Abstract
Ecotropic viral integration site-1 (Evi-1) is a nuclear transcription factor that plays an essential role in the regulation of hematopoietic stem cells. Aberrant expression of Evi-1 has been reported in up to 10% of patients with acute myeloid leukemia and is a diagnostic marker that predicts a poor outcome. Although chromosomal rearrangement involving the Evi-1 gene is one of the major causes of Evi-1 activation, overexpression of Evi-1 is detected in a subgroup of acute myeloid leukemia patients without any chromosomal abnormalities, which indicates the presence of other mechanisms for Evi-1 activation. In this study, we found that Evi-1 is frequently up-regulated in bone marrow cells transformed by the mixed-lineage leukemia (MLL) chimeric genes MLL-ENL or MLL-AF9. Analysis of the Evi-1 gene promoter region revealed that MLL-ENL activates transcription of Evi-1. MLL-ENL–mediated up-regulation of Evi-1 occurs exclusively in the undifferentiated hematopoietic population, in which Evi-1 particularly contributes to the propagation of MLL-ENL–immortalized cells. Furthermore, gene-expression analysis of human acute myeloid leukemia cases demonstrated the stem cell–like gene-expression signature of MLL-rearranged leukemia with high levels of Evi-1. Our findings indicate that Evi-1 is one of the targets of MLL oncoproteins and is selectively activated in hematopoietic stem cell–derived MLL leukemic cells.
Introduction
The ecotropic viral integration site-1 (Evi-1) is a nuclear transcription factor that plays an essential role in the proliferation and maintenance of hematopoietic stem cells (HSCs).1-3 There are 2 major alternative forms generated from the Evi-1 gene, Evi-1a and Mds1-Evi-1 (also called Evi-1c). Mds1-Evi-1 is a fusion variant of Evi-1 generated through intergenic splicing with Mds1,4 a gene located approximately 140 and 500 kb upstream of Evi-1 in the human and mouse genome, respectively. In contrast to Evi-1a, Mds1-Evi-1 possesses the PRDI-BF1-RIZ1 homologous (PR) domain in the N-terminus, which regulates oligomerization of the Evi-1 proteins.5 Both Evi-1a and Mds1-Evi-1 are normally coexpressed in several developing and adult tissues,6 and differences in the normal function between these proteins remain to be elucidated. Like all other PR domain proteins, Evi-1 contains several zinc finger motifs. They are grouped into N-terminal 7 and C-terminal 3 clusters, which are called the first and second zinc finger domain, respectively.7,8 Between these 2 zinc finger domains lie the C-terminal binding protein (CtBP) domain and the repression domain. The first zinc finger, the repression, and the CtBP-binding domains exhibit a growth-promoting effect by blocking transforming growth factor-β signaling.9 The first zinc finger domain also exhibits an antiapoptotic effect by repressing c-Jun N-terminal kinase signaling.10 The second zinc finger domain stimulates proliferation by increasing activator protein-1 activity.11 Thus, Evi-1 possesses diverse functions as an oncoprotein.
Aberrant expression of EVI-1 frequently has been found in myeloid leukemia and in several solid tumors and is associated with poor prognosis of patients with leukemia.12-15 Rearrangements of chromosome 3q26, which contains the EVI-1 gene, lead to overexpression of EVI-1 and are implicated in the development or progression of high-risk acute myeloid leukemia (AML).16 Importantly, EVI-1 is also highly expressed in a subgroup of AML patients without 3q26 rearrangements,12 which indicates the presence of other mechanisms of EVI-1 activation. Recently, several clinical studies revealed a positive correlation between EVI-1 (both EVI-1a and MDS1-EVI-1) overexpression and rearrangements of the mixed-lineage leukemia (MLL) gene located on chromosome 11q23.14,15 Furthermore, we have previously shown that Evi-1 deletion in cells transformed by MLL-ENL, a chimeric gene generated in t(11;19) leukemia, caused a distinct reduction of their proliferative activity.3 These results raise the possibility of functional interaction between Evi-1 and MLL oncoproteins.
The MLL gene encodes a DNA-binding protein that involves the SET [su(var)3-9, enhancer of zeste, and trithorax] domain with histone H3 lysine 4 methyltransferase activity, which regulates gene expression, including multiple Hox genes.17,18 Chromosome translocations involving the MLL gene are associated with aggressive forms of acute leukemia.19 Generation of MLL fusion proteins in leukemia deletes the SET domain that mediates histone H3 lysine 4 methylation and fuses the amino portion of MLL in frame with up to 50 different fusion partners, including ENL, AF9, and AF4.19 It has been shown that several Hox genes are consistently expressed at high levels in MLL-rearranged leukemias, which suggests that MLL oncoproteins inappropriately maintain their expression.20 Hox proteins form hetero-oligomers with TALE (3–amino-acid loop extension) homeobox proteins of the Pbx and Meis families, and Meis1 is also highly expressed in MLL-rearranged leukemias.21,22 A large body of evidence suggests that Hox/Meis genes are crucial targets of MLL oncoproteins in almost all cases of MLL-rearranged leukemias.22,23 However, MLL-rearranged leukemias are biologically and clinically diverse, and additional factors that underlie these differences have been characterized incompletely.
In the present study, we found that Evi-1 is frequently up-regulated by MLL-ENL or MLL-AF9 in a retroviral transduction assay. The reporter assay and chromatin immunoprecipitation analysis (ChIP) revealed that MLL-ENL binds to and activates the promoter of Evi-1. A retroviral transduction assay with defined populations of bone marrow (BM) progenitor cells revealed that MLL-ENL–mediated Evi-1 up-regulation occurs exclusively in HSCs and not in committed myeloid progenitor cells. These results suggest that up-regulation and maintenance of Evi-1 expression are features of MLL oncoproteins that work specifically in undifferentiated HSCs or progenitor cells.
Methods
Plasmid construction
The plasmids pMSCV-neo-Flag-MLL-ENL, pMSCV-neo-MLL-AF9, pMSCV-neo-AML1-ETO, pMXs-neo-E2A-HLF, and pMYs-HoxA9-ires-Meis1 have been described previously.3,24-26 The construction procedure is described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Mice
C57BL/6 mice were purchased from Sankyo Laboratory Service. For the Evi-1 deletion experiments, BM progenitor cells were harvested from wild-type (Evi-1+), loxP-flanked (Evi-1f), and Evi-1–deleted mutant mice (Evi-1+/− and Evi-1f/−).3 Mice were kept at the Center for Disease Biology and Integrative Medicine, University of Tokyo, according to institutional guidelines, and all animal experiments were approved by the University of Tokyo Institutional Animal Care and Use Committee.
Retrovirus transduction
To obtain retrovirus supernatants, Plat-E packaging cells27 were transfected with retrovirus vectors with FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Viral supernatants were collected after 48 hours of culture and used immediately for infection. To produce green fluorescent protein (GFP)–expressing or Cre-GFP–expressing retrovirus, we used ψMP34 packaging cells (Takara) stably transduced with pGCDNsam-eGFP or pGCDNsam-eGFP-iCre. Methods to isolate HSCs/progenitor cells from mice are described in supplemental Methods.28
Myeloid progenitor transformation assay
The myeloid progenitor transformation assay was performed as described previously26 with minor modifications. In brief, retrovirus-infected cells were cultured in MethoCult M3434 (StemCell Technologies) and 1 mg/mL G418 at a density of 1 × 105 cells per 35-mm dish. Colonies were counted weekly, and cells were cultured again at 1 × 104 per plate in M3434 without G418. Colony count scoring and replating were repeated every 7 days. For the Evi-1 deletion experiments, BM progenitor cells from Evi-1+/− or Evi-1f/− mice were used.
In vivo leukemogenesis assay
BM mononuclear cells (BM MNCs) harvested from 5-fluorouracil–injected mice were transduced with MLL-ENL or cMyc/bcl229 under conditions identical to those for the myeloid progenitor transformation assay. Retrovirally transduced BM progenitor cells (1 × 106) were injected into sublethally irradiated (6.5 Gy) recipients. When transplanted mice became moribund, they were euthanized and their BM MNCs isolated.
Luciferase reporter assay
For analysis of luciferase activities, Jurkat cells were seeded in 12-well culture plates at a density of 0.5 × 105 per well. The cells were transfected with 100 ng of pGL4-Luc or an equimolar amount of each reporter construct, together with 100 ng of pME18S or an equimolar amount of each expression plasmid and 5 ng of PSS-LacZ with FuGENE 6. After 48 hours of culture, cells were harvested and luciferase activities were measured in a Lumat LB9507 luminometer (Berthold Technologies) with a PicaGene luminescence kit (Toyo Ink). Each luciferase activity measurement was normalized to that of β-galactosidase, which was measured with Galacton-Plus (Roche). Data are expressed as mean ± SD from 2 or more separate experiments.
Chromatin immunoprecipitation
ChIP analysis was performed as described previously30 with minor modifications. The procedures are described in supplemental Methods.
Bioinformatics analyses
The gene-expression pattern in MLL-rearranged leukemia cells was assessed with use of the data of 285 individuals with AML published by Valk et al14 from the Gene Expression Omnibus (GSE1159). We used 13 cases with MLL rearrangement (after excluding 3 cases with MLL partial tandem duplication) and divided them into 2 groups according to the level of EVI-1 expression: 5 EVI-1–high (GSM20760, 20794, 20838, 20959, and 20966) and 8 EVI-1–low (GSM20757, 20844, 20879, 20891, 20934, 20936, 20938, and 20961) cases. Gene-set enrichment analyses (GSEAs) were performed with GSEA version 2.0 software available from the Broad Institute (http://www.broad.mit.edu/gsea)31 with a Signal2Noise metric for ranking genes and 1000 data permutations. Functional 1892 gene sets (C2) or 8 selected gene sets that represented HSC and progenitor cell clusters32 were evaluated (supplemental Methods).
In addition, gene-expression data of murine c-Kit+, Sca-1+, Lin− (KSL) cells and granulocyte-macrophage progenitor cells (GMPs) were obtained from the Gene Expression Omnibus (GSE3725).33 Dataset comparisons of 13 cases of EVI-1–high or –low leukemias were performed with dChip software (http://www.hsph.harvard.edu/∼cli/complab/dchip/).34 The expression value was calculated by use of a perfect match/mismatch model after transformation into a log2 scale. Differently regulated probe sets in EVI-1–high and –low leukemias were determined by fold change > 1.2, P < .05, and 90% lower confidence-bound criteria. In this way, 120 probes enriched in EVI-1–high leukemias and 192 probes enriched in EVI-1–low leukemias were extracted to make gene sets that represented EVI-1–high and –low MLL-rearranged leukemia, respectively (supplemental Table 2). GSEAs were performed with these gene sets and the gene-expression data of murine KSL cells and GMPs.
Statistics
Data were analyzed by Student t test. P values < .05 were considered significant.
Results
Evi-1 is up-regulated in myeloid progenitor cells immortalized by MLL oncoproteins
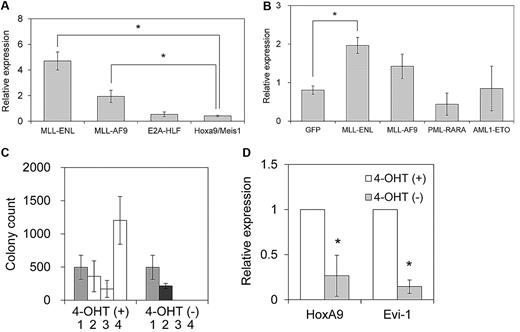
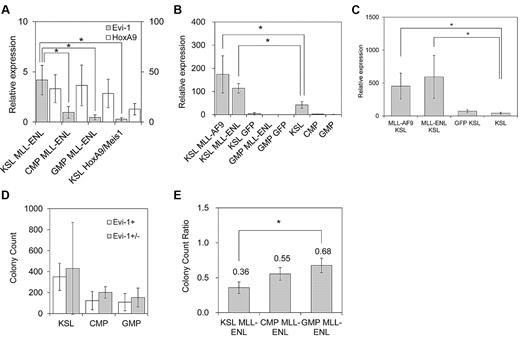
We first assessed Evi-1 expression in myeloid progenitor cells immortalized by various oncogenes. These oncogenes included 2 MLL chimeric genes (MLL-ENL and MLL-AF9), E2A-HLF, and the coexpression of HoxA9 and Meis1. MLL-ENL and MLL-AF9 are major forms of MLL oncoproteins generated in t(11;19) and t(9;11) leukemias, respectively, that contain nuclear proteins as a fusion partner. E2A-HLF is a chimeric gene generated in t(17;19) leukemia, and it transforms myeloid progenitor cells through Hox-independent mechanisms in mice.35 HoxA9 and Meis1 are crucial downstream targets of MLL oncoproteins, and coexpression of HoxA9 and Meis1 is sufficient for myeloid transformation.36 Primary murine hematopoietic progenitor cells (c-Kit+ cells) transduced with these oncogenes, but not those transduced with the empty vector, formed colonies in methylcellulose medium that could be replated through at least 3 rounds of culture (data not shown). After establishment of sustained clonogenic activity after more than 3 rounds of replating, the cells were harvested, and the expression level of Evi-1 was assessed by real-time quantitative polymerase chain reaction analysis. As shown in Figure 1A, Evi-1 was highly expressed in myeloid progenitor cells transformed by MLL-ENL or MLL-AF9 oncoproteins compared with those transformed by HoxA9 and Meis1, which are critical transcriptional targets of MLL oncoproteins. Therefore, HoxA9 and Meis1 appeared unable to complement the transcriptional effect of MLL oncoproteins on Evi-1. On the other hand, Evi-1 was not activated in E2A-HLF–immortalized cells.
Evi-1 is up-regulated in myeloid progenitor cells immortalized by MLL oncoproteins. (A) Murine c-Kit+ BM progenitor cells were retrovirally transduced with pMXs-neo-MLL-ENL, pMXs-neo-MLL-AF9, pMXs-neo-E2A-HLF, or pMYs-HoxA9-ires-Meis1. The expression level of Evi-1 in immortalized cells from the third to fourth round of serial replating in semisolid medium was quantified relative to BM MNCs with real-time polymerase chain reaction. Data are shown as mean ± SD. *P < .05. (B) Murine c-Kit+ BM progenitor cells were retrovirally transduced with leukemia oncogenes. Four types of myeloid leukemia genes cloned into MIG were retrovirally transduced into c-Kit+ BM progenitor cells. Forty-eight hours after initiation of retroviral transduction, GFP-positive cells were isolated and the expression level of Evi-1 was quantified relative to BM MNCs. Data are shown as mean ± SD. *P < .05. (C) Immortalization of c-Kit+ BM progenitor cells by MLL-ENL-ER is dependent on the presence of 4-OHT. Graph indicates the number of colonies, with SD, generated from 104 pMXs-neo-MLL-ENL-ER–transduced BM cells in the presence (□) or absence (■) of 1μM 4-OHT at each round after retroviral transduction. The number of G418-resistant colonies obtained by transduction of MLL-ENL-ER into 104 BM cells in the presence of 4-OHT is shown in the first round ( ). (D) Expression level of HoxA9 or Evi-1 in MLL-ENL-ER–transformed cells cultured with or without 1μM 4-OHT for 72 hours. The averages of the relative expression ratio of 4-OHT− cells (■) to 4-OHT+ cells (□) are shown with SD. *P < .05.
). (D) Expression level of HoxA9 or Evi-1 in MLL-ENL-ER–transformed cells cultured with or without 1μM 4-OHT for 72 hours. The averages of the relative expression ratio of 4-OHT− cells (■) to 4-OHT+ cells (□) are shown with SD. *P < .05.
Evi-1 is up-regulated in myeloid progenitor cells immortalized by MLL oncoproteins. (A) Murine c-Kit+ BM progenitor cells were retrovirally transduced with pMXs-neo-MLL-ENL, pMXs-neo-MLL-AF9, pMXs-neo-E2A-HLF, or pMYs-HoxA9-ires-Meis1. The expression level of Evi-1 in immortalized cells from the third to fourth round of serial replating in semisolid medium was quantified relative to BM MNCs with real-time polymerase chain reaction. Data are shown as mean ± SD. *P < .05. (B) Murine c-Kit+ BM progenitor cells were retrovirally transduced with leukemia oncogenes. Four types of myeloid leukemia genes cloned into MIG were retrovirally transduced into c-Kit+ BM progenitor cells. Forty-eight hours after initiation of retroviral transduction, GFP-positive cells were isolated and the expression level of Evi-1 was quantified relative to BM MNCs. Data are shown as mean ± SD. *P < .05. (C) Immortalization of c-Kit+ BM progenitor cells by MLL-ENL-ER is dependent on the presence of 4-OHT. Graph indicates the number of colonies, with SD, generated from 104 pMXs-neo-MLL-ENL-ER–transduced BM cells in the presence (□) or absence (■) of 1μM 4-OHT at each round after retroviral transduction. The number of G418-resistant colonies obtained by transduction of MLL-ENL-ER into 104 BM cells in the presence of 4-OHT is shown in the first round ( ). (D) Expression level of HoxA9 or Evi-1 in MLL-ENL-ER–transformed cells cultured with or without 1μM 4-OHT for 72 hours. The averages of the relative expression ratio of 4-OHT− cells (■) to 4-OHT+ cells (□) are shown with SD. *P < .05.
). (D) Expression level of HoxA9 or Evi-1 in MLL-ENL-ER–transformed cells cultured with or without 1μM 4-OHT for 72 hours. The averages of the relative expression ratio of 4-OHT− cells (■) to 4-OHT+ cells (□) are shown with SD. *P < .05.
Next, we assessed immediate changes in the expression level of Evi-1 in hematopoietic cells induced by transduction of myeloid leukemia genes. We used 2 chimeric genes frequently found in AML: AML1-ETO and PML-RARA. The former is generated in t(8;21) leukemia, whereas the latter is generated in t(15;17) leukemia. We retrovirally transduced these myeloid leukemia genes into BM myeloid progenitor cells. Transduced cells were isolated 48 hours later, and the expression level of Evi-1 was assessed. As shown in Figure 1B, Evi-1 expression was higher in MLL-ENL–transduced BM cells than in GFP-transduced cells, whereas it was not enhanced in either PML-RARA– or AML1-ETO–transduced cells, which are found in the most common forms of myeloid leukemia. Evi-1 was also not significantly up-regulated in MLL-AF9–transduced BM cells.
To determine the dependency of Evi-1 activation on MLL-ENL, we constructed MLL-ENL fused to the estrogen receptor (MLL-ENL-ER), which rendered the transcriptional and transforming properties of MLL-ENL strictly dependent on the presence of 4-hydroxy-tamoxifen (4-OHT). Consistent with a previous report,23 MLL-ENL-ER–transduced hematopoietic progenitor cells required 4-OHT for myeloid transformation (Figure 1C). Using this system, we quantified the expression of Evi-1 and HoxA9 in MLL-ENL-ER–immortalized cells. By 72 hours after 4-OHT withdrawal, the expression of Evi-1 and HoxA9 was reduced significantly compared with that seen in 4-OHT–positive cells. Thus, inactivation of MLL-ENL results in down-regulation of Evi-1 (Figure 1D), which again suggests a potential relationship between a distinct expression of Evi-1 and MLL-ENL.
Expression of Evi-1 in MLL fusion-transformed leukemic cells in vivo
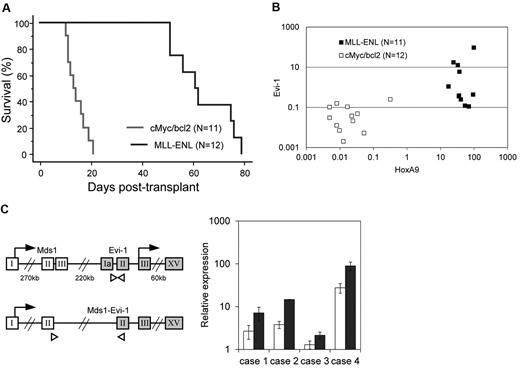
We next assessed Evi-1 expression in leukemic cells transformed by MLL-ENL using a mouse leukemia model. As a control, we used cMyc/bcl2-induced biphenotypic leukemia, the BM infiltration of which consists of a large number of myeloblasts and a small number of lymphoblasts. We harvested BM MNCs from mice treated with 5-fluorouracil. These cells were transduced with MLL-ENL or cMyc/bcl2 and then intravenously injected into sublethally irradiated recipient mice. Mice transplanted with MLL–ENL– or cMyc/bcl2–transduced cells developed leukemia within 85 or 26 days, respectively, which is consistent with previous reports (Figure 2A).29,37 Leukemic cells were isolated from BM of moribund mice, and the expression level of Evi-1 was determined along with that of HoxA9, a well-known target of MLL-ENL. As shown in Figure 2B, Evi-1 was distinctly up-regulated in MLL-transformed leukemic cells of 4 of the 11 mice but was never activated in cMyc/bcl2–transformed cells (P = .037). The expression level of Evi-1 varied considerably among individuals, whereas that of HoxA9 was similar (Figure 2B). These results suggest that the regulation of Evi-1 is independent of HoxA9. Because the Evi-1 gene gives rise to 2 major alternative forms, Evi-1a and Mds1-Evi-1 (Evi-1c), we then assessed the expression of those Evi-1 isoforms using specific primers to detect respective forms. Interestingly, in all 4 individuals with high Evi-1 expression, the expression of both isoforms, Evi-1a and Mds1-Evi-1, was up-regulated (Figure 2C).
Evi-1 is frequently up-regulated in leukemic cells transformed by MLL fusion protein in vivo. (A) Survival curves of sublethally irradiated recipients transplanted with BM cells transduced with either MSCV-neo-MLL-ENL (n = 11; blue) or MSCV-cMyc-ires-bcl2 (n = 12; red) are shown. (B) Expression levels of Evi-1 and HoxA9 in leukemic cells transformed by MLL fusion genes (MLL-ENL; n = 11; ■) or cMyc/bcl2 (n = 12; □) are indicated. Expression levels of Evi-1 and HoxA9 relative to BM MNCs are shown as squares. (C left) Gene structures of Evi-1a and Mds1-Evi-1 and positions of primer sets for quantitative real-time polymerase chain reaction are shown. The exons, start codons, and primers are depicted in boxes, with arrows, and with white triangles, respectively. Sequences of primers are presented in the supplemental Methods. (Right) Expression levels of Evi-1a (□) and Mds1-Evi-1 ( ) in leukemic cells from 4 mice with high Evi-1 expression relative to BM MNCs are shown with SD.
) in leukemic cells from 4 mice with high Evi-1 expression relative to BM MNCs are shown with SD.
Evi-1 is frequently up-regulated in leukemic cells transformed by MLL fusion protein in vivo. (A) Survival curves of sublethally irradiated recipients transplanted with BM cells transduced with either MSCV-neo-MLL-ENL (n = 11; blue) or MSCV-cMyc-ires-bcl2 (n = 12; red) are shown. (B) Expression levels of Evi-1 and HoxA9 in leukemic cells transformed by MLL fusion genes (MLL-ENL; n = 11; ■) or cMyc/bcl2 (n = 12; □) are indicated. Expression levels of Evi-1 and HoxA9 relative to BM MNCs are shown as squares. (C left) Gene structures of Evi-1a and Mds1-Evi-1 and positions of primer sets for quantitative real-time polymerase chain reaction are shown. The exons, start codons, and primers are depicted in boxes, with arrows, and with white triangles, respectively. Sequences of primers are presented in the supplemental Methods. (Right) Expression levels of Evi-1a (□) and Mds1-Evi-1 ( ) in leukemic cells from 4 mice with high Evi-1 expression relative to BM MNCs are shown with SD.
) in leukemic cells from 4 mice with high Evi-1 expression relative to BM MNCs are shown with SD.
MLL oncoproteins specifically up-regulate Evi-1 through 5′ promoter regions
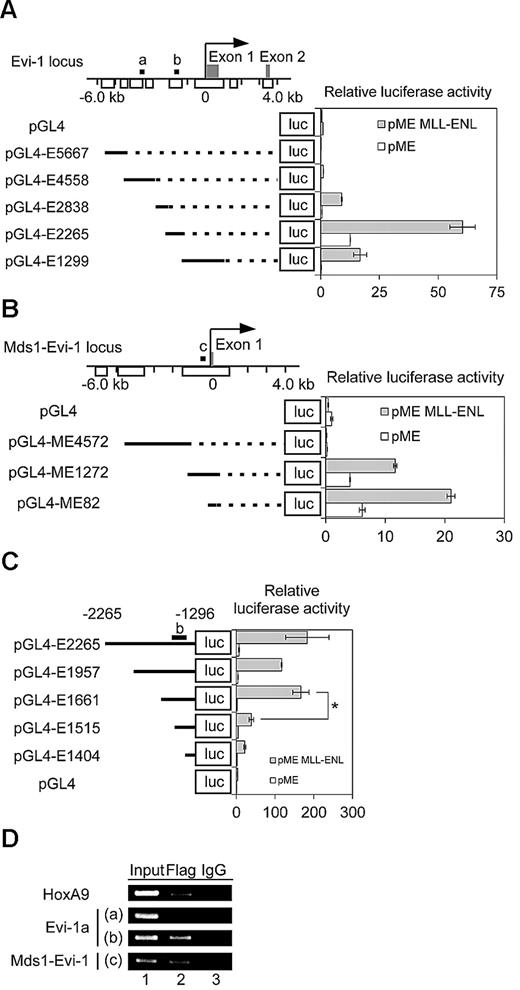
To determine whether MLL-ENL regulates the transcription of Evi-1, we performed a luciferase reporter assay. Because the genomic region 5.7 kb upstream of the transcription start site (TSS) of Evi-1a is highly conserved among species (supplemental Figure 1), we divided the region into 5 fragments and inserted them upstream of luciferase cDNA in the pGL4-Basic vector (Figure 3A). Each reporter plasmid was transfected into Jurkat cells along with the MLL-ENL expression plasmid. MLL-ENL exhibited the greatest increase in luciferase activity with pGL4-E2265 (Figure 3A). These data suggest that MLL-ENL up-regulates Evi-1a through the region that lies between −2.3 and −1.3 kb of the TSS. We next cloned 3 fragments within the genomic region around the Mds1-Evi-1 TSS, which is also evolutionally conserved (Figure 3B; supplemental Figure 1). The luciferase reporter assay revealed that MLL-ENL activated Mds1-Evi-1 transcription mainly through the region between −0.1 and 0.3 kb of the TSS (Figure 3B).
MLL-ENL binds to the promoter regions of both Evi-1a and Mds1-Evi-1. (A left) Five segments of the Evi-1a promoter were inserted upstream of the luciferase cassette of the pGL4-Luc vector to generate luciferase reporter constructs. Arrows, gray boxes, white boxes, solid lines, and dashed lines represent TSS, exons, highly conserved regions between human and mice, DNA sequences cloned into pGL4-Luc, and connection of each DNA segment and luciferase gene, respectively. (Right) Graph shows relative luciferase activity of Evi-1a promoter reporter constructs in Jurkat cell lysates with transiently transfected MLL-ENL (shaded bars) compared with that without MLL-ENL (open bars). Data shown are mean ± SD from 3 independent experiments. (B left) 3 segments of Mds1-Evi-1 promoter were inserted upstream of the luciferase cassette of pGL4-Luc. (Right) Graph shows relative luciferase activity of Mds1/Evi-1 promoter reporter constructs in Jurkat cell lysates with transiently transfected MLL-ENL (shaded bars) compared with that without MLL-ENL (open bars). Data shown are mean ± SD from 3 independent experiments. (C left) Serial deletions of pGL4-E2265 were constructed. The pGL4-E1957 through pGL4-E1404 constructs are named according to the base length between the N-terminal residue of inserted fragments and TSS of Evi-1a on murine genome. The DNA fragment inserted in pGL4-E2265 corresponds to the genomic region that is between 2265 and 1296 bp upstream of the TSS of Evi-1a. (Right) Experiments were performed as described in panel A. Data are representative of 3 independent experiments and shown as mean ± SD. *P < .05. (D) Enrichment of MLL-ENL to the promoter of Evi-1a and Mds1-Evi-1 was detected by ChIP. Genomic DNA fragments were immunoprecipitated with anti-Flag antibody (lane 2) or normal mouse IgG (lane 3) from formaldehyde-fixed leukemic cells transduced with Flag-MLL-ENL. DNA fragments containing the indicated promoter regions of Evi-1a or Mds1-Evi-1 were amplified by polymerase chain reaction. The positions of the amplified regions in Evi-1a or Mds1-Evi-1 promoters (labeled a, b, and c) are shown in Figure 3A, B, or C, respectively. For controls, each genomic region was amplified from 1% of purified DNA after formaldehyde fixation and sonication (input, lane 1). Representative data of 4 experiments are shown.
MLL-ENL binds to the promoter regions of both Evi-1a and Mds1-Evi-1. (A left) Five segments of the Evi-1a promoter were inserted upstream of the luciferase cassette of the pGL4-Luc vector to generate luciferase reporter constructs. Arrows, gray boxes, white boxes, solid lines, and dashed lines represent TSS, exons, highly conserved regions between human and mice, DNA sequences cloned into pGL4-Luc, and connection of each DNA segment and luciferase gene, respectively. (Right) Graph shows relative luciferase activity of Evi-1a promoter reporter constructs in Jurkat cell lysates with transiently transfected MLL-ENL (shaded bars) compared with that without MLL-ENL (open bars). Data shown are mean ± SD from 3 independent experiments. (B left) 3 segments of Mds1-Evi-1 promoter were inserted upstream of the luciferase cassette of pGL4-Luc. (Right) Graph shows relative luciferase activity of Mds1/Evi-1 promoter reporter constructs in Jurkat cell lysates with transiently transfected MLL-ENL (shaded bars) compared with that without MLL-ENL (open bars). Data shown are mean ± SD from 3 independent experiments. (C left) Serial deletions of pGL4-E2265 were constructed. The pGL4-E1957 through pGL4-E1404 constructs are named according to the base length between the N-terminal residue of inserted fragments and TSS of Evi-1a on murine genome. The DNA fragment inserted in pGL4-E2265 corresponds to the genomic region that is between 2265 and 1296 bp upstream of the TSS of Evi-1a. (Right) Experiments were performed as described in panel A. Data are representative of 3 independent experiments and shown as mean ± SD. *P < .05. (D) Enrichment of MLL-ENL to the promoter of Evi-1a and Mds1-Evi-1 was detected by ChIP. Genomic DNA fragments were immunoprecipitated with anti-Flag antibody (lane 2) or normal mouse IgG (lane 3) from formaldehyde-fixed leukemic cells transduced with Flag-MLL-ENL. DNA fragments containing the indicated promoter regions of Evi-1a or Mds1-Evi-1 were amplified by polymerase chain reaction. The positions of the amplified regions in Evi-1a or Mds1-Evi-1 promoters (labeled a, b, and c) are shown in Figure 3A, B, or C, respectively. For controls, each genomic region was amplified from 1% of purified DNA after formaldehyde fixation and sonication (input, lane 1). Representative data of 4 experiments are shown.
To further confirm the crucial region for Evi-1a activation by MLL-ENL, we generated a series of pGL4-E2265 deletions and performed a luciferase reporter assay (supplemental Methods). We observed no significant changes of luciferase activity between pGL4-E2265 and E1661 (Figure 3C). In contrast, a remarkable reduction of luciferase activity was observed with the deletion of the N-terminal 146 bases from pGL4-E1661 (Figure 3C). These data suggest that the responsive elements for MLL-ENL are within 1.7 and 1.5 kb upstream of the Evi-1a TSS.
To determine whether MLL-ENL binds to these genomic regions in vivo, we performed ChIP with lysates from MLL-ENL–transformed cells collected from the mice with leukemia. The ChIP assay demonstrated that MLL-ENL bound to the 5′ promoter regions of both Evi-1a and Mds1-Evi-1, which are responsible for activation in the reporter assay, but not to the irrelevant region (Figure 3D).
MLL oncoproteins, but not wild-type MLL, activate the promoter of Evi-1
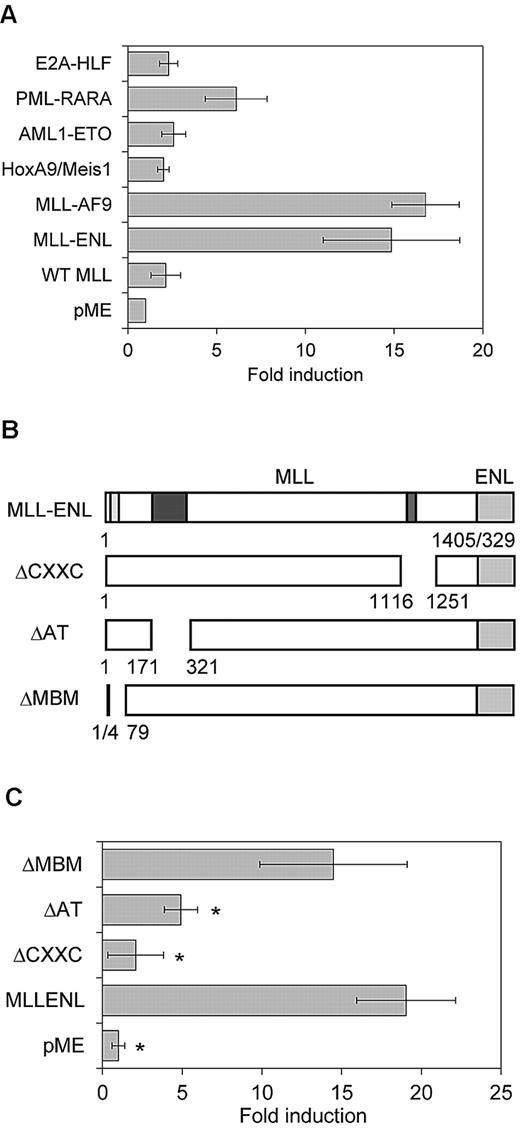
We then performed the luciferase assay using another MLL-associated gene, MLL-AF9, and several other leukemia-associated genes (PML-RARA, AML1-ETO, E2A-HLF, and the combination of HoxA9 and Meis1). MLL-AF9 exhibited transcriptional activity comparable with that of MLL-ENL on the Evi-1 promoter (Figure 4A). In contrast, PML-RARA, AML1-ETO, E2A-HLF, and HoxA9/Meis1 exhibited minimal or no transcriptional activity on the Evi-1 promoter (Figure 4A), which is in agreement with the results of the expression analysis of Evi-1 in BM cells (Figure 1A-B).
Reporter assays using MLL-ENL deletion mutants and other oncoproteins. (A) Transactivation of pGL4-E2265 induced by MLL-AF9, AML1-ETO, PML-RARA, E2A-HLF, HoxA9/Meis1, or wild-type MLL (WT MLL) is shown. Data are presented as a relative fold increase in mean luciferase activity, with SD, after adjustment for β-galactosidase activity. (B) Schematics represent the composition of MLL-ENL deletion mutants. Numbers denote amino acid positions in wild-type MLL and ENL. Positions of CXXC DNA binding motif (red), AT-hooks DNA binding motif (AT; blue), and menin-binding motif (MBM; yellow) are shown in the schematics of intact MLL-ENL. (C) Transactivation of pGL4-E2265 induced by intact MLL-ENL or its deletion mutants is shown. Data are presented as described in (A). *P < .05 versus MLL-ENL.
Reporter assays using MLL-ENL deletion mutants and other oncoproteins. (A) Transactivation of pGL4-E2265 induced by MLL-AF9, AML1-ETO, PML-RARA, E2A-HLF, HoxA9/Meis1, or wild-type MLL (WT MLL) is shown. Data are presented as a relative fold increase in mean luciferase activity, with SD, after adjustment for β-galactosidase activity. (B) Schematics represent the composition of MLL-ENL deletion mutants. Numbers denote amino acid positions in wild-type MLL and ENL. Positions of CXXC DNA binding motif (red), AT-hooks DNA binding motif (AT; blue), and menin-binding motif (MBM; yellow) are shown in the schematics of intact MLL-ENL. (C) Transactivation of pGL4-E2265 induced by intact MLL-ENL or its deletion mutants is shown. Data are presented as described in (A). *P < .05 versus MLL-ENL.
Because wild-type MLL also transcriptionally activates its targets such as Hox genes, we tested its transcriptional activity on the Evi-1 promoter. We observed no significant transcriptional activities of wild-type MLL on the Evi-1 promoter, which suggests that wild-type MLL by itself is not sufficient for activation of Evi-1 (Figure 4A).
Next, we determined the domain contribution of MLL-ENL in the activation of Evi-1 using a series of MLL-ENL mutants (Figure 4B). The CXXC domain of MLL mediates binding to nonmethylated CpG DNA and is essential for myeloid transformation.38 The AT-hook motifs of MLL are thought to facilitate binding to AT-rich DNA in the minor grove but are dispensable for myeloid transformation.38 Consistent with its contribution to the transforming activity of MLL-ENL, the CXXC domain was essential for Evi-1 activation (Figure 4C). Unexpectedly, deletion of the AT-hook motifs affected reporter activity, although the AT-hook motif was dispensable for MLL-ENL–mediated myeloid transformation and up-regulation of Hox genes, and the menin-binding motif was not required for Evi-1 activation (Figure 4C).39
Evi-1 is up-regulated by MLL-ENL exclusively in HSC-derived transformed cells
Although clinical studies revealed a positive correlation between high Evi-1 expression and MLL rearrangements, there exists a subset of MLL-rearranged leukemia with normal Evi-1 expression levels.14,15 We also observed that the expression level of Evi-1 in MLL cells varied considerably among the individual mice (Figure 2B). Because Evi-1 is preferentially expressed in HSCs and the expression level decreased on differentiation,2 we hypothesized that Evi-1 expression in MLL cells depends on their cellular origin.
To test this hypothesis, we transduced MLL-ENL into the defined hematopoietic populations, including KSL cells, which contain HSCs, myeloid-restricted common myeloid progenitor cells (CMPs), and GMPs.28 Consistent with a previous report, MLL-ENL immortalized committed myeloid progenitor cells (CMPs and GMPs), as well as KSL cells (data not shown).40 After 3 rounds of replating in semisolid medium, we compared expression levels of Evi-1 and HoxA9 in MLL-ENL–immortalized cells derived from KSL cells, CMPs, and GMPs. Notably, Evi-1 expression was significantly high in KSL-derived MLL-ENL–immortalized cells compared with CMP- and GMP-derived cells immortalized by MLL-ENL (Figure 5A). In contrast, HoxA9 was similarly up-regulated in the 3 populations (Figure 5A).
MLL-ENL up-regulates Evi-1 expression exclusively in HSCs. (A) Defined hematopoietic populations were transduced with pMXs-neo-MLL-ENL or pMYs-HoxA9-ires-Meis1 and replated in semisolid medium. The expression level of Evi-1 (shaded bars; scale on the left) and HoxA9 (open bars; scale on the right) in MLL-ENL–transformed cells from each population (KSL MLL-ENL, CMP MLL-ENL, or GMP MLL-ENL lanes) and HoxA9/Meis1-transformed KSL cells (KSL HoxA9/Meis1 lane) was quantified relative to BM MNCs. Data are shown as mean ± SD from 2 independent experiments. *P < .05 vs CMP MLL-ENL, GMP MLL-ENL, or KSL HoxA9/Meis1, respectively. (B) KSL cells and GMPs were transduced with MIG (KSL/GMP GFP lanes), MIG-MLL-ENL (KSL/GMP MLL-ENL lanes), or MIG-MLL-AF9 (KSL MLL-AF9 lane). After 48 hours of transduction, the expression level of Evi-1 in GFP-positive cells was quantified relative to BM MNCs by real-time polymerase chain reaction and was compared with that of freshly isolated KSL cells (KSL lane), CMPs, and GMPs. Data shown are mean ± SD from 3 independent experiments. *P < .05. (C) BM progenitor cells from 5-fluorouracil–treated mice were transduced with MIG (GFP KSL lane), MIG-MLL-ENL (MLL-ENL KSL lane), or MIG-MLL-AF9 (MLL-AF9 KSL lane). After 36 hours of transduction, the expression level of Evi-1 in GFP-positive cells isolated from the KSL population was quantified relative to BM MNCs and compared with that of freshly isolated KSL cells (KSL lane). Data are mean ± SD from 3 independent experiments. *P < .05. (D) BM KSL cells, CMPs, and GMPs were isolated from Evi-1+ (open bars) and Evi-1+/− (shaded bars) mice and transformed by MLL-ENL in the same way as in the myeloid progenitor transformation assay. Bar graph shows mean colony numbers ± SD in the third round of serial replating from 2 independent experiments. *P < .05. (E) BM KSL cells, CMPs, and GMPs were isolated from Evi-1f/− mice. After they were transformed by MLL-ENL as in (D), they were transduced with either pGCDNsam-eGFP or pGCDNsam-eGFP-iCre. GFP-positive cells were isolated, and colony-forming activity after Evi-1 deletion was assessed in the next round of plating. Bar graph shows colony count ratio of iCre-GFP–transduced cells compared with GFP-transduced cells. Data are mean ± SD from 2 independent experiments. *P < .05.
MLL-ENL up-regulates Evi-1 expression exclusively in HSCs. (A) Defined hematopoietic populations were transduced with pMXs-neo-MLL-ENL or pMYs-HoxA9-ires-Meis1 and replated in semisolid medium. The expression level of Evi-1 (shaded bars; scale on the left) and HoxA9 (open bars; scale on the right) in MLL-ENL–transformed cells from each population (KSL MLL-ENL, CMP MLL-ENL, or GMP MLL-ENL lanes) and HoxA9/Meis1-transformed KSL cells (KSL HoxA9/Meis1 lane) was quantified relative to BM MNCs. Data are shown as mean ± SD from 2 independent experiments. *P < .05 vs CMP MLL-ENL, GMP MLL-ENL, or KSL HoxA9/Meis1, respectively. (B) KSL cells and GMPs were transduced with MIG (KSL/GMP GFP lanes), MIG-MLL-ENL (KSL/GMP MLL-ENL lanes), or MIG-MLL-AF9 (KSL MLL-AF9 lane). After 48 hours of transduction, the expression level of Evi-1 in GFP-positive cells was quantified relative to BM MNCs by real-time polymerase chain reaction and was compared with that of freshly isolated KSL cells (KSL lane), CMPs, and GMPs. Data shown are mean ± SD from 3 independent experiments. *P < .05. (C) BM progenitor cells from 5-fluorouracil–treated mice were transduced with MIG (GFP KSL lane), MIG-MLL-ENL (MLL-ENL KSL lane), or MIG-MLL-AF9 (MLL-AF9 KSL lane). After 36 hours of transduction, the expression level of Evi-1 in GFP-positive cells isolated from the KSL population was quantified relative to BM MNCs and compared with that of freshly isolated KSL cells (KSL lane). Data are mean ± SD from 3 independent experiments. *P < .05. (D) BM KSL cells, CMPs, and GMPs were isolated from Evi-1+ (open bars) and Evi-1+/− (shaded bars) mice and transformed by MLL-ENL in the same way as in the myeloid progenitor transformation assay. Bar graph shows mean colony numbers ± SD in the third round of serial replating from 2 independent experiments. *P < .05. (E) BM KSL cells, CMPs, and GMPs were isolated from Evi-1f/− mice. After they were transformed by MLL-ENL as in (D), they were transduced with either pGCDNsam-eGFP or pGCDNsam-eGFP-iCre. GFP-positive cells were isolated, and colony-forming activity after Evi-1 deletion was assessed in the next round of plating. Bar graph shows colony count ratio of iCre-GFP–transduced cells compared with GFP-transduced cells. Data are mean ± SD from 2 independent experiments. *P < .05.
We next evaluated the effect of HoxA9/Meis1 on the expression of Evi-1 in the 3 hematopoietic populations noted above. HoxA9/Meis1 transformed KSL cells (data not shown), whereas CMPs and GMPs were not transformed in our experiments. Consistent with our results in the myeloid progenitor transformation assay, KSL cells transformed by HoxA9/Meis1 exhibited a low level of expression of Evi-1 (Figure 5A). These data indicate that Evi-1 is not a transcriptional target of HoxA9 or Meis1 even in KSL cells.
To identify whether Evi-1 transcription in KSL cells is activated or maintained by MLL-ENL, we first assessed immediate changes in the Evi-1 expression level induced by MLL-ENL. We retrovirally transduced MLL-ENL into KSL cells and GMPs. Transduced cells were isolated 48 hours later, and the expression level of Evi-1 was assessed. Remarkably, Evi-1 expression in MLL-ENL–transduced KSL cells was significantly higher than that in freshly isolated KSL cells (Figure 5B). On the other hand, MLL-ENL–transduced GMPs showed low expression of Evi-1 compared with freshly isolated GMPs (Figure 5B).
To further examine the transcriptional regulation of Evi-1 in undifferentiated hematopoietic cells, we retrovirally transduced MLL-ENL into BM MNCs from 5-fluorouracil–treated mice in which HSCs were propagated. Thirty-six hours later, transduced cells were isolated from the KSL population. Consistent with the results shown in Figure 5B, MLL-ENL–transduced KSL population cells exhibited significantly higher expression of Evi-1 than freshly isolated KSL cells (Figure 5C).
We also examined the transcriptional regulation of Evi-1 by MLL-AF9, because MLL-AF9 exhibited a transcriptional activity comparable with that of MLL-ENL on the Evi-1 promoter (Figure 4A). Transduction analysis with undifferentiated hematopoietic cells clearly demonstrated that MLL-AF9 activates Evi-1 expression in the same manner as MLL-ENL (Figure 5B-C). Collectively, our results show that Evi-1 transcription is not only maintained but also activated by MLL-ENL or MLL-AF9 exclusively in undifferentiated hematopoietic populations such as KSL cells.
Propagation of MLL-ENL–immortalized HSCs is highly dependent on Evi-1
Previous studies showed that Evi-1 is required for efficient propagation of MLL-ENL–immortalized BM cells.3 However, it had not been assessed whether the requirement for Evi-1 differs depending on the cellular origin that is immortalized by MLL-ENL. To address this issue, BM progenitor cells from Evi-1+/− and Evi-1f/− mice were sorted into KSL cells, CMPs, and GMPs. Then, they were transduced with MLL-ENL and immortalized via a myeloid transformation assay. First, using Evi-1+/− cells, we revealed that deletion of 1 Evi-1 allele had no significant impact on the clonogenic activity of each hematopoietic population (Figure 5D). Next, using Evi-1f/− cells and Cre-GFP retrovirus, we completely disrupted Evi-1 alleles in MLL-ENL–immortalized cells derived from the defined populations (supplemental Figure 2). Then, GFP- or Cre-GFP–infected cells were sorted and cultured for another round in semisolid medium to compare the effects of Evi-1 deletion on the clonogenic activity among populations. Notably, colony formation of MLL-ENL–immortalized cells derived from KSL cells was most severely attenuated by disruption of Evi-1, compared with cells derived from CMPs or GMPs (Figure 5E). On subsequent replating of Evi-1–deleted cells, however, we observed no significant difference in colony counts among populations (supplemental Figure 2). These results indicate that MLL-ENL–immortalized cells are heterogenous with regard to dependency on Evi-1 for proliferation, even if they are derived from KSL cells. The frequency of Evi-1–dependent cells should be highest in HSC-derived cells and low in progenitor cell–derived cells. Thus, if colony counts decrease immediately after Evi-1 deletion in HSC-derived cells, the residual cells, most of which are no longer dependent on Evi-1, would show almost the same clonogenic activity as progenitor cell–derived cells in the next round.
HSC genes are enriched in EVI-1–high cases of MLL-rearranged leukemia
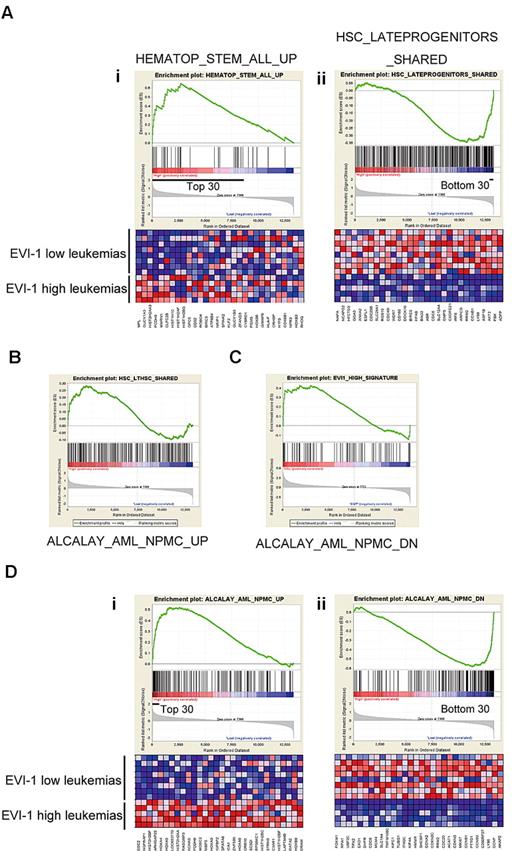
On the basis of the finding that up-regulation of Evi-1 in MLL fusion-transformed cells is related to their origin, we hypothesized that the gene-expression pattern in human cases of MLL-rearranged leukemia would also reflect their origin. To address this issue, we extracted the gene-expression data of 13 MLL-rearranged AML patients reported by Valk et al from the Gene Expression Omnibus14 and divided them into 2 groups: 5 EVI-1–high cases and 8 EVI-1–low cases. We then applied GSEA to identify functional gene sets (C2) correlated with EVI-1 expression and found that 2 and 68 gene sets were particularly enriched in the EVI-1–high and EVI-1–low groups, respectively (false-discovery rate < 0.01; gene sets consisting of < 30 genes were excluded; supplemental Table 1). Of note, GSEA revealed a strong correlation of genes up-regulated in EVI-1–high leukemias with the gene set that identifies HSCs (Figure 6Ai). Conversely, the genes down-regulated in EVI-1–high leukemias were strongly correlated with a gene set that is highly expressed in progenitor cells (Figure 6Aii). We also found another gene set that contained long-term HSC-enriched genes was significantly correlated to genes up-regulated in EVI-1–high leukemias (Figure 6B).
Gene-expression profiles of Evi-1–high leukemias with MLL rearrangement revealed a stem cell–like character. (A) A strong correlation of up-regulated genes in EVI-1–high leukemias with a gene set representing HSCs and of down-regulated genes in EVI-1–high leukemias with a gene set typical of progenitor cells is shown. GSEA of gene expression in human EVI-1–high AMLs with MLL rearrangement (n = 5) was compared with EVI-1–low AMLs with MLL rearrangement (n = 8) using functional gene sets (C2). (Ai,ii) GSEA enrichment plots of the selected gene sets. (Ai) Up-regulated genes in human HSCs.41 (Aii) Up-regulated genes in hematopoietic late progenitor cells.32 The corresponding heat maps represent expression of the 30 leading genes of the respective gene sets. Up-regulated and down-regulated genes are shown in red and blue, respectively. (B) GSEA plots show that expression of genes representing long-term HSCs (LTHSC) is enriched in EVI-1–high AMLs with MLL rearrangement compared with EVI-1–low AMLs with MLL rearrangement. Normalized enrichment score = 1.33; false-discovery rate q = .141. (C) GSEA of gene expression in 5 normal KSL samples and 4 normal GMP samples using a gene set representing EVI-1 high leukemia with MLL rearrangement. GSEA plots show that expression of genes representing EVI-1–high leukemia is enriched in KSL cells compared with GMPs. Normalized enrichment score = 1.46; false-discovery rate q = .025. (D) A strong correlation of up-regulated genes in EVI-1–high leukemias with those in cytoplasmic nucleophosmin–positive (NPMc+) leukemias existed. GSEA of gene expression in human EVI-1–high AMLs with MLL rearrangement (n = 5) was compared with EVI-1–low AMLs with MLL rearrangement (n = 8) using functional gene sets (C2). (Di-ii) GSEA enrichment plots of the selected gene sets. (Di) Up-regulated genes in NPMc+ leukemias.42 (Dii) Down-regulated genes in NPMc+ leukemias.42
Gene-expression profiles of Evi-1–high leukemias with MLL rearrangement revealed a stem cell–like character. (A) A strong correlation of up-regulated genes in EVI-1–high leukemias with a gene set representing HSCs and of down-regulated genes in EVI-1–high leukemias with a gene set typical of progenitor cells is shown. GSEA of gene expression in human EVI-1–high AMLs with MLL rearrangement (n = 5) was compared with EVI-1–low AMLs with MLL rearrangement (n = 8) using functional gene sets (C2). (Ai,ii) GSEA enrichment plots of the selected gene sets. (Ai) Up-regulated genes in human HSCs.41 (Aii) Up-regulated genes in hematopoietic late progenitor cells.32 The corresponding heat maps represent expression of the 30 leading genes of the respective gene sets. Up-regulated and down-regulated genes are shown in red and blue, respectively. (B) GSEA plots show that expression of genes representing long-term HSCs (LTHSC) is enriched in EVI-1–high AMLs with MLL rearrangement compared with EVI-1–low AMLs with MLL rearrangement. Normalized enrichment score = 1.33; false-discovery rate q = .141. (C) GSEA of gene expression in 5 normal KSL samples and 4 normal GMP samples using a gene set representing EVI-1 high leukemia with MLL rearrangement. GSEA plots show that expression of genes representing EVI-1–high leukemia is enriched in KSL cells compared with GMPs. Normalized enrichment score = 1.46; false-discovery rate q = .025. (D) A strong correlation of up-regulated genes in EVI-1–high leukemias with those in cytoplasmic nucleophosmin–positive (NPMc+) leukemias existed. GSEA of gene expression in human EVI-1–high AMLs with MLL rearrangement (n = 5) was compared with EVI-1–low AMLs with MLL rearrangement (n = 8) using functional gene sets (C2). (Di-ii) GSEA enrichment plots of the selected gene sets. (Di) Up-regulated genes in NPMc+ leukemias.42 (Dii) Down-regulated genes in NPMc+ leukemias.42
In addition, we extracted the expression data of murine normal KSL cells and GMPs and applied GSEA using 2 gene sets that represented EVI-1–high and EVI-1–low MLL-rearranged leukemias, respectively (supplemental Table 2). As expected, GSEA showed a significant correlation of genes enriched in EVI-1–high leukemias with those in KSL cells (Figure 6C), although the reverse correlation between genes representing EVI-1–low leukemias and GMPs was not significant (supplemental Figure 3). These results are consistent with our findings that Evi-1 is up-regulated by MLL oncoproteins in HSCs and suggest that EVI-1–high MLL-rearranged leukemias are derived from HSCs. GSEA also demonstrated strong correlation between the up-regulated genes in AMLs with aberrant cytoplasmic localization of nucleophosmin (NPMc+ AMLs) and those in EVI-1–high cases of MLL-rearranged AMLs (Figure 6Di-ii; supplemental Table 1).
Discussion
Despite the established role of Evi-1 in leukemogenesis, the molecular mechanisms for Evi-1 activation in leukemic cells have been poorly understood. Recently, several clinical studies revealed a positive correlation between EVI-1 overexpression and MLL rearrangements in AML patients.14,15 Furthermore, we have previously shown that Evi-1 deletion in MLL-ENL–immortalized cells caused a distinct reduction of their colony-forming capacity, which suggests a functional interaction between Evi-1 and MLL oncoproteins.3 In the present study, we demonstrated that MLL oncoproteins activate transcription of the Evi-1 gene in hematopoietic cells. Importantly, this MLL-mediated Evi-1 activation occurs exclusively in HSCs and not in committed myeloid progenitor cells.
Mds1 is located approximately 140 and 500 kb upstream of the first exon of Evi-1 in the human and mouse genome, respectively; therefore, the expression of Evi-1a and Mds1-Evi-1 is regulated by different promoters. Nevertheless, MLL oncoproteins bind to the promoters of both Evi-1a and Mds1-Evi-1 and activate their transcription (Figure 3A-B,D). These findings are consistent with the clinical observations that showed that expression of both EVI-1a and Mds1-EVI-1 is frequently enhanced in MLL-rearranged leukemia.15 Although some evidence suggests that Evi-1a is oncogenic and Mds1-Evi-1 contributes to tumor suppression,12,43 several reports showed that the activating retroviral insertions in the Mds1/Evi-1 locus were involved in long-term dominance in hematopoiesis, which suggests a similar function of Mds1/Evi-1 and Evi-1.44,45 The specific roles of Evi-1a and Mds1-Evi-1 in MLL-rearranged leukemia should be clarified in future studies.
We found a genomic region, 146 bp in length, that is thought to be crucial for Evi-1a activation by MLL-ENL (Figure 3C). The ChIP assay revealed that DNA binding of MLL-ENL was enriched near this genomic region, which also suggests the importance of this region for MLL-ENL to regulate Evi-1 (Figure 3D). To identify the precise genomic DNA sequence to which MLL binds, we performed an electrophoresis mobility shift assay using 3 probes from the genomic region with purified His-tagged protein that contained the MLL-ENL CXXC domain. In this setting, we observed a sequence-specific shifted band using 1 of 3 probes; however, it was not supershifted by the addition of His antibodies (data not shown). These results may be due to some technical difficulties in protein-antibody binding in native conditions. Therefore, it remains to be determined whether this region fragment is sufficient for MLL-ENL to bind to DNA or whether other regions are also involved.
Several studies showed that both HSCs and committed myeloid progenitor cells could be transformed by retroviral transduction of MLL oncoproteins, and they could develop immunophenotypically similar AML.40 Of note, we found that Evi-1 was activated by MLL-ENL or MLL-AF9 exclusively when it was transduced into KSL cells (Figure 5A-C). Previously, Chen et al46 reported that HSCs from MLL-AF9 knock-in mice express high levels of Evi-1. In light of that report and the present results, the cellular milieu provided by HSCs appears necessary for Evi-1 up-regulation by MLL oncoproteins. One possibility that accounts for these phenomena is that MLL-ENL can bind to the promoter region of Evi-1 only in HSCs. However, using ChIP assay, we found that MLL-ENL bound to the promoter regions of Evi-1a and Mds1-Evi-1 even in leukemic cells with low Evi-1 expression (data not shown). Therefore, binding to the Evi-1 promoter alone is not sufficient for activation of the transcription of Evi-1. The methylation status of the Evi-1 promoter can affect the expression of Evi-1. To address this issue, we analyzed the methylation status of the Evi-1 promoter in MLL-ENL–transformed cells using bisulfite DNA sequencing. However, the methylation status at CpGs was largely low in the Evi-1 promoter, regardless of Evi-1 expression levels (supplemental Figure 4). On the basis of these findings, it is unlikely that the expression of Evi-1 is shut down in progenitor cells by DNA methylation in the promoter. Another possibility is that undifferentiated HSCs irreversibly lose some key factors that contribute to activation of Evi-1 along with hematopoietic cell differentiation. Significant in this regard is that menin is required for some targets to be activated by MLL oncoproteins.39 In the present case, however, menin itself was not likely to be a key factor in the MLL-mediated activation of Evi-1, because the menin-binding motif was not required for activation of the Evi-1 promoter in the luciferase assay (Figure 4C).
In the reporter assay used in the present study, Jurkat cells provided a condition sufficient for MLL-ENL to activate the Evi-1 promoter. However, only an exogenous Evi-1 promoter was activated by MLL-ENL in Jurkat cells, given that endogenous Evi-1 expression was not concurrently activated (data not shown). The cellular milieu provided by HSCs ultimately appears necessary for activation of the endogenous Evi-1 promoter.
It has been shown that both wild-type MLL and Evi-1 are crucial for proliferation and maintenance of HSCs.2,3,47,48 Because wild-type MLL and MLL oncoproteins share some transcriptional targets, such as HoxA9, we assessed the transcriptional activity of MLL on the Evi-1 promoter. The luciferase reporter assays used in the present study showed no significant transcriptional activities of wild-type MLL on the Evi-1 promoter, which suggests that the wild-type MLL by itself is not sufficient for the activation of Evi-1 (Figure 4A). Given that Evi-1 expression decreases along with normal hematopoietic cell differentiation in spite of the preserved expression of MLL, the physiologic expression level of wild-type MLL may not be able to activate the expression of Evi-1 by itself.
GSEA analysis with gene-expression data of AML samples revealed that EVI-1–high MLL-rearranged leukemias exhibit HSC-like signatures, whereas genes involved in more differentiated hematopoietic progenitor cells are enriched in EVI-1–low MLL-rearranged leukemias. Considering that HSCs are more efficient targets for leukemogenic transformation by MLL oncoproteins,40,46 up-regulated Evi-1 may contribute to the propagation of leukemia stem cells in MLL-rearranged leukemias. In support of this concept is our finding that Evi-1 deletion reduces clonogenic activity most severely in KSL-derived cells (Figure 5E).
In addition to the enrichment of HSC genes in Evi-1–high leukemias, GSEA also revealed that the gene-expression signature of NPMc+ AMLs resembles that of Evi-1–high MLL-rearranged AMLs. Because NPMc+ AMLs display a specific gene-expression profile dominated by an HSC molecular signature,42 the results probably indicate that HSC genes are enriched in Evi-1–high leukemias. Alternatively, Evi-1 overexpression and cytoplasmic NPM may cooperatively contribute to leukemia development, and this possibility should be investigated in the future.
Given that Evi-1 plays an essential role in the proliferation and maintenance of HSCs in normal hematopoiesis,3 it is tempting to speculate that activated expression of Evi-1 by MLL oncoproteins results in the propagation of leukemia stem cells that is associated with therapeutic resistance and disease progression. In support of this is a recent report that the adverse effect of EVI-1 positivity on prognosis was clinically observed in AML patients with MLL rearrangement.49 We showed that MLL-ENL–transformed cells with up-regulated Evi-1 expression are derived from HSCs (Figure 5A-B) and that their clonogenic potential is highly dependent on Evi-1 (Figure 5E). Collectively, our findings suggest that Evi-1 is an attractive therapeutic target in the treatment of Evi-1–high MLL-rearranged leukemias. Putative key factors collaborating with MLL oncoproteins in undifferentiated hematopoietic cells at the Evi-1 promoter remain unknown. Some clues may be found from the clinical experience that high expression of Evi-1 is frequently observed in leukemias with another MLL rearrangement, MLL-AF6.14 MLL-AF6, as well as MLL-ENL and MLL-AF9, aberrantly recruits AF4 and ENL family proteins to its transcriptional target promoters to cause sustained target-gene expression.50 These functions, which are common in major MLL oncoproteins, may be involved in activation of Evi-1. Further investigation would clarify how Evi-1 is activated not only in MLL-rearranged leukemias but also in other leukemias or normal hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank T. Kitamura for Plat-E packaging cells, H. Nakauchi for pGCDNsam-eGFP retroviral vector, R. Ono and T. Nosaka for MLL-ENL cDNA, J.L. Hess for MLL-AF9 cDNA, T. Nakamura for HoxA9/Meis1 cDNA, T. Inaba for E2A-HLF cDNA, M.H. Tomasson for cMyc/bcl2 cDNA, I. Kitabayashi for PML-RARA cDNA, Y. Ishii and Y. Shimamura for expert technical assistance, and Kyowa Kirin for cytokines.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and by Health and Labor Sciences Research grants from the Ministry of Health, Labor and Welfare.
Authorship
Contribution: S.A., S.G., and M.K. designed the experiments and the study; S.A., M.N., Y.I., S.G., and M.K. wrote the manuscript; S.A., A.Y., and S.G. performed experiments and collected and analyzed data; and M.S. and M.I. provided important reagents and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mineo Kurokawa, Department of Hematology and Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-8655, Japan; e-mail: kurokawa-tky@umin.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal