 | F | 65 | 8.4 | Gluten-free diet, no clinical disease | Dapsone 25 mg/day |
| DH2 | M | 40 | 30 | Normal diet, no clinical disease | Dapsone 175 mg/day |
| MS01 | F | 72 | 54 | SPMS, EDSS 6.5, not clinically active | None |
| MS02 | M | 62 | 24 | RRMS, EDSS 6.5, clinically active | BIFN |
| MS03 | M | 33 | 2 | RRMS, EDSS 1.0, disease not clinically active | BIFN |
| MS04 | M | 75 | 29 | SPMS, EDSS 8.0, disease not clinically active | ITMTX |
| MS05 | M | 52 | 24 | PPMS, EDSS 6.5, disease clinically active | MMF, pulse steroids |
| MS06 | M | 55 | 25 | PPMS, EDSS 7.5, disease clinically active | ITMTX |
| MS07 | F | 39 | 16 | SPMS, EDSS 7.0, disease not clinically active | Natalizumab (2 months earlier) |
| MS08 | F | 51 | 7 | SPMS, EDSS 5.5, disease not clinically active | BIFN |
| Diagnosis no. . | Sex . | Age, y . | Disease duration, y . | Autoantibody/clinical status . | Immunosuppressive therapy . |
|---|---|---|---|---|---|
| RA01 | M | 54 | 11 | RF = 600 IU/mL; anti-CCP = 68.5 U/mL | MTX, ADA, Pred 5 mg/day |
| RA02 | F | 44 | 4 | RF = 352 IU/mL; anti-CCP>100 U/mL | MTX |
| RA03 | F | 54 | 14 | RF = 146 IU/mL | MTX, LEF |
| RA04 | F | 85 | 18 | RF = negative | MTX, IFX |
| RA05 | M | 69 | 19 | RF = 208 IU/mL; anti-CCP>100 U/mL | MTX, Pred 5 mg/day |
| RA06 | F | 71 | 8 | RF = negative | MTX |
| RA07 | F | 67 | 13 | RF = 333 IU/mL | ETN |
| RA08 | F | 58 | 25 | RF = 53 IU/mL | MTX, ETN, Pred 3 mg/day |
| RA09 | F | 68 | 7 | RF = 339 IU/mL | MTX, Pred 3 mg/day |
| RA10 | M | 75 | 13 | RF = 420 IU/mL; anti-CCP = 18.9 U/mL | MTX, LEF, Pred 10 mg/day |
| RA11 | F | 73 | 7 | RF and anti-CCP = negative | MTX, LEF |
| RA12 | M | 61 | 27 | RF = 107 IU/mL | MTX, LEF |
| RA13 | F | 66 | 6 | RF = positive | ADA |
| RA14 | F | 84 | 11 | RF = 275 IU/mL; anti-CCP = 28 U/mL | ETN |
| RA15 | F | 52 | 3 | RF = negative; anti-CCP>100 U/mL | ETN |
| RA16 | F | 76 | 18 | RF = positive | MTX |
| RA17 | M | 63 | 11 | RF = negative; anti-CCP>100 U/mL | MTX, SSZ, Pred 1 mg/day |
| RA18 | F | 43 | 15 | RF = 23 IU/mL | MTX, IFX |
| RA19 | F | 62 | 30 | RF = 148 IU/mL; anti-CCP>100 U/mL | LEF; Pred 5 mg/day |
| SLE01 | F | 65 | 11 | ANA = 1:2560; IgG CL and anti-dsDNA = positive | HCQ, LEF, Pred 5 mg/day |
| SLE02 | M | 31 | 3 | ANA = 1:640; anti-RNP, anti-Sm, and anti-Ro = positive | HCQ, Pred 3 mg/day |
| SLE03 | M | 63 | 32 | ANA = 1:640; anti-dsDNA and IgG anti-CL = positive | HCQ, Pred 5 mg/day |
| SLE04 | F | 37 | 5 | ANA = 1:2560; anti-Ro = positive | None |
| SLE05 | F | 43 | 15 | ANA = 1:160; RF = 36 IU/mL | HCQ, MMF |
| SLE06 | F | 46 | 8 | ANA = positive; anti-Ro = positive | MMF 2 g/day, Pred 10 mg/day |
| SLE07 | M | 31 | 23 | ANA = 1:160; anti-dsDNA and IgG anti-CL = positive | HCQ |
| SLE08 | F | 47 | 10 | ANA = 1:2560; anti-dsDNA, anti-Ro and anti-La = positive; RF = 103 IU/mL | None |
| SLE09 | F | 37 | 16 | ANA = positive; anti-dsDNA, anti-IgM and IgG CL = positive | HCQ; Pred 10 mg/day |
| SLE10 | F | 48 | 7 | ANA = 1:2560; anti-dsDNA, anti-Ro and anti-La = positive | Pred 5 mg/day |
| SLE11 | F | 37 | 25 | ANA = 1:640 | HCQ |
| SLE12 | F | 49 | 6 | ANA = 1:640; anti-dsDNA and anti-Ro = positive | HCQ |
| SLE13 | F | 48 | 20 | ANA = 1:640; anti-dsDNA and anti-Ro = positive | Pred 5 mg/day |
| SLE14 | F | 58 | 13 | ANA = positive; anti-dsDNA, anti-Ro and anti-La = positive | MMF, HCQ, Pred 10 mg/day |
| SjS01 | F | 52 | 1 | ANA = 1:2560; RF = 354 IU/mL; anti-Ro and anti-La = positive | None |
| SjS02 | F | 65 | 15 | ANA = 1:2560; RF = 22 IU/mL; anti-Ro = positive | HCQ |
| SjS03 | F | 57 | 37 | ANA = 1:160; anti-Ro = positive | MMF, Pred 40 mg/day |
| SjS04 | F | 67 | 22 | ANA = 1:2560; RF 110 IU/mL, anti-Ro and anti-La = positive | HCQ |
| SjS05 | F | 60 | 9 | ANA = 1:2560; RF = 126 IU/mL, anti-Ro = positive | HCQ |
| SjS06 | F | 58 | 21 | ANA = 1:2560; anti-Ro and anti-La = positive | None |
| SjS07 | F | 41 | 13 | ANA = 1:2560; anti-Ro and anti-La = positive | HCQ |
| SjS08 | F | 59 | 8 | ANA = 1:2560; RF = 508 IU/mL; anti-Ro = positive | None |
| SjS09 | F | 42 | 4 | ANA = 1:2560; RF = 110 IU/mL; anti-Ro and anti-La = positive | HCQ |
| SjS10 | F | 58 | 5 | ANA = 1:2560; anti-Ro and anti-La = positive | HCQ, Pred 3 mg/day |
| SjS11 | M | 66 | 5 | ANA = 1:2560; anti-Ro and anti-La = positive | None |
| SjS12 | F | 76 | 13 | ANA = 1:2560; anti-Ro = positive; RF = 32 IU/mL | None |
| SjS13 | M | 51 | 1 | ANA = 1:160 | HCQ |
| SjS14 | F | 68 | 28 | ANA = 1:2560; anti-Ro = positive | HCQ |
| SjS15 | F | 66 | 12 | ANA = 1:2560 | None |
| SjS16 | F | 64 | 8 | ANA = 1:2560; anti-Ro = positive | None |
| SjS17 | F | 78 | 13 | ANA = positive; anti-Ro = positive | HCQ |
| BP01 | M | 72 | 0.3 | Anti-BP180 = 84 U/mL; anti-BP230 = 115 U/mL, no clinical disease | Pred 60 mg/day |
| BP02 | M | 54 | 1.2 | Anti-BP180 = 72 U/mL; anti-BP230 = negative, no clinical disease | MMF, Pred 12 mg/day |
| BP03 | F | 56 | 2 | Anti-BP180 = 51 U/mL; anti-BP230 = negative, no clinical disease | Pred 20 mg/day |
| BP04 | M | 75 | 4.3 | Anti-BP180 = 45 U/mL; anti-BP230 = 3, mild disease | None |
| BP05 | F | 66 | 1.8 | Anti-BP180 = 96 U/mL; anti-BP230 = 131, severe disease | None |
| BP06 | M | 77 | 0.5 | Anti-BP180 = 5 U/mL; anti-BP230 = 95, mild disease | None |
| BP07 | F | 67 | 17 | Anti-BP180 = 46 U/mL; anti-BP230 = negative, mild disease | RTX (20 months earlier) |
| BP08 | F | 54 | 2 | Anti-BP180 = 22 U/mL; anti-BP230 = 7, no clinical disease | RTX (13 months earlier), Pred 15 mg/day |
| BP09 | F | 62 | 5.4 | Anti-BP180 = 30 U/mL; anti-BP230 = 3, mild disease | RTX (52 months earlier) |
| BP10 | M | 73 | 0.25 | Anti-BP180 = 222 U/mL; anti-BP230 = 7, no clinical disease | Pred 50 mg/day |
| PF01 | M | 54 | 8.6 | Anti-DSG1 = 134 U/mL; anti-DSG3 = negative, mild disease activity | AZA |
| PF02 | M | 55 | 9.8 | Anti-DSG1 = negative; anti-DSG3 = negative, mild disease activity | RTX (30 months earlier) |
| PF03 | M | 46 | 6.6 | Anti-DSG1 = 416; anti-DSG3 = negative, moderate disease activity | None |
| PF04 | M | 50 | 5.3 | Anti-DSG1 = 1906; anti-DSG3 = negative, moderate disease activity | MMF |
| PF05 | M | 72 | 2.8 | Anti-DSG1 = 118; anti-DSG3 = negative, no clinical disease | Dapsone 100 mg/day |
| PF06 | F | 47 | 0.4 | Anti-DSG1 = 211; anti-DSG3 = negative, moderate clinical disease activity | Pred 40 mg/day |
| PV02 | M | 43 | 3 | Anti-DSG1 = negative; anti-DSG3 = 213 U/mL, moderate oral disease activity | MMF, Pred 20 mg/day |
| PV03 | M | 73 | 3.3 | Anti-DSG1 = negative; anti-DSG3 = 948 U/mL, mild oral disease | Pred 12 mg/day |
| PV04 | F | 55 | 4.7 | Anti-DSG1 = negative; anti-DSG3 = 406 U/mL, moderate oral disease activity | RTX (15 months earlier) |
| PV05 | M | 59 | 8.3 | Anti-DSG1 = negative; anti-DSG3 = 50 U/mL, no disease activity | AZA |
| PV06 | F | 48 | 8.6 | Anti-DSG1 = negative; anti-DSG3 = 25 U/mL, no disease activity | None |
| PV07 | M | 45 | 0.25 | Anti-DSG1 = 968; anti-DSG3 = 735, severe disease involving 20% of the skin | MMF; Pred 80 mg/day |
| PV08 | M | 84 | 0.2 | Anti-DSG1 = 75; anti-DSG3 = 146 U/mL, moderate disease activity | None |
| PV09 | M | 64 | 9.8 | Anti-DSG1 = 14; anti-DSG3 = 115 U/mL, mild disease activity | Pred 20 mg/day; IFX |
| PV10 | M | 59 | 3.8 | Anti-DSG1 = 1; anti-DSG3 = 49 U/mL, no disease activity | AZA |
| PV11 | M | 55 | 6 | Anti-DSG1 = 34; anti-DSG3 = 35 U/mL, mild oral disease activity | Pred 20 mg/day; MMF |
| PV12 | F | 58 | 9.5 | Anti-DSG1 = 1; anti-DSG3 = 43 U/mL, mild oral disease activity | AZA |
| PV13 | M | 75 | 5 | Anti-DSG1 = 1; anti-DSG3 = 42 U/mL, mild disease | RTX (6 months earlier), Pred 1 mg/day |
| PV14 | M | 70 | 8.4 | Anti-DSG1 = 110; anti-DSG3 = 113 U/mL, no clinical disease | AZA |
| PV15 | F | 37 | 0.3 | Anti-DSG1 = 104; anti-DSG3 = 810 U/mL, moderate disease activity | Pred 40 mg/day |
| PV16 | M | 78 | 15 | Anti-DSG1 = 297; anti-DSG3 = 447 U/mL, moderate disease activity | None |
| DH1 | F | 65 | 8.4 | Gluten-free diet, no clinical disease | Dapsone 25 mg/day |
| DH2 | M | 40 | 30 | Normal diet, no clinical disease | Dapsone 175 mg/day |
| MS01 | F | 72 | 54 | SPMS, EDSS 6.5, not clinically active | None |
| MS02 | M | 62 | 24 | RRMS, EDSS 6.5, clinically active | BIFN |
| MS03 | M | 33 | 2 | RRMS, EDSS 1.0, disease not clinically active | BIFN |
| MS04 | M | 75 | 29 | SPMS, EDSS 8.0, disease not clinically active | ITMTX |
| MS05 | M | 52 | 24 | PPMS, EDSS 6.5, disease clinically active | MMF, pulse steroids |
| MS06 | M | 55 | 25 | PPMS, EDSS 7.5, disease clinically active | ITMTX |
| MS07 | F | 39 | 16 | SPMS, EDSS 7.0, disease not clinically active | Natalizumab (2 months earlier) |
| MS08 | F | 51 | 7 | SPMS, EDSS 5.5, disease not clinically active | BIFN |
Normal values are as follows: anti-BP = 180, anti-BP = 230, anti-DSG1 and anti-DSG3 antibodies < 9 IU/mL.
RA indicates rheumatoid arthritis; RF, rheumatoid factor; CCP, cyclic citrullinated peptide; MTX, methotrexate; ADA, adalimumab; Pred, prednisone; LEF, leflunomide; IFX, infliximab; ETN, etanercept; SLE, systemic lupus erythematosus; SSZ, sulfasalazine; ANA, antinuclear antibody; CL, cardiolipin; dsDNA, double-stranded DNA; HCQ, hydroxychloroquine; RNP, ribonucleoprotein; MMF, mycophenolate mofetil; SjS, primary Sjögren syndrome; BP, bullous pemphigoid; RTX, rituximab; DSG, desmoglein; PF, pemphigus foliaceus; PV, pemphigus vulgaris; AZA, azathioprine; DH, dermatitis herpetiformis; SPMS, secondary progressive multiple sclerosis; EDSS, disability scale (from 0 = normal to 10 = death); RRMS, relapsing remitting multiple sclerosis; BIFN, β-interferon; ITMTX, intrathecal methotrexate; and PPMS, primary progressive multiple sclerosis.
Mean B10 + B10pro-cell frequencies from patients with autoimmune disease were significantly higher than controls after either CD40L + LPS or CD40L + CpG stimulation (Figure 7A-B). Multiple patients had significantly higher B10 + B10pro-cell frequencies, including 2 patients not undergoing therapy (SLE04, PF03; Table 1). No patients expressed significantly lower B10 + B10pro-cell frequencies relative to age-matched controls. B10 + B10pro-cell frequencies increased after either LPS or CPG stimulation, but the scatter of the results was broad, suggesting inherently different patient sensitivities to LPS and CpG stimulation (Figure 7C right panel). Patients with high blood B10-cell frequencies did not necessarily have high B10 + B10pro-cell frequencies after either LPS or CpG stimulation (data not shown). Likewise, B10 or B10 + B10pro-cell frequencies did not correlate with CD27+ B-cell frequencies (data not shown). Relative B10 and B10 + B10pro-cell frequencies did correlate with the intensity of cytoplasmic IL-10 expression, but only one patient generated significantly higher (P < .05) cytoplasmic IL-10 expression levels on a per-cell basis relative to controls and other patients (Figure 7D). Thus, blood B10 and B10pro cell numbers were not decreased in patients with systemic or organ-specific autoimmune disease compared with healthy controls but were significantly increased in some patients.
Discussion
These studies demonstrate the existence of human IL-10–competent B10 cells, which were readily identified by their ability to express cytoplasmic IL-10 after in vitro stimulation for 5 hours (Figure 1). Moreover, IL-10 production by human B10 cells regulated cytokine production by monocytes in vitro, demonstrating a functional link between regulatory B cells and the innate immune system (Figure 6B). Peripheral blood B10-cell frequencies were characteristically low in most persons, consistent with their low frequencies in mice. Human B10pro cells were also identified at low frequencies by their ability to express IL-10 after in vitro maturation during 48-hour cultures. Remarkably, the adaptive and innate activation pathways that induced human B10 and B10pro cell generation, maturation, cytoplasmic IL-10 expression, and IL-10 secretion were similar to those used to characterize mouse regulatory B10 cells. Specifically, human B10 cells responded to phorbol ester/ionomycin, LPS, and CpG stimulation, with B10pro cell maturation in response to CD40-, LPS-, and CpG-induced signals. Previous studies of IL-10 production by human B cells have predominantly studied bulk B-cell populations using stimulation and assay conditions that were not optimized for quantifying or characterizing individual B cells that were competent to express IL-10.25,26,39-43 Nonetheless, the results herein demonstrate that rare B10 and B10pro cells that are competent to express IL-10 exist in human blood and can be quantified in vitro.
IL-10–competence remains the best phenotypic marker for defining human B10 cells. However, freshly isolated blood B10 and B10pro cells were also predominantly CD24hiCD27+, with approximately 60% also expressing CD38 (Figure 5A,C). Others have found similar total numbers of IL-10+ B cells in the CD24hiCD38hi and CD24intCD38int B-cell fractions,28 in agreement with the current findings (Figure 5C). B10 cells also expressed CD48 and CD148 at high levels (Figure 5A). CD48 is up-regulated on activated B cells,44 and CD148 is considered a marker for human memory B cells.45 CD27 expression is also a well-characterized marker for memory B cells, although some memory B cells may be CD27−.38,46,47 The CD27+ B-cell subpopulation can also expand during the course of autoimmunity and may serve as a marker for disease activity.38,47 However, B10-cell frequencies did not parallel the size of the blood CD27+ memory B-cell pool in normal donors or patients (data not shown). Thus, the CD24hiCD27+ phenotype of B10 and B10pro cells may indicate their selection into the memory B-cell pool during development, or they may represent a distinct B-cell subset that shares common cell surface markers with memory B cells. Consistent with a memory phenotype, the proliferative capacity of blood B10 cells in response to mitogen stimulation was higher than that for other B cells (Figure 5F), as is seen for mouse B10 cells.21 Moreover, IL-10 was also predominantly secreted by ex vivo CD24hiCD27+ B cells (Figure 5G). However, most CD24lowCD27− B cells were not IL-10 competent, even after 48 hours of LPS or CpG stimulation along with CD40 ligation. Human transitional B cells are also rare (2%-3% of B cells) in adult blood and are generally CD10+CD24hiCD38hi cells that are also IgD+CD27−.48,49 Given that CD10 expression is a well-accepted marker for most cells within the transitional B-cell pool,50 its absence on B10 cells suggests that these cells are not recent emigrants from the bone marrow. Thereby, in addition to inherent IL-10 competence, B10 cells demonstrate elevated proliferative responses that may reflect prior antigen stimulation.
Because IL-10 is critical for B cell–regulatory activity in mice, the current studies demonstrate that B10 cells were functionally competent to express IL-10 in healthy persons and 91 autoimmune disease patients (Figure 7A-B). Blood B10-cell frequencies in most patients with lupus, rheumatoid arthritis, Sjögren syndrome, autoimmune skin disease, and multiple sclerosis were not significantly different from those observed in healthy controls, although mean B10 + B10pro-cell frequencies were significantly increased. Multiple patients also had significantly higher B10 and/or B10 + B10pro-cell frequencies, including systemic lupus and pemphigus patients with either untreated or severe disease (Table 1). Consistent with this, IL-10 production by blood B cells is reported to be higher in patients with rheumatoid arthritis, lupus, and systemic sclerosis.23,24,26 Moreover, elevated B10/B10pro-cell frequencies in humans parallel what has been found during inflammation12,13 and autoimmunity in mice.14,21 Although patient cohorts with recent-onset disease and clinically active disease across multiple organ systems will be needed to fully assess the relationship of blood B10-cell numbers with clinical, laboratory, and treatment status, none of the patients or patient groups had significantly lower blood B10-cell numbers than age-matched healthy controls. Because multiple sclerosis and lupus patient's B cells are reported to produce decreased amounts of IL-10,27,28 the current studies demonstrate that a careful enumeration of IL-10–competent B10-cell frequencies within patients will be required to interpret experimental results, particularly when mixed populations of B cells are assayed functionally. Blair et al have also functionally characterized human blood CD24hiCD38hi B cells stimulated in vitro with CD40L expressing CHO cells and shown that they reduce CD4+ T-cell expression of interferon-γ and TNF-α after T-cell stimulation with CD3 mAb.28 By contrast, we find that both activated CD24hiCD27+ and CD24lowCD27− B cells reduce TNF-α expression after CD4+ T-cell stimulation with CD3 mAb through IL-10–independent pathways (Figure 6A). Nonetheless, we have found that mouse spleen CD1dhiCD5+ B cells can inhibit CD4+ T-cell interferon-γ and TNF-α expression, which is completely dependent on IL-10 expression, but this requires B10-cell activation and is only observed with antigen-specific T-cell activation.22 Furthermore, mouse spleen CD1dhiCD5+ B cells regulate the antigen-presenting capacity of dendritic cells in vitro and can thereby also regulate CD4+ T-cell activation indirectly.22 Moreover, the ability of human B10 cells to influence innate monocyte function (Figure 6B) expands their regulatory role during immunity and disease. Thus, it is probable that IL-10 produced by human B10 cells will have pleiotrophic regulatory effects on the immune system, as occurs in mice.
In conclusion, the current findings demonstrate the existence of a small but significant subset of CD24hiCD27+ B cells that is preprogrammed in vivo to express IL-10 after ex vivo maturation/stimulation. Monitoring the numbers and ability of individual B10 cells to produce IL-10 will become even more informative once the in vivo physiologic triggers of B10 cell-regulatory activity are identified. The identification of antigen-specific B10 cells may also facilitate a further understanding of their relevance to immune responsiveness because antigen receptor specificity is important for mouse B10-cell development and in vivo functional activity in the regulation of inflammation and autoimmunity.8,12,13
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Robert Streilein, Edna Scarlett, and Yen-Yu Lin for help with patient procurement and analysis, Jacquie Bryant for statistical analysis, and Dr Max Cooper for helpful advice.
This work was supported by the National Institutes of Health (AI56363) and the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (grant U54 AI057157, T.F.T.; grants CA122645 and CA130805, S.H.B.; and grants CA132110 and HL091749, P.M.S.).
National Institutes of Health
Authorship
Contribution: Y.I., T.M., M.H., D.J.D., K.Y., G.M.V., and T.F.T. made substantial contributions to conception and design of the study and acquisition of the data, and drafted the article; P.M.S., S.H.B., C.M.M., A.D.W., R.P.H., and E.W.S.C. provided critical patient materials, information, and input into the design of the study; and all authors reviewed the data and manuscript critically for intellectual content, analysis, and interpretation of the data, and have given final approval of the version to be published.
Conflict-of-interest disclosure: T.F.T. is a paid consultant for MedImmune Inc and a paid consultant and shareholder for Angelica Therapeutics Inc. The remaining authors declare no competing financial interests.
Correspondence: Thomas F. Tedder, Department of Immunology, Box 3010, Duke University Medical Center, Durham, NC 27710; e-mail: thomas.tedder@duke.edu.
References
Author notes
Y.I. and T.M. contributed equally to this study and share first authorship.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal