Abstract
In a recessive ENU mutagenesis screen for embryonic lethality, we identified a mouse pedigree with a missense mutation of SHIP1 (SHIP1el20) leading to an amino acid substitution I641T in the inositol-5′-phosphatase domain that represses phosphatidylinositol-3-kinase signaling. Despite detectable expression of functional SHIP1 protein, the phenotype of homozygous SHIP1el20/el20 mice was more severe than gene-targeted SHIP1-null (SHIP1−/−) mice. Compared with age-matched SHIP1−/− mice, 5-week-old SHIP1el20/el20 mice had increased myeloid cells, serum IL-6 levels, marked reductions in lymphoid cells, and died by 7 weeks of age with infiltration of the lungs by activated macrophages. Bone marrow transplantation demonstrated that these defects were hematopoietic-cell-autonomous. We show that the el20 mutation reduces expression in SHIP1el20/el20 macrophages of both SHIP1 and s-SHIP, an isoform of SHIP1 generated by an internal promoter. In contrast, SHIP1−/− macrophages express normal levels of s-SHIP. Compound heterozygous mice (SHIP1−/el20) had the same phenotype as SHIP1−/− mice, thus providing genetic proof that the more severe phenotype of SHIP1el20/el20 mice is probably the result of concomitant loss of SHIP1 and s-SHIP. Our results suggest that s-SHIP synergizes with SHIP1 for suppression of macrophage activation, thus providing the first evidence for a role of s-SHIP in adult hematopoiesis.
Introduction
Inositol-5′-phosphatases play a critical role in negative regulation of a diverse range of cellular processes, including cell proliferation, survival, and activation. Accordingly, loss of function of these phosphatases is implicated in many disease states, such as autoimmune disease, allergy, and cancer.1 Germline deletion in the mouse of the hematopoietic-restricted Src homology 2-containing inositol-5′-phosphatase (SHIP1) leads to hyperactive myeloid cells and a shortened life span because of lung infiltration by activated macrophages.2-4 SHIP1 expression in myeloid cells is particularly important for regulating endotoxin tolerance.5 Studies of SHIP1−/− mice have also demonstrated important roles for SHIP1 in regulating the development and function of B and T cells, mast cells, and basophils.6-12 However, the analysis of the cellular abnormalities observed in SHIP1−/− mice is complicated by the inflammatory response, which can lead to secondary effects, such as loss of B cells through increased production of IL-6.12,13 Tissue-specific deletion of SHIP1 in macrophages has suggested that loss of B cells in SHIP1−/− mice may be secondary to the inflammatory environment.14,15
The SHIP1 phosphatase domain inhibits the phosphatidylinositol-3-kinase (PI3K) pathway by hydrolyzing the 5′-phosphate group of the second messenger phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3), a lipid that promotes cell survival and proliferation through activation of downstream pleckstrin homology domain-containing enzymes, such as AKT and BTK.16 Consistent with the 5′-phosphatase function of SHIP1, macrophages and B cells from SHIP1−/− mice have increased PI3K signaling as measured by phosphorylation of Akt at Ser473. SHIP1 can also inhibit the Ras/mitogen-activated protein kinase and nuclear factor-κB pathways either by cross-talk with the PI3K pathway or by recruitment of adaptor proteins, such as Shc, Grb2, and Doks to its N-terminal SH2 domain or the C-terminal NPXY motifs and proline-rich regions.17,18 Both the phosphatase and PI3K-independent activities of SHIP1 may be important for regulation of the inflammatory response.19,20
Multiple isoforms of SHIP1 exist, including one identified in embryonic stem cells (s-SHIP) that is transcribed from an internal promoter driving expression in stem and progenitor cells.21-23 s-SHIP lacks the N-terminal SH2 domain required for binding to Shc but retains the catalytic phosphatase domain and can bind to Grb2, presumably via its C-terminal proline-rich region.16,21 Importantly, SHIP1−/− mice generated by targeting of exon 1 retain expression of s-SHIP; therefore, it has been postulated that s-SHIP expression might moderate the phenotype of SHIP1−/− mice.24,25 However, as yet, a function for s-SHIP in adult hematopoiesis has not been determined.
In an ENU mutagenesis screen, we have identified a point mutant of SHIP1 that results in loss of expression of both SHIP1 and s-SHIP. Mice lacking both SHIP1 and s-SHIP have a severe inflammatory phenotype with macrophage infiltration of the lungs and lethality by 7 weeks of age. This provides the first functional evidence that s-SHIP synergizes with SHIP1 for suppression of macrophage activation.
Methods
Gene mapping and mice
A G3 pedigree with anemia and shortened life span was identified in an ENU mutagenesis screen on a 129Sv background established as previously described.26 Mapping of the mutation was performed by breeding with wild-type C57BL/6J mice. Genomic DNA was collected from affected and unaffected G3 mice on weaning and genotyped using a panel of simple sequence-length polymorphism markers. The candidate interval for the mutation was refined to a 3-Mb region of chromosome 1. Subsequent sequencing of candidate genes led to identification of a T to C base substitution at nucleotide position 2099 of the Inpp5 days gene (GenBank Accession NM_010566), which encodes the 5′-phosphatase SHIP1. SHIP1−/− mice were also used and have been described.12 All mice used for phenotypic analyses were backcrossed onto a C57BL/6J background for at least 5 generations. For repopulation assays, female CD45.1 (B6.SJL(Ptprca[Ly5.1])) mice were purchased from the Walter and Eliza Hall Institute of Medical Research Animal House Facility. The Animal Ethics Committees of the Walter and Eliza Hall Institute and University of Melbourne approved all animal procedures.
Genotyping
SHIP1 point mutant mice were genotyped by an initial polymerase chain reaction (PCR) using primers: forward, (5′-GACCAACTGCTCCTGGAGAG-3′) and reverse, (5′-TCTGCTTCGTGTATGCATACTTG-3′); followed by a restriction digest of the PCR product with DpnII to isolate the T2099C base substitution. SHIP1−/− mice were genotyped by 2-allele, 3-primer PCR using primers: forward, (5′-GCCTACGCACTCTGCGTG-3′), reverse, (5′-CTCTGTCCCTGCTCCTGC-3′) and Mk3.3 (5′-GACCGCTTCCTCGTGCTTTACGGT-3′).
Sample collection and preparation
For flow cytometric analysis, bone marrow cells were harvested by flushing femurs and tibiae with mouse tonicity-phosphate-buffered saline/2% fetal calf serum and running eluant through a 40-μm nylon strainer. Whole organs were filtered through a 40-μm nylon strainer to generate single-cell suspensions. For whole blood counts, 250 μL of blood was collected from the retro-orbital plexus into tubes containing potassium ethylenediaminetetraacetic acid (Sarstedt) and analyzed using an Advia 2120 automated hematologic analyzer (Bayer). For serum analysis, peripheral blood was obtained immediately after death by cardiac puncture and serum prepared using standard procedures. Alveolar macrophages were collected from 4- to 6-week-old mice by bronchoalveolar lavage (BAL). The cellular component of BAL was then lyzed immediately in 100mM NaCl, 10mM Tris-HCl (pH 7.5), 1% glycerol, 2mM ethylenediaminetetraacetic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 50mM NaF, 1mM pervanadate, and Complete protease inhibitors (Roche Diagnostics) for further biochemical studies.
Flow cytometric analysis and sorting
All antibodies used were purchased from BD Biosciences PharMingen or Invitrogen Biosource and were: FITC-conjugated B220 (RA3-6B2), CD8a (53-6.7), CD45.2(104), CD71 (C2/C2F2), IgM (AF6-78), Ly6C (AL-21), and Sca-1 (E13-161-7); phycoerythrin-conjugated B220 (RA3-6B2), CD3 (145-2C11), CD4 (GK1.5), CD19 (1D3), CD45 (30-F11), CD45.1 (A20), c-Kit (ACK45), Ly6G/Gr-1 (1A8), Mac-1a (M1/70), and ter119 (TER-119); BIO-conjugated B220 (RA3-6B2), CD3a (145-2C11), CD8a (53-6.7), CD25 (7D4), Ly6G/Gr-1 (1A8), Mac-1a (M1/70), and ter119 (TER-119); and allophycocyanin-conjugated B220 (RA3-6B2), CD4 (OX-35), and c-Kit (2B8). Second-stage fluorescent reagents used were either SAv-allophycocyanin or SAv-peridinin chlorophyll protein-Cy5.5. The appropriate conjugated rat antimouse isotypes were used as negative controls. Cell viability was determined by exclusion of propidium iodide-positive cells (Sigma-Aldrich). Antibody-labeled cell suspensions were collected on a FACSCalibur flow cytometer (BD Biosciences) or sorted using a FACSVantage SE system (BD Biosciences) and CellQuest software Version 5.2.1 (BD Biosciences). For isolation of lineage−c-Kit+Sca-1+ (LKS) and lineage−c-Kit+Sca-1− (LK) cell fractions, cell sorting of immature BM cells and immunomagnetic bead depletion of lineage marker+ cells were performed before sorting. All data were analyzed using a Walter and Eliza Hall Institute of Medical Research in-house program, Weasel Version 2.6.2.
Transplantation assays
Bone marrow cells from 3-week-old SHIP1+/+ and SHIP1el20/el20 mice were harvested for transplantation by flushing femurs and tibiae with MT-PBS/5% fetal calf serum. Viability of single-cell suspensions was checked by trypan blue exclusion. For competitive transplantations, age- and sex-matched B6.SJL mice (Ly5.1) were used as recipient mice. Recipients were lethally irradiated with 2 doses of 5.5 Gy administered 3 hours apart from a 60Co γ source at a dose rate of 0.45 Gy/minute 2 to 4 hours before transplantation, and transplanted with 2 × 106 donor cells simultaneously with 2 × 106 competitor (Ly5.1) BM cells. A minimum of 3 recipients was used for each donor inoculum. For reciprocal transplantations, 3-week-old SHIP1+/+ and SHIP1el20/el20 mice were sublethally irradiated with 2 doses of 3.3 Gy and transplanted with 1.5 × 106 Ly5.1 BM cells. All transplanted mice were fed antibiotic-treated water and housed in hooded cages for the first 3 weeks.
5′-Phosphatase assays
For PtdIns(3,4,5)P3 5′-phosphatase assays, full-length SHIP1 and SHIP1el20 cDNA sequences were subcloned into the pCGN mammalian expression vector in-frame with the N-terminal HA-tag, after which the resultant recombinant plasmids overexpressed in COS-1 cells. Forty-eight hours after transfection, cell lysates were prepared and recombinant proteins immunoprecipitated using HA antibodies (Sigma-Aldrich). 5′-Phosphatase assays were performed on immunoprecipitates as described previously,27 and parallel immunoprecipitates were immunoblotted with HA antibodies.
Quantitative PCR
Total RNA was extracted from organs, sorted cells, and cultured bone marrow-derived macrophages (BMMΦs) using TRIzol (Invitrogen), and genomic DNA removed using Turbo DNA-free kit (Ambion). Reverse transcription of 1 μg of RNA into cDNA was carried out in 20-μL reactions using the Transcriptor First-strand cDNA Synthesis kit (Roche Diagnostics) with Anchored-oligo(dT)18 primers as per the manufacturer's instructions. Quantitative PCR was performed in a LightCycler 480 II (Roche Diagnostics) using 10 μL of 2× SYBR Green PCR Master Mix (Applied Biosystems), 1 μL of 10μM specific primers, and 1 μL of cDNA. Primer sequences used were as follows: hypoxanthine phosphoribosyl transferase (HPRT): forward, 5′-GCTGGTGAAAAGGACCTCT-3′; reverse, 5′-CACAGGACTAGAACACCTGC-3′ (248 bp); SHIP1: forward, 5′-GGCTCCAGCAACCTCCCTCAC-3′; reverse, 5′-TTCTCC GTCTCCACCAAAATCACC-3′ (491 bp); and s-SHIP: forward, 5′-GTTCCCACTAGATGTTGA ACTTTAC-3′; reverse, 5′-TCTCCTTCTCCGTCTCCACC-3′ (442 bp). For HPRT, amplification was performed at 95°C for 5 minutes, followed by 40 cycles of 15 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 82°C. For SHIP1 and s-SHIP, touchdown amplification was performed at 94°C for 5 minutes, followed by 43 cycles of 15 seconds at 94°C, 30 seconds at 62°C to 42°C (decreasing by 0.5°C/cycle) and 30 seconds at 72°C. All results were normalized to HPRT and expressed as relative gene expression for each target gene.
Histologic analysis
Whole lungs were fixed in 10% neutral buffered formalin and embedded in paraffin for sectioning. Sections were prepared and stained with hematoxylin and eosin at the Walter and Eliza Hall Institute. Images were acquired using a Nikon Eclipse 80i; microscope equipped with a 40×/0.75 NA objective lens. Images were captured with a Nikon XM1200C digital camera, and figures were created using Adobe Photoshop CS4, Version 11.0.1 (Adobe Systems Inc).
IL-6 ELISA
Serum samples and tissue culture supernatants of resting or stimulated BMMΦs were assessed for protein levels of IL-6 by ELISA (BD Biosciences) according to the manufacturer's instructions.
Tissue culture
To obtain BMMΦs, bone marrow cells were sterilely collected from femurs and tibiae of 4- to 6-week-old mice and allowed to adhere overnight at 37°C, 5% CO2 in BMMΦ medium (DMEM + 15% fetal calf serum + penicillin/streptomycin + 20% L cell conditioned medium [as a source of macrophage colony stimulating factor]). Nonadherent cells were then transferred to fresh tissue culture plates and incubated at 37°C, 5% CO2 for 5 days in BMMΦ medium with the addition of 10% volume L cell conditioned medium every second day.
LPS treatments
For IL-6 analysis, BMMΦs at 1 × 106 cells/mL were serum-starved overnight then incubated with 100 ng/mL lipopolysaccharide (LPS) for various time points before supernatants harvested and used for ELISA.
Western blots
Whole cell lysates of resting BMMΦs were prepared by solubilization in lysis buffer containing 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.6), 250mM Nonidet P-40, 5mM ethylenediaminetetraacetic acid, and Complete protease inhibitors (Roche Diagnostics) for 1 hour on ice. Lysates were clarified by centrifugation at 13 000g for 20 minutes at 4°C and protein levels determined by Bradford assay. Lysates were boiled for 5 minutes and then fractionated by SDS-PAGE electrophoresis before being transferred onto Hybond-P polyvinylidene difluoride or nitrocellulose membranes (GE Healthcare). Immunoblotting for specific primary antibodies was performed as per the manufacturer's instructions and with the appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare). Protein levels were detected using an ECL kit (GE Healthcare). Antibodies against the following were used: SHIP1 and tubulin (Santa Cruz Biotechnology); and Akt, phospho-Akt (Ser473), S6, and phospho-pS6 (Cell Signaling Technologies). In experiments assessing the phosphorylation state of proteins in alveolar macrophages, blots were analyzed using the Odyssey Infrared Imaging System and fluorescence-conjugated secondary antibodies (LI-COR Biosciences), and the fluorescence signal converted to grayscale for figures using Odyssey Application software Version 2.0.
Results
Generation of a SHIP1 mutant mouse by ENU mutagenesis
In a recessive ENU mouse mutagenesis screen established to identify mutants with early lethality,26 we identified a G3 pedigree (el20) with runting and pancytopenia. The mutation was mapped to a 3-Mb region on chromosome 1, and sequencing of candidate genes led to identification of a T to C base substitution at nucleotide position 2099 of Inpp5d. The Inpp5d locus encodes the inositol 5′-phosphatase SHIP1, and the point mutation is predicted to cause an amino acid substitution of an evolutionarily conserved isoleucine residue to threonine at amino acid position 641 (Figure 1A). This mutant allele is referred to as SHIP1el20. The change from a hydrophobic to hydrophilic residue located within the α7-β9 loop region of the SHIP1 catalytic 5′-phosphatase domain was predicted to alter enzymatic activity. An in vitro 5′-phosphatase assay demonstrated that this mutant showed enzyme activity, but this was reduced compared with wild-type SHIP1 protein, which hydrolyzed all PtdIns(3,4,5)P3 present to form PtdIns(3,4)P2 (Figure 1B). Western blot analyses of cell lysates generated from homozygous mutant (SHIP1el20/el20) BMMΦs demonstrated less than 10% of the normal SHIP1 protein levels (Figure 1C). This reduced protein expression could be explained by reduced mRNA levels as determined by quantitative real-time PCR of cDNA isolated from whole bone marrow, thymus, freshly isolated LK progenitors and cultured BMMΦs (Figure 1D). Thus, the SHIP1el20 mutation led to markedly reduced levels of SHIP1 protein, presumably because of the mutation affecting RNA stability.
Identification of a missense point mutation of SHIP1. (A) Schematic of the isoleucine to threonine amino acid substitution in the 5′-phosphatase domain of SHIP1 and the corresponding conserved hydrophobic residue of other inositol 5′-phosphatases. (B) 5′-Phosphatase assay showing the conversion of PtdIns(3,4,5)P3 substrate to PtdIns(3,4)P2 product by immunoprecipitated recombinant wild-type or mutant SHIP1. The HA vector sample is included as a transfection control, and results are representative of 2 individual experiments. The HA-immunoblot of lysates and parallel HA-immunoprecipitates, on which the 5′-phosphatase assays were performed, demonstrates similar loading of SHIP1 protein. (C) Western blot for SHIP1 expression in resting SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 BMMΦs. Western blot is representative of 3 independent samples for each genotype. (D) Quantitative RT-PCR for SHIP1 mRNA expression in whole bone marrow (BM), thymus (THY), sorted lineage−c-Kit+ (LK) BM cells and resting BMMΦs. All values have been normalized to expression of HPRT. Bar graphs represent the mean ± SD of 3 samples for each genotype, with the exception of LK cells, which were obtained from sorts of 3 mice for each genotype. **P < .01, 2-tailed Mann-Whitney test. ***P < .001, 2-tailed Mann-Whitney test. ND indicates not detected.
Identification of a missense point mutation of SHIP1. (A) Schematic of the isoleucine to threonine amino acid substitution in the 5′-phosphatase domain of SHIP1 and the corresponding conserved hydrophobic residue of other inositol 5′-phosphatases. (B) 5′-Phosphatase assay showing the conversion of PtdIns(3,4,5)P3 substrate to PtdIns(3,4)P2 product by immunoprecipitated recombinant wild-type or mutant SHIP1. The HA vector sample is included as a transfection control, and results are representative of 2 individual experiments. The HA-immunoblot of lysates and parallel HA-immunoprecipitates, on which the 5′-phosphatase assays were performed, demonstrates similar loading of SHIP1 protein. (C) Western blot for SHIP1 expression in resting SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 BMMΦs. Western blot is representative of 3 independent samples for each genotype. (D) Quantitative RT-PCR for SHIP1 mRNA expression in whole bone marrow (BM), thymus (THY), sorted lineage−c-Kit+ (LK) BM cells and resting BMMΦs. All values have been normalized to expression of HPRT. Bar graphs represent the mean ± SD of 3 samples for each genotype, with the exception of LK cells, which were obtained from sorts of 3 mice for each genotype. **P < .01, 2-tailed Mann-Whitney test. ***P < .001, 2-tailed Mann-Whitney test. ND indicates not detected.
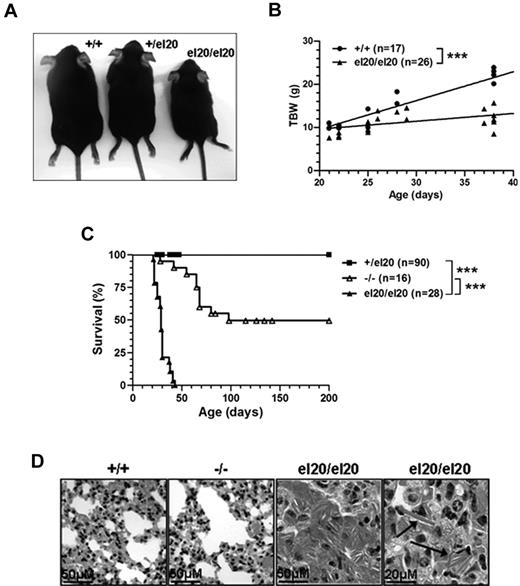
SHIP1el20/el20 mice exhibit failure to thrive and early lung disease
Heterozygous mutant mice (SHIP1+/el20) displayed no significant abnormalities when examined up to 12 months of age (data not shown). At weaning, homozygous SHIP1el20/el20 mice were present at expected Mendelian ratios but exhibited runting and a subsequent failure to thrive (Figure 2A-B). Survival of SHIP1el20/el20 mice was significantly impaired, with no mutant mice living beyond 7 weeks of age compared with 50% survival of SHIP1-null (SHIP1−/−) mice at 6 months of age (Figure 2C). These differences in survival could not be attributed to differences in genetic background or the environment as both SHIP1el20/el20 mice and SHIP1−/− mice were maintained on a C57BL/6 background in similar housing conditions. Death of SHIP1−/− mice is the result of respiratory failure from alveoli filled with lipid-laden macrophages containing eosinophilic Ym1 crystals, a product of activated macrophages.2,3 Histologic examination of lungs from 4-week-old SHIP1el20/el20 mice revealed severe lung pathology similar to that reported in older SHIP1−/− mice (Figure 2D). As expected, lungs of 4-week-old SHIP1−/− mice were relatively unaffected. Thus, the significantly shorter survival of SHIP1el20/el20 mice appears to be explained by a more rapid onset of respiratory failure from accumulation of activated macrophages in the lungs.
Homozygous SHIP1el20/el20 mice have runting, early lethality, and severe lung disease. (A) Five-week-old sex-matched littermates showing runting of a homozygous SHIP1el20/el20 mouse compared with wild-type SHIP1+/+ and heterozygous SHIP1+/el20 mice. (B) Total body weight (TBW) of male SHIP1el20/el20 mice compared with male SHIP1+/+ controls. ***P < .001 using 1-way ANOVA. (C) Survival curves of SHIP1el20/el20 mice compared with SHIP1−/− and SHIP1+/el20 mice. ***P < .0001 using log-rank (Mantel Cox) test. (D) Hematoxylin and eosin–stained histologic lung sections of 4-week-old SHIP1el20/el20 mice showing alveolar airspace consolidation of lungs with macrophages containing eosinophilic Ym1 crystals (arrows). Normal lungs from age-matched SHIP1+/+ and SHIP1−/− mice are shown.
Homozygous SHIP1el20/el20 mice have runting, early lethality, and severe lung disease. (A) Five-week-old sex-matched littermates showing runting of a homozygous SHIP1el20/el20 mouse compared with wild-type SHIP1+/+ and heterozygous SHIP1+/el20 mice. (B) Total body weight (TBW) of male SHIP1el20/el20 mice compared with male SHIP1+/+ controls. ***P < .001 using 1-way ANOVA. (C) Survival curves of SHIP1el20/el20 mice compared with SHIP1−/− and SHIP1+/el20 mice. ***P < .0001 using log-rank (Mantel Cox) test. (D) Hematoxylin and eosin–stained histologic lung sections of 4-week-old SHIP1el20/el20 mice showing alveolar airspace consolidation of lungs with macrophages containing eosinophilic Ym1 crystals (arrows). Normal lungs from age-matched SHIP1+/+ and SHIP1−/− mice are shown.
SHIP1el20/el20 mice have more marked hematopoietic abnormalities than SHIP1−/− mice
Given that SHIP1el20/el20 mice express detectable levels of SHIP1 protein, albeit with reduced catalytic activity (Figure 1B-C), the more dramatic lung phenotype was surprising. We analyzed hematopoiesis in SHIP1el20/el20 mice at 4 to 5 weeks of age, a time point before the development of major systemic illness, to determine whether the hematopoietic phenotype was also more severe than age-matched SHIP1−/− mice. Peripheral blood analysis of 5-week-old SHIP1el20/el20 mice demonstrated significantly reduced numbers of circulating lymphocytes compared with age-matched SHIP1−/− mice (Table 1). SHIP1el20/el20 mice also displayed anemia and thrombocytopenia, but these abnormalities were of similar severity to SHIP1−/− mice (Table 1).
Hematopoietic cell numbers in SHIP1el20/el20 mice
| . | SHIP1+/+ . | SHIP1el20/el20 . | SHIP1−/− . |
|---|---|---|---|
| Peripheral blood | |||
| Hematocrit, % | 44.7 ± 1.0 | 40.5 ± 1.1* | 39.6 ± 4.2* |
| White cells, × 109/L | 10.9 ± 0.8 | 7.2 ± 0.2*† | 9.9 ± 0.7 |
| Lymphocytes, × 109/L | 8.8 ± 0.6 | 5.1 ± 0.2*† | 7.2 ± 0.6 |
| Neutrophils, × 109/L | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.7 ± 0.4 |
| Platelets, × 109/L | 1016 ± 81 | 499 ± 52* | 607 ± 78* |
| Bone marrow, cells × 106 | 61 ± 4 | 49 ± 5*† | 71 ± 5 |
| Pre-B | 9.4 ± 1.7 | 0.2 ± 0.1*† | 9.0 ± 0.2 |
| Mature B | 2.5 ± 0.9 | 0.2 ± 0.1*† | 2.8 ± 0.2 |
| Mac-1+ | 18 ± 0.4 | 23 ± 3.6 | 22 ± 1.4* |
| Mac-1+Gr-1+ | 7.3 ± 0.8 | 21 ± 2.3* | NA |
| Pro-erythroblast | 23 ± 1.6 | 20 ± 4.8 | 23 ± 2.3 |
| Late erythroblast | 10 ± 1.2 | 5.7 ± 0.7* | 15 ± 0.4* |
| Spleen, cells × 106 | 107 ± 8 | 224 ± 4*† | 98 ± 14 |
| B220+ | 17 ± 5 | 9 ± 3† | 23 ± 3 |
| CD4+ | 12 ± 3 | 6 ± 1* | 9 ± 1 |
| CD8+ | 7 ± 1 | 4 ± 1* | 5 ± 1 |
| Mac-1+ | 18 ± 4 | 23 ± 01 | 14 ± 6 |
| Mac-1+Gr-1+ | 16 ± 2 | 22 ± 2* | NA |
| Pro-erythroblast | 9 ± 2 | 143 ± 26*† | 14 ± 2* |
| Late erythroblast | 24 ± 4 | 28 ± 0.7 | 19 ± 1 |
| Thymus, cells × 106 | 91 ± 13 | 6.5 ± 2.7* | 103 ± 3 |
| DN | 3.1 ± 0.6 | 1.4 ± 0.7*† | 3.1 ± 0.3 |
| DP | 76 ± 12 | 2.7 ± 0.7*† | 80 ± 10 |
| CD4+ | 8.4 ± 1.4 | 1.7 ± 0.3*† | 8.9 ± 1.1 |
| CD8+ | 2.8 ± 0.5 | 0.7 ± 0.2*† | 4.5 ± 0.5* |
| . | SHIP1+/+ . | SHIP1el20/el20 . | SHIP1−/− . |
|---|---|---|---|
| Peripheral blood | |||
| Hematocrit, % | 44.7 ± 1.0 | 40.5 ± 1.1* | 39.6 ± 4.2* |
| White cells, × 109/L | 10.9 ± 0.8 | 7.2 ± 0.2*† | 9.9 ± 0.7 |
| Lymphocytes, × 109/L | 8.8 ± 0.6 | 5.1 ± 0.2*† | 7.2 ± 0.6 |
| Neutrophils, × 109/L | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.7 ± 0.4 |
| Platelets, × 109/L | 1016 ± 81 | 499 ± 52* | 607 ± 78* |
| Bone marrow, cells × 106 | 61 ± 4 | 49 ± 5*† | 71 ± 5 |
| Pre-B | 9.4 ± 1.7 | 0.2 ± 0.1*† | 9.0 ± 0.2 |
| Mature B | 2.5 ± 0.9 | 0.2 ± 0.1*† | 2.8 ± 0.2 |
| Mac-1+ | 18 ± 0.4 | 23 ± 3.6 | 22 ± 1.4* |
| Mac-1+Gr-1+ | 7.3 ± 0.8 | 21 ± 2.3* | NA |
| Pro-erythroblast | 23 ± 1.6 | 20 ± 4.8 | 23 ± 2.3 |
| Late erythroblast | 10 ± 1.2 | 5.7 ± 0.7* | 15 ± 0.4* |
| Spleen, cells × 106 | 107 ± 8 | 224 ± 4*† | 98 ± 14 |
| B220+ | 17 ± 5 | 9 ± 3† | 23 ± 3 |
| CD4+ | 12 ± 3 | 6 ± 1* | 9 ± 1 |
| CD8+ | 7 ± 1 | 4 ± 1* | 5 ± 1 |
| Mac-1+ | 18 ± 4 | 23 ± 01 | 14 ± 6 |
| Mac-1+Gr-1+ | 16 ± 2 | 22 ± 2* | NA |
| Pro-erythroblast | 9 ± 2 | 143 ± 26*† | 14 ± 2* |
| Late erythroblast | 24 ± 4 | 28 ± 0.7 | 19 ± 1 |
| Thymus, cells × 106 | 91 ± 13 | 6.5 ± 2.7* | 103 ± 3 |
| DN | 3.1 ± 0.6 | 1.4 ± 0.7*† | 3.1 ± 0.3 |
| DP | 76 ± 12 | 2.7 ± 0.7*† | 80 ± 10 |
| CD4+ | 8.4 ± 1.4 | 1.7 ± 0.3*† | 8.9 ± 1.1 |
| CD8+ | 2.8 ± 0.5 | 0.7 ± 0.2*† | 4.5 ± 0.5* |
Analyses of peripheral blood and hematopoietic organs were performed on 5-week-old SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 mice. Values are the mean ± SD of at least 3 mice for each genotype. BM cell numbers were calculated per femur.
Pre-B indicates B220loCD25+; mature B, B220hiIgM+; Pro-erythroblast, ter119+CD71hi; Late erythroblast, ter119+CD71−; and NA, not assessed.
P < .05 by 2-tailed Mann-Whitney test versus SHIP1+/+ controls.
P < .05 by 2-tailed Mann-Whitney test versus SHIP1−/− mice.
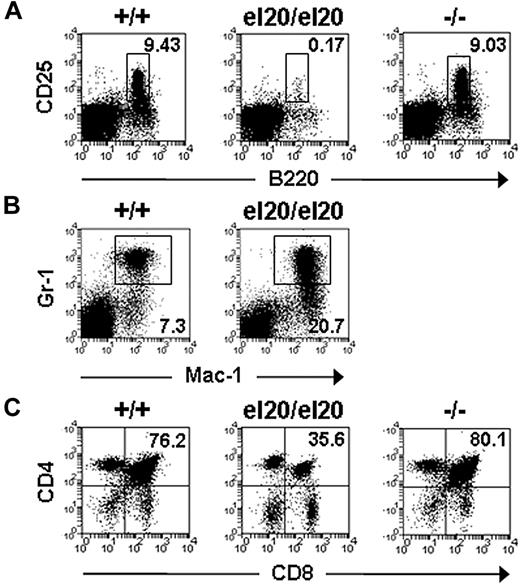
The bone marrow cellularity of SHIP1el20/el20 mice was significantly reduced because of a marked (50-fold) reduction of pre-B cells (B220loCD25+) and mature B cells (Table 1; Figure 3A). A similar, but less severe, reduction of B cells was observed in the BM of older (10-week) SHIP1−/− mice (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Consistent with a negative regulatory role for SHIP1 in B-cell antigen receptor signaling,28 SHIP1el20/el20 mice had a 53-fold increase in serum IgM levels (supplemental Figure 1A). Levels of serum IgG were not significantly altered, although there was a trend toward increased IgG2a (supplemental Figure 1B), which has been reported in SHIP1−/− mice.29 Consistent with a role for SHIP1 in the repression of myeloid immunoregulatory macrophages,8 SHIP1el20/el20 mice displayed a 3-fold increase in the numbers of Mac-1+Gr-1+ cells within the BM (Table 1; Figure 3B). SHIP1el20/el20 mice also had increased spleen size because of markedly increased erythropoiesis and, to a lesser extent, Mac-1+Gr-1+ cells (Table 1).
Hematopoietic defects of SHIP1el20/el20 mice. Representative dot plots of (A) bone marrow B220loCD25+ pre-B cells, (B) bone marrow Mac-1+Gr-1+ myeloid cells, and (C) thymic T-cell subsets from 5-week-old mice. The mean proportion for each subset was gated on live hematopoietic cells and calculated from at least 3 mice of each genotype.
Hematopoietic defects of SHIP1el20/el20 mice. Representative dot plots of (A) bone marrow B220loCD25+ pre-B cells, (B) bone marrow Mac-1+Gr-1+ myeloid cells, and (C) thymic T-cell subsets from 5-week-old mice. The mean proportion for each subset was gated on live hematopoietic cells and calculated from at least 3 mice of each genotype.
In addition to the marked B-cell deficiency, SHIP1el20/el20 mice also exhibited very small thymi (10- to 20-fold smaller than controls), affecting all T-cell subsets but most notably the CD4+CD8+ double-positive (DP) fraction (Table 1; Figure 3C). In contrast, SHIP1−/− mice had normal numbers of thymocytes, even at 10 weeks of age (supplemental Table 1).
Cell-autonomous and non–cell-autonomous hematopoietic defects of SHIP1el20/el20 mice
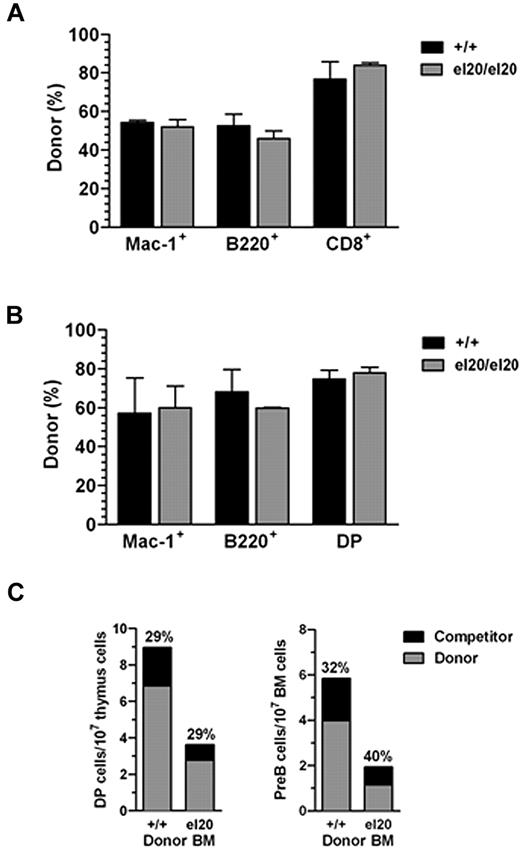
SHIP1 expression is predominantly hematopoietic, although expression has also been demonstrated in the testis, fibroblasts, endothelial cells, and osteoblasts.30-32 We transplanted BM cells from SHIP1el20/el20 mice into lethally irradiated wild-type mice to determine whether the more severe phenotype, including impaired survival of SHIP1el20/el20 mice, was intrinsic to the hematopoietic cells. Within 6 weeks of transplantation, mice reconstituted with SHIP1el20/el20 BM exhibited hallmarks of the SHIP1el20/el20 phenotype, including splenomegaly and reduced DP thymocytes (supplemental Figure 2) with death of most mice within 12 weeks of transplantation because of macrophage infiltration of the lungs. The phenotype was not as rapid or severe as that observed in SHIP1el20/el20 mice, which may be explained by the slower kinetics of tissue macrophage turnover, especially in the lungs.33 Conversely, transplantation of wild-type BM cells into 3-week-old SHIP1el20/el20 mice rescued the hematopoietic phenotype (supplemental Figure 2) and their survival, with the majority of mice surviving beyond 12 weeks after transplantation. A small number of SHIP1el20/el20 mice died, but analysis revealed expansion of endogenous SHIP1el20/el20 BM cells surviving the irradiation (data not shown). Thus, the hematopoietic defects and impaired survival of SHIP1el20/el20 mice were hematopoietic cell-autonomous.
The role of SHIP1 in hematopoietic stem cell (HSC) function is complex, with reports of either normal or increased activity in SHIP1−/− mice.30,31,34 We performed competitive BM transplantation assays to evaluate the effect of the SHIP1el20 mutation on HSC function. Donor CD45.2+ BM cells from wild-type or SHIP1el20/el20 mice were transplanted with equal numbers of wild-type competitor CD45.1+ BM cells into lethally irradiated CD45.1+ wild-type mice. Evaluation of the proportions of donor and competitor hematopoietic cells in the peripheral blood (Figure 4A) and hematopoietic organs (Figure 4B) demonstrated similar contributions by donor wild-type and SHIP1el20/el20 HSCs, indicating that the SHIP1el20 mutation had no significant effect on HSC activity. However, mice receiving SHIP1el20/el20 donor BM cells exhibited significantly reduced thymus size and numbers of BM pre-B cells because of loss of both donor SHIP1el20/el20 and wild-type competitor cells (Figure 4C). This suggested that the lymphoid defects in SHIP1el20/el20 mice are mediated in part by a non–cell-autonomous mechanism.
Reduced number of lymphoid cells in SHIP1el20/el20 mice is non–cell-autonomous. (A) Donor contribution of myeloid (Mac-1+), B-cell (B220+), and T-cell (CD8+) lineages in peripheral blood of recipient mice 4 weeks after transplantation with a 1:1 ratio of donor (SHIP1+/+ or SHIP1el20/el20) and wild-type CD45.1 competitor BM cells. Bar graphs represent the mean ± SD calculated from 3 recipients of each donor genotype. (B) Donor contribution 12 weeks after transplantation. Mice were killed to determine donor contribution to the BM myeloid (Mac-1+), BM B-cell (B220+), and thymic T-cell (DP) lineages. Bar graphs represent the mean ± SD calculated from 3 recipients of each donor genotype. (C) Total numbers of DP thymocytes and pre-B cells in mice 12 weeks after transplantation with a 1:1 ratio of donor (SHIP1+/+ or SHIP1el20/el20) and wild-type CD45.1 competitor BM cells. The relative contribution of competitor cells is shown.
Reduced number of lymphoid cells in SHIP1el20/el20 mice is non–cell-autonomous. (A) Donor contribution of myeloid (Mac-1+), B-cell (B220+), and T-cell (CD8+) lineages in peripheral blood of recipient mice 4 weeks after transplantation with a 1:1 ratio of donor (SHIP1+/+ or SHIP1el20/el20) and wild-type CD45.1 competitor BM cells. Bar graphs represent the mean ± SD calculated from 3 recipients of each donor genotype. (B) Donor contribution 12 weeks after transplantation. Mice were killed to determine donor contribution to the BM myeloid (Mac-1+), BM B-cell (B220+), and thymic T-cell (DP) lineages. Bar graphs represent the mean ± SD calculated from 3 recipients of each donor genotype. (C) Total numbers of DP thymocytes and pre-B cells in mice 12 weeks after transplantation with a 1:1 ratio of donor (SHIP1+/+ or SHIP1el20/el20) and wild-type CD45.1 competitor BM cells. The relative contribution of competitor cells is shown.
SHIP1el20/el20 macrophages have increased IL-6 production
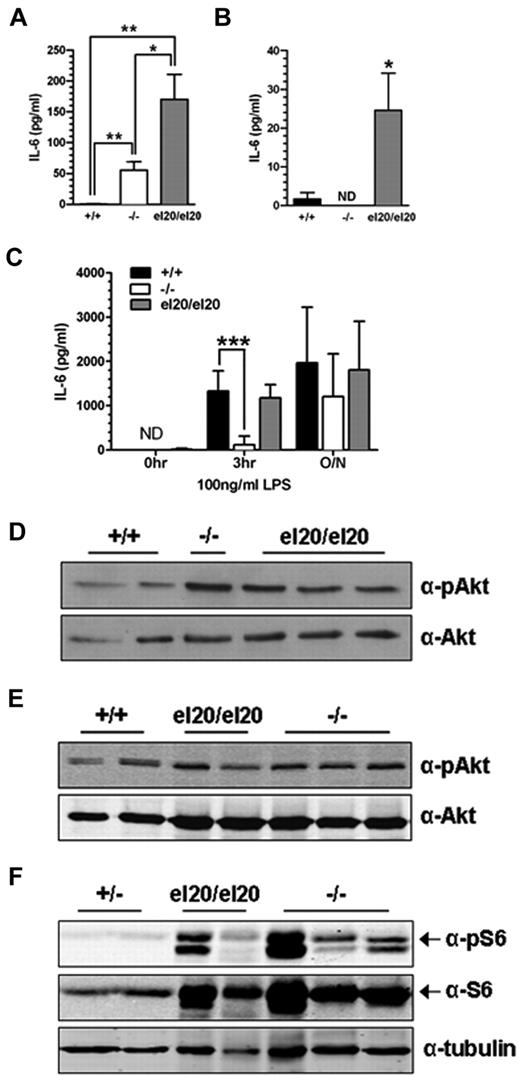
Elevated levels of circulating IL-6 secreted by activated macrophages are a proposed non–cell-autonomous mechanism of reduced B-cell numbers in SHIP1−/− mice.13,35 Therefore, we measured serum IL-6 levels in age-matched SHIP1el20/el20 and SHIP1−/− mice. Serum IL-6 levels in 4- to 5-week-old SHIP1el20/el20 mice were 100-fold greater than in control mice and 3-fold greater than in age-matched SHIP1−/− mice (Figure 5A). We examined IL-6 production by BMMΦs to address a potential source of the elevated serum IL-6. SHIP1el20/el20 macrophages displayed markedly elevated basal levels of IL-6 compared with control BMMΦs (Figure 5B). Secretion of IL-6 by resting SHIP1−/− macrophages was not increased, consistent with the recent finding that peritoneal and splenic macrophages are the major source of IL-6 in SHIP1−/− mice.36 SHIP1 has also been implicated in the LPS response, although reports of its effects on IL-6 production by BMMΦs are conflicting.5,37 In our hands, SHIP1−/− BMMΦs had a blunted IL-6 response to LPS at the 3-hour time point as previously reported.5,37 In contrast, SHIP1el20/el20 BMMΦs had a normal LPS response (Figure 5C). Thus, cultured SHIP1el20/el20 macrophages had increased LPS response compared with SHIP1−/− macrophages, consistent with the increased serum IL-6 levels and the more severe lung inflammatory phenotype of SHIP1el20/el20 mice.
Increased IL-6 production and PI3K/mTOR signaling in SHIP1el20/el20 mice. (A) IL-6 levels in serum from 4- to 5-week-old mice. Bar graph represents the mean ± SD of at least 3 mice for each genotype. *P < .05, 2-tailed Mann-Whitney test. **P < .01, 2-tailed Mann-Whitney test. (B) IL-6 levels in resting BMMΦs. Bar graph represents the mean ± SD of at least 6 samples for each genotype. *P < .05, 2-tailed Mann-Whitney test compared with SHIP1+/+ control. ND indicates not detected. (C) IL-6 levels in resting BMMΦs after stimulation with LPS for the indicated times. Bar graph represents the mean ± SD of 6 independent samples for each genotype. ***P < .001, 2-tailed Mann-Whitney test compared with SHIP1+/+ control. ND indicates not detected. (D) Western blot for pAkt Ser473 and total Akt in resting BMMΦs. (E) Western blot for pAkt Ser473 and total Akt in freshly isolated BAL cells from the same mice used for panel D. (F) Western blot for pS6 and total S6 in freshly isolated BAL cells from the same mice used for panel D except for the controls, which were from SHIP1+/− heterozygous mice.
Increased IL-6 production and PI3K/mTOR signaling in SHIP1el20/el20 mice. (A) IL-6 levels in serum from 4- to 5-week-old mice. Bar graph represents the mean ± SD of at least 3 mice for each genotype. *P < .05, 2-tailed Mann-Whitney test. **P < .01, 2-tailed Mann-Whitney test. (B) IL-6 levels in resting BMMΦs. Bar graph represents the mean ± SD of at least 6 samples for each genotype. *P < .05, 2-tailed Mann-Whitney test compared with SHIP1+/+ control. ND indicates not detected. (C) IL-6 levels in resting BMMΦs after stimulation with LPS for the indicated times. Bar graph represents the mean ± SD of 6 independent samples for each genotype. ***P < .001, 2-tailed Mann-Whitney test compared with SHIP1+/+ control. ND indicates not detected. (D) Western blot for pAkt Ser473 and total Akt in resting BMMΦs. (E) Western blot for pAkt Ser473 and total Akt in freshly isolated BAL cells from the same mice used for panel D. (F) Western blot for pS6 and total S6 in freshly isolated BAL cells from the same mice used for panel D except for the controls, which were from SHIP1+/− heterozygous mice.
We next investigated the effect of the SHIP1el20 mutation on the PI3K/mammalian target of rapamycin signaling cascade. After a 5-hour serum starvation, SHIP1el20/el20 BMMΦs had mildly increased pAkt relative to wild-type, although not significantly greater than that seen in SHIP1−/− BMMΦs (Figure 5D). pAkt was similar to wild-type in freshly isolated BAL cells obtained from 5-week-old SHIP1el20/el20 and 10-week-old SHIP1−/− mice (Figure 5E). In contrast to minimal changes in pAkt levels, there was marked increase in total S6 and pS6, a downstream target of the mTORC1 complex (Figure 5F). Thus, the SHIP1el20 mutation leads to activation of the PI3K/mTOR pathway as measured by pS6, although not different from that observed in SHIP1−/− BAL cells.
The more severe inflammatory phenotype of SHIP1el20/el20 mice is the result of loss of both SHIP1 and s-SHIP
Gene targeting of exon 1 of the Inpp5d locus does not prevent expression of the shorter s-SHIP isoform, which is initiated from an alternate promoter within intron 5.21 Although s-SHIP expression is reported to be limited to embryonic stem cells and HSCs, s-SHIP mRNA was detectable in lineage-restricted hematopoietic cells, including Mac-1+ myeloid cells at levels comparable with embryonic stem cells (Figure 6A). We examined s-SHIP mRNA expression in BMMΦs generated from wild-type SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 mice to determine whether s-SHIP expression was affected by the SHIP1el20 mutation (Figure 6B). As predicted by the targeting strategy used to generate the SHIP1−/− mice, s-SHIP mRNA was expressed at normal levels in SHIP1−/− macrophages. In contrast, SHIP1el20/el20 macrophages expressed 20-fold less s-SHIP mRNA. Thus, the SHIP1el20 mutation leads to loss of both SHIP1 and s-SHIP mRNA expression in macrophages.
The el20 mutation leads to loss of expression of both SHIP1 and the s-SHIP isoform. (A) Quantitative RT-PCR for s-SHIP mRNA expression in wild-type sorted hematopoietic cell subsets. Sorted cell subsets were obtained from pools of at least 3 mice. All values have been normalized to expression of HPRT. LKS indicates lineage−c-Kit+Sca-1+; LK, lineage−c-Kit+; DN, CD4−CD8− double-negative thymocytes; and DP, CD4+CD8+ double-positive thymocytes. (B) Quantitative RT-PCR for s-SHIP and SHIP1 mRNA expression in BMMΦs from wild-type SHIP1+/+, SHIP1−/−, SHIP1el20/el20, and compound heterozygous SHIP1−/el20 mice. All values have been normalized to expression of HPRT. Bar graphs represent the mean ± SD of 4 samples for each genotype. ***P < .0001 using log-rank (Mantel Cox) test compared with SHIP1+/+ control. (C) Representative dot plots of bone marrow pre-B cells (top panel) and thymic T-cell subsets (bottom panel). The mean proportion for each subset was gated on live hematopoietic cells and calculated from at least 3 mice of each genotype. (D) IL-6 levels in serum of age-matched SHIP1−/el20 mice compared with SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 mice shown in Figure 5A. **P < .01 by 2-tailed Mann-Whitney test. (E) IL-6 production by resting SHIP1−/el20 BMMΦs compared with SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 samples shown in Figure 5B. *P < .05, 2-tailed Mann-Whitney test. ND indicates not detected. (F) Hematoxylin and eosin-stained histologic sections showing normal lungs at 6 weeks of age but consolidation of alveolar airspaces in 11-week-old SHIP1−/el20 lungs. (G) Improved survival of SHIP1−/el20 mice compared with SHIP1el20/el20 mice. Results for all other mice reproduced from Figure 2C. ***P < .0001 using log-rank (Mantel Cox) test. NS indicates not significant.
The el20 mutation leads to loss of expression of both SHIP1 and the s-SHIP isoform. (A) Quantitative RT-PCR for s-SHIP mRNA expression in wild-type sorted hematopoietic cell subsets. Sorted cell subsets were obtained from pools of at least 3 mice. All values have been normalized to expression of HPRT. LKS indicates lineage−c-Kit+Sca-1+; LK, lineage−c-Kit+; DN, CD4−CD8− double-negative thymocytes; and DP, CD4+CD8+ double-positive thymocytes. (B) Quantitative RT-PCR for s-SHIP and SHIP1 mRNA expression in BMMΦs from wild-type SHIP1+/+, SHIP1−/−, SHIP1el20/el20, and compound heterozygous SHIP1−/el20 mice. All values have been normalized to expression of HPRT. Bar graphs represent the mean ± SD of 4 samples for each genotype. ***P < .0001 using log-rank (Mantel Cox) test compared with SHIP1+/+ control. (C) Representative dot plots of bone marrow pre-B cells (top panel) and thymic T-cell subsets (bottom panel). The mean proportion for each subset was gated on live hematopoietic cells and calculated from at least 3 mice of each genotype. (D) IL-6 levels in serum of age-matched SHIP1−/el20 mice compared with SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 mice shown in Figure 5A. **P < .01 by 2-tailed Mann-Whitney test. (E) IL-6 production by resting SHIP1−/el20 BMMΦs compared with SHIP1+/+, SHIP1−/−, and SHIP1el20/el20 samples shown in Figure 5B. *P < .05, 2-tailed Mann-Whitney test. ND indicates not detected. (F) Hematoxylin and eosin-stained histologic sections showing normal lungs at 6 weeks of age but consolidation of alveolar airspaces in 11-week-old SHIP1−/el20 lungs. (G) Improved survival of SHIP1−/el20 mice compared with SHIP1el20/el20 mice. Results for all other mice reproduced from Figure 2C. ***P < .0001 using log-rank (Mantel Cox) test. NS indicates not significant.
We hypothesized that the more dramatic phenotype of SHIP1el20/el20 mice was the result of concomitant loss of SHIP1 and s-SHIP expression. To address this hypothesis, we crossed heterozygous SHIP1+/− mice with heterozygous SHIP1+/el20 mice to generate compound heterozygous SHIP1−/el20 mice, which should express a single allele of s-SHIP from the SHIP1− targeted locus. Quantitative real-time PCR confirmed expression of s-SHIP in compound heterozygous SHIP1−/el20 BMMΦs (Figure 6B). Examination of SHIP1−/el20 mice at 6 weeks of age revealed complete rescue of the pre-B-cell and thymocyte phenotype (Figure 6C). In addition, serum IL-6 levels and IL-6 production by BMMΦs were restored to that observed in SHIP1−/− mice and BMMΦs, respectively (Figure 6D-E). Histologic examination of the lungs from SHIP1−/el20 mice was also normal at 6 weeks of age but, similar to SHIP1−/− mice, showed evidence of abnormal macrophage deposition by 11 weeks of age (Figure 6F). Most importantly, SHIP1−/el20 mice had a similar survival to SHIP1−/− mice (Figure 6G). Thus, the SHIP1− targeted locus, which expresses s-SHIP, was able to restore the more dramatic phenotype of SHIP1el20/el20 mice back to the SHIP1−/− phenotype.
Discussion
We present the phenotype of a point mutant of SHIP1 resulting from ENU mutagenesis. Although the phenotype of SHIP1el20/el20 mice was similar to SHIP1−/− mice, it was much more dramatic with early lethality (death by 7 weeks of age) associated with marked macrophage infiltration of the lungs and marked reductions of pre-B cells and thymocytes. We provide several pieces of evidence that the accentuated phenotype was the result of loss of expression of s-SHIP, an isoform of SHIP1 with no previously known function. First, we show that s-SHIP mRNA is expressed in mature hematopoietic cells at levels comparable with embryonic stem cells despite previous reports that it is stem cell-restricted.21,22 Second, we show that s-SHIP is expressed in SHIP1−/− BMMΦs but not SHIP1el20/el20 BMMΦs. Third, by generating compound heterozygous SHIP1−/el20 mice, we show that expression of s-SHIP rescues the SHIP1el20/el20 phenotype back to the SHIP1−/− phenotype, providing genetic evidence that concomitant loss of SHIP1 and s-SHIP is the most likely explanation for the more severe phenotype. This provides the first evidence for a functional role of s-SHIP in adult hematopoiesis.
Once we identified the mutation in the SHIP1 allele, we did not sequence the other 22 genes within the 3-Mb interval. Therefore, we cannot completely exclude the possibility that the more severe phenotype is the result of a combination of the SHIP1el20 mutation plus an independent mutation in one of the other 22 genes. However, this appears unlikely because extensive molecular studies by multiple groups indicate that, at the dose used, ENU induces single base pair changes at a rate of 1 per 1 to 2 Mb.38 Given that the coding region of the 22 other genes is only 56 kb, the chances of additional coding sequence mutations would be extremely small.
Overall, the functions of s-SHIP appear to be synergistic with SHIP1 because the phenotype of SHIP1el20/el20 mice is similar but more severe and occurs at an earlier age than SHIP1−/− mice. The major differences between SHIP1el20/el20 and SHIP1−/− mice are shortened survival, marked reductions in B and T cells, and higher levels of circulating IL-6. SHIP1el20/el20 mice with identical genetic background and housed in the same environment as SHIP1−/− mice develop an identical lung phenotype but at a much earlier age. The lung infiltrate of SHIP1−/− mice has been attributed to an activated macrophage phenotype.2,3 It is likely that the more severe lung phenotype of SHIP1el20/el20 mice reflects increased macrophage activation, as demonstrated by increased serum IL-6 levels compared with age-matched SHIP1−/− mice. The impaired survival of mice lacking SHIP1 has been attributed to an inflammatory infiltrate of the lungs, although inflammation of other organs, such as the bowel, may contribute to the runting phenotype seen in SHIP1el20/el20 mice.39 Therefore, it appears likely that s-SHIP functions synergistically with SHIP1 to suppress activation of macrophages.
The reduction in B and T cells in SHIP1el20/el20 mice was the other major difference with SHIP1−/− mice. Determining the lineage-specific functions of s-SHIP is complicated by the inflammatory phenotype of SHIP1el20/el20 mice. In the case of SHIP1, lineage-specific deletion or transplantation studies have suggested that the B-cell deficit is partly non–cell-autonomous.14,31 Furthermore, SHIP1−/− mice generated on an IL-6-null background can partially rescue the loss of B cells.13 Because we were unable to generate lineage-specific deletion of s-SHIP, we used a competitive transplantation assay to distinguish cell-autonomous from non–cell-autonomous effects. Although the overall phenotype, including impaired survival and lung disease, was transplantable and therefore intrinsic to the hematopoietic system, the competitive transplantation results suggested that the loss of lymphoid cells in SHIP1el20/el20 mice was mediated, at least in part, by a non–cell-autonomous mechanism. Like SHIP1−/− mice, this may in part be the result of elevated inflammatory cytokines, such as IL-6, which induces apoptosis of B cells at the pre-B-cell stage of development.40 The competitive transplantations also suggest that the concomitant loss of SHIP1 and s-SHIP does not impair HSC activity. Some, but not all, studies have suggested a role for SHIP1 in HSC function.30,34 However, these effects on HSCs may be indirect through defects of the cell niche or osteoporosis from hyperactive osteoclasts, which may require longer to establish than the life span of SHIP1el20/el20 mice.31,41
The biochemical functions of SHIP1 are complex.42 Although the predominant function of SHIP1 is through its 5′-phosphatase activity to convert PtdIns(3,4,5)P3 to PtdIns(3,4)P2, it also has phosphatase-independent activities by interaction with key signaling intermediates through its C-terminal domain.43,44 Given that s-SHIP has identical 5′-phosphatase and C-terminal domains, it is likely that s-SHIP has similar complex biochemical functions. Although s-SHIP lacks the SH2 domain important for membrane localization of SHIP1, it is still capable of recruitment to the cell membrane through binding of adaptor proteins, such as Grb2 to its polyproline C-terminus.21 Our preliminary biochemical studies were unable to demonstrate any clear difference in PI3K/mTOR signaling between macrophages that lack SHIP1 with those lacking both SHIP1 and s-SHIP. This may reflect the artificial conditions of in vitro culture. Alternatively, s-SHIP may synergize with SHIP1 in other signaling pathways that regulate the inflammatory response, such as the mitogen-activated protein kinase or nuclear factor-κB pathways.19,45,46 In support of this possibility, the human homolog of s-SHIP, SIP-110, can bind Grb2 and inhibit phosphorylation of mitogen-activated protein kinase in a phosphatase-independent manner.47 Two possible interactions that may be impaired by loss of both SHIP1 and s-SHIP are with the Dok proteins and the Src kinases, Lyn and Hck. Mice deficient of both SHIP1 and Dok-1 have a severe phenotype very similar to SHIP1el20/el20 mice with failure to thrive, shortened life span, and marked T- and B-cell loss.48 Alternatively, loss of both SHIP1 and s-SHIP may lead to loss of function of the Src kinases Lyn and Hck, which in part require SHIP1 for their repressive functions.49,50
Generation of mouse mutants by ENU mutagenesis can provide new insights into gene function that are not afforded by traditional germline targeting. Study of an ENU mouse mutant of SHIP1 provides the first functional evidence that the s-SHIP isoform of the Inpp5d locus synergizes with SHIP1 for suppression of macrophage activation. Generation of mice lacking only the s-SHIP isoform will be required to define any nonredundant role of s-SHIP in adult hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Melanie Howell, Marc Sacco, Lan Ta, Rebecca Bowyer, Lara Mizhiritsky, and Jenny Davis for animal husbandry; Jason Corbin for blood analyses; Dean Hewish and Catherine Li for flow cytometry; and David Tarlinton for serum immunoglobulin measurements.
This work was supported by the National Health & Medical Research Council of Australia (project grant 435107; fellowships, D.J.C., S.M.J., M.L.H., M.J.M., and D.J.H.) and the Australian Research Council (QEII fellowship; B.T.K.).
Authorship
Contribution: N.Y.N.N., M.L.H., C.A.M., D.J.H., S.M.J., and D.J.C. designed research; N.Y.N.N., M.J.M., L.M.O., E.M.D., A.A.H., J.E.C., B.T.K., D.J.H., and D.J.C. performed research; D.J.H. and M.L.H. contributed critical reagents; N.Y.N.N., M.J.M., L.M.O., E.M.D., D.J.H., A.A.H., B.T.K., M.L.H., C.A.M., and D.J.C. analyzed data; and N.Y.N.N. and D.J.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David J. Curtis, Bone Marrow Research Laboratories, Royal Melbourne Hospital, Victoria 3050, Australia; e-mail: dcurtis@wehi.edu.au.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal