Abstract
Macrophages and neutrophils play important roles during the innate immune response, phagocytosing invading microbes and delivering antimicrobial compounds to the site of injury. Functional analyses of the cellular innate immune response in zebrafish infection/inflammation models have been aided by transgenic lines with fluorophore-marked neutrophils. However, it has not been possible to study macrophage behaviors and neutrophil/macrophage interactions in vivo directly because there has been no macrophage-only reporter line. To remove this roadblock, a macrophage-specific marker was identified (mpeg1) and its promoter used in mpeg1-driven transgenes. mpeg1-driven transgenes are expressed in macrophage-lineage cells that do not express neutrophil-marking transgenes. Using these lines, the different dynamic behaviors of neutrophils and macrophages after wounding were compared side-by-side in compound transgenics. Macrophage/neutrophil interactions, such as phagocytosis of senescent neutrophils, were readily observed in real time. These zebrafish transgenes provide a new resource that will contribute to the fields of inflammation, infection, and leukocyte biology.
Introduction
After injury or infection, the first line of defense against invading pathogens is the innate immune system. During the inflammatory response, interaction between the cells of the innate immune system is critical for correct coordination of phases of cellular influx and resolution.1
The zebrafish has proven to be a very useful tool for modeling innate immune responses.2 Leukocyte behavior can be directly observed in optically transparent zebrafish embryos and larvae, and transgenic zebrafish strains have enabled particular cellular behaviors to be visualized in vivo with an ease and precision unparalleled in other vertebrate models. Detailed examination of inflammation and host-pathogen interactions at the cellular level has been enabled by several transgenic strains marking embryonic and adult neutrophils, such as Tg(mpx:EGFP)3,4 and lyz-driven transgenes.5,6 Such lines have been used to study neutrophil responses and fluxes during infection7,8 and during acute and chronic inflammation.3,4,9 Combinations of these approaches have provided some important new insights (eg, regarding the role of leukocytes in mycobacterial infection).10-13
In contrast to the availability of neutrophil-reporter zebrafish lines for more than 5 years, no line has yet been developed that specifically labeled macrophages, one of the most important cell types of the innate immune system. Direct in vivo imaging of embryonic and larval macrophages has been possible by identifying cells on the basis of size, location, motility, and behavior.10,14-17 The Tg(fli1a:EGFP) line18 expresses in primitive leukocytes as well as endothelial cells, and this has been exploited to examine primitive macrophage behavior in wounded embryos.19 In Tg(mpx:EGFP) zebrafish, a subpopulation of cells with duller fluorescence than its more brightly fluorescent neutrophils, has been characterized as macrophages.20 The absence of a specific reporter line for macrophages per se has been an ongoing hindrance to comprehensive analyses of the cellular innate immune response in this model organism.
Macrophage expressed gene 1 (mpeg1) was first identified as a gene with expression tightly restricted to human and murine macrophages,21 and has subsequently been used as a marker for this cell lineage in mammalian systems22 and zebrafish.23 These studies present transgenic constructs and lines based on the zebrafish mpeg1 promoter that drive transgene expression specifically in zebrafish embryonic macrophages, allowing visualization of macrophage behavior in vivo. Specificity of transgene expression is demonstrated in transient and stable transgenic embryos. Transgene utility is demonstrated by imaging and quantifying several macrophage behavioral dynamics and by a comparative analysis of macrophage and neutrophil behaviors and interactions after wounding. The exchange of large cytoplasmic portions from living neutrophils to living macrophages in vivo is described for the first time.
Methods
Zebrafish
Adults and embryos were maintained and studied at 28°C. The AB strain was used for transgenesis. Other zebrafish lines used were Tg(mpx:EGFP)i114,3 Tg(lyz:EGFP)nz117,5 Tg(lyz:dsRed)nz50,5, and Tg(UAS-E1b:Kaede)s1999t.24 Animal protocols were approved by the Walter and Eliza Hall Institute Animal Ethics Committee. All embryos were treated with 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) from 8 hpf.
Gene expression analysis
mpeg1 promoter identification and cloning
Zebrafish genome assembly Zv8 (http://www.ensembl.org/Danio_rerio/) informed primer design for amplification of 30437143–30438999 nt from chromosome 8, generating 1.86 kb of sequence corresponding to the immediately proximal mpeg1 5′-untranslated region. Cloning used Gateway methods and vectors.30 Supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) lists promoter and riboprobe primer sequences.
Microinjection and transgenesis
mpeg1 transgenesis was performed using Gateway protocols for Tol2-mediated recombination.30 Antisense morpholino oligonucleotides were obtained from GeneTools (www.gene-tools.com) and microinjected at 200 to 500μM into 1- or 2-cell embryos at 1.7 nL per bolus (morpholino oligonucleotide sequences in supplemental Table 1).
Leukocyte function studies
For wounding, tails were transected coronally by scalpel blade at 3 dpf29 and embryos mounted in 1.5% low melting agarose. Imaging commenced approximately 4 minutes after injury. For neutral red staining, embryos were incubated in egg water with 2.5 μg/mL neutral red for 12 hours.31 To demonstrate phagocytosis, Penicillium marneffei spores were prepared as described.32 Spores were heat-killed at 70°C for 15 minutes. Calcofluor staining was performed by incubation of spores in 10mM Calcofluor White (Sigma-Aldrich) for 15 minutes, followed by several rounds of washing and resuspension in normal saline. A total of 10 to 20 spores/embryo were microinjected into the somatic muscle of 3 dpf embryos and imaging commenced 20 minutes later.
Microscopy and image analysis
For dissecting microscopy (Figures 1, 2A-B,E), an Olympus SZX16 microscope with a DP71 camera and 1× (Figures 1, 2A-B) and 2× (Figure 2E) objectives was used. Filter sets used: enhanced green fluorescent protein/unconverted Kaede: SZX2-FGFPHQ (excitation, 460-480 nm; emission, 495-540 nm); dsRed/photoconverted Kaede/Neutral Red: SZX2-RFP2 (excitation, 540-580 nm; emission, 610 nm); and Kaede photoconversion: CFP CHR-U-N49001 (Chroma; excitation, 434-438 nm, emission 440-520 nm, exposure, 15-20 minutes). Original images were 1360 × 1024 RGB color (cropped).
For single photon confocal microscopy (Figure 2C-D), an Olympus FV1000-BX61WI upright multiphoton with an XLUMPlan F1 20×, water immersion, 0.95 NA objective was used. Excitation wavelengths used were 473 nm for EGFP and 559 nm for Kaede. Original image details were, for Figure 2Ci, xyz: 512 × 512 × 37 pixels, maximum intensity projection (cropped); for Figure 2Cii, xyz: 512 × 512 × 34 pixels, maximum intensity projection (cropped); for Figure 2D, (head) xyz: 512 × 512 × 113 pixels, maximum intensity projection; for Figure 2D, (ICM) xyz: 512 × 512 × 105 pixels, maximum intensity projection; and for Figure 2D (Tail) xyz: 512 × 512 × 140 pixels, maximum intensity projection.
Line-scanning confocal microscopy (Figures 2F, 3A, and Figure 4), was performed using a Zeiss LSM 5 Live inverted microscope with a Plan-Apochromat 20×, 0.8 NA objective. Wavelengths used were: 405 nm for Calcofluor; 488 nm for EGFP; and 561 nm for photoconverted Kaede. Original image details were, for Figure 2F, xyzt: 512 × 512 × 4 (pixels) × 100 (2 frames per minute) maximum intensity projection (cropped, deconvoluted); for Figure 3A, xyzt: 512 × 512 × 1 (pixels) × 1138 (2 frames per minute); for Figure 4A, xyzt: 512 × 512 × 1 (pixels) × 1138 (2 frames per minute; cropped); and for Figure 4B-D, xyzt: 512 × 512 × 1 (pixels) × 1800 (2 frames per minute) (cropped).
Wide-field microscopy for cell-tracking studies (Figure 3) was performed using a Nikon Ti-E inverted microscope with a Plan Fluor 10×, 0.3 NA objective. Wavelength filters used were FITC (Nikon; excitation 465-495 nm; emission, 515-565) for EGFP and TRITC (Nikon; excitation 515-565 nm; emission 550-660) for photoconverted Kaede. Original image details were Figure 3B-H, xyzt: 512 × 512 × 4 (pixels, 2 × binning) × 1080, 1 frame per minute). Deconvolution and maximum intensity projection were performed before tracking analysis.
Images were processed in Adobe Creative Suite CS4 and ImageJ, Version 1.4q, programs for presentation. Quantitative leukocyte behavior measurements were extracted using Metamorph (Series 7.5). The “meandering index” is as previously defined (linear distance between start and finish points divided by actual path length).33 The “in-wound persistence index” was defined as the percentage of remaining time in a time lapse that a cell spent at the wound edge after arrival.
Statistics
Descriptive and analytical statistics were prepared in Prism 5, Version 5.0a (GraphPad Software). Data are mean ±SD unless other-wise stated.
Results
Myeloid expression of mpeg1
In a search of an online database25 for genes with potentially leukocyte-restricted expression in zebrafish whose promoters might be useful for making new transgenic lines, mpeg1 attracted interest in for its expression at 20 hours post fertilization (hpf) to 5 days post fertilization (dpf) in cells dispersed in a typical pattern for leukocytes.29
Our initial whole-mount in situ hybridization characterization of mpeg1 expression until 78 hpf confirmed this pattern (supplemental Figure 1A). MO knockdown studies demonstrated that, as expected of a gene expressed in myeloid cells, mpeg1 expression was dependent on spi1/pu.134 and csf3r35 signaling (supplemental Figure 1B). On this basis, fragments of its promoter were cloned. A recent independent study has also characterized zebrafish mpeg1 as a gene with highly restricted, specific expression in macrophages up to 48 hpf.23
These data collectively indicate that mpeg1 is expressed in dispersed early zebrafish leukocytes with macrophage-lineage restriction. The macrophage lineage specificity of mpeg1 expression has been further demonstrated by its promoter activity as characterized in “The mpeg1 promoter drives transgene expression in macrophages.”
The mpeg1 promoter drives transgene expression in macrophages
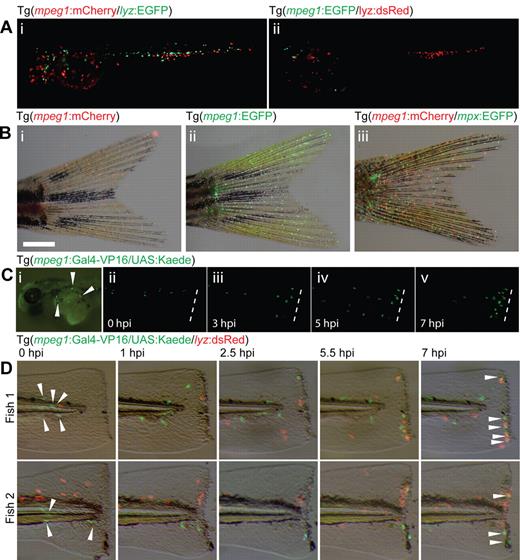
Several mpeg1-driven, tol2-flanked transgene constructs were built for recapitulating mpeg1 expression in live animals: Tg(mpeg1:EGFP); Tg(mpeg1:mCherry); and Tg(mpeg1:Gal4-VP16). In F0-microinjected embryos, transgene expression was observed in a dispersed population of mobile, putative myeloid cells (Figure 1A-C). Inflammation assays using F0 Tg(mpeg1:Gal4-VP16/UAS:Kaede) embryos demonstrated Kaede-expressing cells migrating in response to wounding (Figure 1C-D), a characteristic leukocyte behavior. Encouragingly, in F0s, no overlap of fluorophore expression was observed between mpeg1-driven transgenes and characterized neutrophil-specific Tg(mpx:EGFP) and lyz-driven transgenes (Figure 1A-B,D), although these F0 embryos were potentially mosaic.
A 1.86-kb mpeg1 promoter fragment drives transient transgene expression in macrophages. (A) Transient mosaic transgene expression in embryos injected with tol2-flanked Tg(mpeg1:mCherry) (i) and Tg(mpeg1:EGFP) (ii) DNA constructs into Tg(lyz:EGFP) and Tg(lyz:dsRed) transgenic backgrounds, respectively. The experiment was conducted on Tg(lyz:EGFP) and Tg(lyz:dsRed) embryos to provide comparison and nonoverlap of expression with neutrophils. (B) Transgene expression in F0 adults injected with tol2-flanked Tg(mpeg1:mCherry) (i,iii) and Tg(mpeg1:EGFP) (ii) DNA constructs. (iii) Tg(mpeg1:mCherry) was introduced onto the Tg(mpx:EGFP) background to provide comparison and nonoverlap of expression with neutrophils. (C) Transient transgene expression resulting from delivery of a tol2-flanked Tg(mpeg1:Gal4-VP16) construct into Tg(UAS:Kaede) embryos results in expression of Kaede in dispersed cells (i) that migrate in response to wounding (tail transection) (ii-v). Time: hours post injury (hpi). (D) Delivery of tol2-flanked Tg(mpeg1:Gal4-VP16) into Tg(UAS:Kaede/lyz:dsRed) embryos allows comparison of macrophage (green, arrowheads) and neutrophil populations (red) migrating in response to wounding (tail transection). Time: hours post injury (hpi).
A 1.86-kb mpeg1 promoter fragment drives transient transgene expression in macrophages. (A) Transient mosaic transgene expression in embryos injected with tol2-flanked Tg(mpeg1:mCherry) (i) and Tg(mpeg1:EGFP) (ii) DNA constructs into Tg(lyz:EGFP) and Tg(lyz:dsRed) transgenic backgrounds, respectively. The experiment was conducted on Tg(lyz:EGFP) and Tg(lyz:dsRed) embryos to provide comparison and nonoverlap of expression with neutrophils. (B) Transgene expression in F0 adults injected with tol2-flanked Tg(mpeg1:mCherry) (i,iii) and Tg(mpeg1:EGFP) (ii) DNA constructs. (iii) Tg(mpeg1:mCherry) was introduced onto the Tg(mpx:EGFP) background to provide comparison and nonoverlap of expression with neutrophils. (C) Transient transgene expression resulting from delivery of a tol2-flanked Tg(mpeg1:Gal4-VP16) construct into Tg(UAS:Kaede) embryos results in expression of Kaede in dispersed cells (i) that migrate in response to wounding (tail transection) (ii-v). Time: hours post injury (hpi). (D) Delivery of tol2-flanked Tg(mpeg1:Gal4-VP16) into Tg(UAS:Kaede/lyz:dsRed) embryos allows comparison of macrophage (green, arrowheads) and neutrophil populations (red) migrating in response to wounding (tail transection). Time: hours post injury (hpi).
Stable germline transgenic Tg(mpeg1:Gal4-VP16) zebrafish were generated to provide opportunity for this promoter to drive a range of effector gene functionalities (Figure 2). Initial transgenesis was performed on a Tg(UAS:Kaede) background. mpeg1-transgene-marked cells were present into early larval stages of F0, F1 (Figure 2A), and F2 embryos (Figure 2Bii-iii). However, transgene expressing cells were observed only rarely in Tg(mpeg1:Gal4-VP16/UAS:Kaede) F1 and F2 adults (Figure 2Bi,iv). In contrast, Tg(mpeg1:mCherry) and Tg(mpeg1:EGFP) F0 adults showed strong transgene expression (Figure 1B).
Macrophage ontology, morphology, and behavior in stable mpeg1 transgenic zebrafish. (A) Unconverted Kaede expression (green) in dispersed macrophages in Tg(mpeg1:Gal4-VP16/UAS:Kaede) F1 embryos from 28 to 144 hpf. Images at 28, 50, and 120 hpf are composites assembled from photographs of the same embryo taken in 2 focal planes. (B) Loss of transgene expression occurs in young F1 and F2 Tg(mpeg1:Gal4-VP16/UAS:Kaede) adults (i,iv), demonstrated by the absence of dispersed fluorescent macrophages in the tail fin; compare with macrophages in the tail fins of Tg(mpeg1:mCherry) and Tg(mpeg1:EGFP) F0 animals in Figure 1B. However, the direct offspring of an outcross of the F1 adult (i) still shows strong embryonic transgene expression in dispersed macrophages (ii,iii). (I,iv) Arrowheads indicate autofluorescent iridophores. Bar represents 1 mm. (C) Dendritic morphology (arrowheads) of photoconverted Kaede (red) marked cells in Tg(mpeg1:Gal4-VP16/UAS:Kaede) F1 embryos. Bar represents 20 μm. (D) No overlap of fluorophore expression was observed between photoconverted Kaede and EGFP in F1 Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) compound transgenic embryos, demonstrating that the mpeg1 promoter drives expression in an entirely separate myeloid cell population to that of the mpx promoter (green represents neutrophils; and red, macrophages). Bar represents 50 μm. (E) Macrophage pinocytosis leads to accumulation of neutral red staining in vacuoles of unconverted Tg(mpeg1:Gal4-VP16/UAS:Kaede) positive cells (green) in the brain (arrowheads) of F1 embryos. Bar represents 50 μm. (F) Phagocytosis of heat-killed Penicillium marneffei spores (calcofluor-labeled, blue, arrowhead) by macrophages (red represents photoconverted Kaede) and neutrophils (green represents EGFP). Note phagocytosis of lower fungal spore by macrophage (bottom filled arrowhead) and migration of neutrophil with intracellular spore (open arrowhead). Stills from supplemental Video 1. Bar represents 50 μm. Time: minutes after infection.
Macrophage ontology, morphology, and behavior in stable mpeg1 transgenic zebrafish. (A) Unconverted Kaede expression (green) in dispersed macrophages in Tg(mpeg1:Gal4-VP16/UAS:Kaede) F1 embryos from 28 to 144 hpf. Images at 28, 50, and 120 hpf are composites assembled from photographs of the same embryo taken in 2 focal planes. (B) Loss of transgene expression occurs in young F1 and F2 Tg(mpeg1:Gal4-VP16/UAS:Kaede) adults (i,iv), demonstrated by the absence of dispersed fluorescent macrophages in the tail fin; compare with macrophages in the tail fins of Tg(mpeg1:mCherry) and Tg(mpeg1:EGFP) F0 animals in Figure 1B. However, the direct offspring of an outcross of the F1 adult (i) still shows strong embryonic transgene expression in dispersed macrophages (ii,iii). (I,iv) Arrowheads indicate autofluorescent iridophores. Bar represents 1 mm. (C) Dendritic morphology (arrowheads) of photoconverted Kaede (red) marked cells in Tg(mpeg1:Gal4-VP16/UAS:Kaede) F1 embryos. Bar represents 20 μm. (D) No overlap of fluorophore expression was observed between photoconverted Kaede and EGFP in F1 Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) compound transgenic embryos, demonstrating that the mpeg1 promoter drives expression in an entirely separate myeloid cell population to that of the mpx promoter (green represents neutrophils; and red, macrophages). Bar represents 50 μm. (E) Macrophage pinocytosis leads to accumulation of neutral red staining in vacuoles of unconverted Tg(mpeg1:Gal4-VP16/UAS:Kaede) positive cells (green) in the brain (arrowheads) of F1 embryos. Bar represents 50 μm. (F) Phagocytosis of heat-killed Penicillium marneffei spores (calcofluor-labeled, blue, arrowhead) by macrophages (red represents photoconverted Kaede) and neutrophils (green represents EGFP). Note phagocytosis of lower fungal spore by macrophage (bottom filled arrowhead) and migration of neutrophil with intracellular spore (open arrowhead). Stills from supplemental Video 1. Bar represents 50 μm. Time: minutes after infection.
To assess overlap between expression of the new mpeg1-driven transgene and a standard neutrophil marker in nonmosaic F1s, F0 founders were crossed into the Tg(mpx:EGFP) line3 and compound transgenic Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) F1 embryos were imaged after photoconversion of Kaede. No overlap at all was observed between cell populations in the embryos at 2, 3, or 7 dpf (number of scored cells expressing mpeg1/mpx/both: at 2 dpf, 50/125/0; at 3 dpf, 60/306/0; at 7 dpf, 222/401/0; scores collected from multiple fields in ≥ 2 embryos), establishing that in this line the mpeg1 promoter drives expression exclusively in a nonneutrophil cell population (Figure 2D).
The fluorescent cells in Tg(mpeg1:Gal4-VP16/UAS:Kaede) embryos had several defining morphologic and functional attributes characteristic of macrophage-lineage cells. They were large cells with extensive cytoplasmic extensions (50-100 μm including branches, Figure 2C). Incubation with neutral red led to accumulation of stain via pinocytosis (Figure 2E), as previously described for primitive zebrafish macrophages.31 The cells were phagocytic: injection of calcofluor-stained fungal conidia into the trunk led to migration of marked cells to the wound site and phagocytosis of microbes (Figure 2F; supplemental Video 1).
During inflammation, macrophages migrate with different kinetics to neutrophils
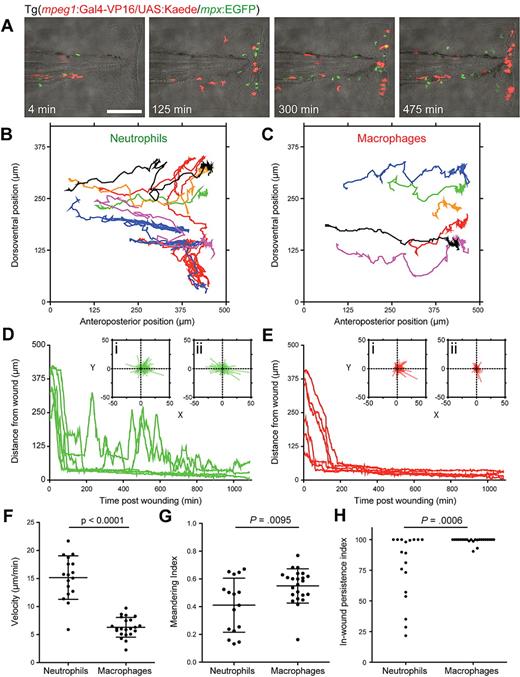
F1 Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) transgenic embryos were wounded and imaged, using photoconverted Kaede (red) to discriminate macrophages from EGFP-expressing (green) neutrophils (Figure 3, supplemental Video 2).
Comparative descriptive and quantitative analysis of macrophage and neutrophil behavior after wounding. (A) Frames extracted from 11.5 hours of time lapse microscopy (supplemental Video 2) after tail transection in a Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) F1 embryo (red represents macrophages; and green, neutrophils). Bar represents 100 μm. (B-C) Cell migration paths for the first 6 neutrophils (B) and 6 macrophages (C) to arrive at the wound, followed for 18 hours after tail transection, extracted from a video of another wounded Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) F1 transgenic embryo. (D-H) Graphing of distance to wound edge for the same 6 neutrophils (D) and macrophages (E) as in panels B and C demonstrates an overall difference in migratory and dwelling behaviors. All macrophages migrate directly to the wound and remain near the wound edge for the remainder of the time course, whereas approximately 30% of neutrophils resume a roaming behavior from 3 to 6 hours after wounding. Insets i and ii for each panel present collected x/y movement vectors before (i) and after (ii) wound arrival. Strong directionality is demonstrated by both cell types before arrival, but macrophages move with a slower prearrival velocity (F) and with a stronger directionality reflected by their higher meandering index (direct path length/actual path length) (G). In-wound persistence index, the percentage of remaining time spent at the wound edge after arrival (H), demonstrates the propensity of macrophages to remain in the wound margin for long periods after their arrival, whereas neutrophils show a significantly larger spread of behaviors. Descriptive statistics: (B-E) Data for the same 6 cells of each type. (F-H) Data points are individual cells from 4 embryos imaged for 18 hours after wounding. (F,G) Bars represent mean plus or minus SD. Tests of significance: (F,G) t test, 2-tailed. (H) Mann-Whitney test, 2-tailed.
Comparative descriptive and quantitative analysis of macrophage and neutrophil behavior after wounding. (A) Frames extracted from 11.5 hours of time lapse microscopy (supplemental Video 2) after tail transection in a Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) F1 embryo (red represents macrophages; and green, neutrophils). Bar represents 100 μm. (B-C) Cell migration paths for the first 6 neutrophils (B) and 6 macrophages (C) to arrive at the wound, followed for 18 hours after tail transection, extracted from a video of another wounded Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) F1 transgenic embryo. (D-H) Graphing of distance to wound edge for the same 6 neutrophils (D) and macrophages (E) as in panels B and C demonstrates an overall difference in migratory and dwelling behaviors. All macrophages migrate directly to the wound and remain near the wound edge for the remainder of the time course, whereas approximately 30% of neutrophils resume a roaming behavior from 3 to 6 hours after wounding. Insets i and ii for each panel present collected x/y movement vectors before (i) and after (ii) wound arrival. Strong directionality is demonstrated by both cell types before arrival, but macrophages move with a slower prearrival velocity (F) and with a stronger directionality reflected by their higher meandering index (direct path length/actual path length) (G). In-wound persistence index, the percentage of remaining time spent at the wound edge after arrival (H), demonstrates the propensity of macrophages to remain in the wound margin for long periods after their arrival, whereas neutrophils show a significantly larger spread of behaviors. Descriptive statistics: (B-E) Data for the same 6 cells of each type. (F-H) Data points are individual cells from 4 embryos imaged for 18 hours after wounding. (F,G) Bars represent mean plus or minus SD. Tests of significance: (F,G) t test, 2-tailed. (H) Mann-Whitney test, 2-tailed.
During the arrival phase, both neutrophils and macrophages migrated toward the site of injury with strong directionality, demonstrated by their tracks (Figure 3B-C) and arrival movement vectors (Figure 3Di,Ei) during migration. Compared with the swift initial neutrophil migration toward the wound averaging 15 μm/minute, macrophages were significantly tardier, extending more pseudopodia and migrating more slowly (6 μm/minute; Figure 3F; supplemental Video 2). Macrophages took a more direct route to the wound margin, reflected in a significantly higher meandering index (0.55) than neutrophils (0.4; Figure 3G). Macrophages arriving at the wound in the first 24 hours remained for as long as 4 days after wounding, a behavior proven by localized photoconversion of Kaede at the wound site and monitoring cell location thereafter (supplemental Figure 2B-C), substantially extending previous observations.36,37 Additional macrophages continued to arrive until at least 48 hours after wounding (supplemental Figure 2C). In contrast, neutrophils discontinued directional migration toward the wound after 6 hours (Figure 3A; supplemental Video 2).
Neutrophils and macrophages also behaved differently after arrival at the wound. During 18- hour time lapses, approximately 30% of neutrophils that arrived at the wound margin resumed a roaming behavior from 3 to 6 hours after wounding (Figure 3D,H), whereas more than 90% macrophages dwelt persistently at the wound margin after their arrival (Figure 3E,H).
These data demonstrate the utility of this new transgenic zebrafish line, in combination with other transgenes, to observe, compare, and quantify distinctive macrophage behaviors.
Live cell imaging of macrophage-neutrophil interactions during inflammation
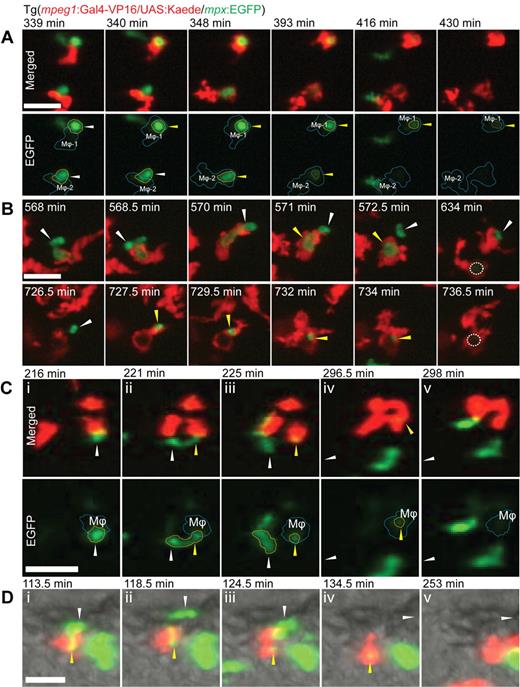
With both neutrophils and macrophages marked by distinguishable fluorophores, neutrophil/macrophage interactions in vivo can be observed dynamically. One such behavior is the phagocytosis of apoptotic neutrophils during inflammation resolution, previously investigated statically in fixed zebrafish.37 After tail transection, this interactive behavior was readily observed in living Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) embryos, after photoconversion of Kaede to red fluorescence (Figure 4A; supplemental Video 3). On neutrophil arrival at the wound margin, some take on the rounded morphology previously attributed to an apoptotic cell.37 With the subsequent arrival of macrophages, some engaged in direct, complex, intertwining, behavior with such neutrophils, resulting in their apparent phagocytosis with loss of neutrophil EGFP fluorescence. Sometimes only part of the neutrophil cytoplasm detached and disappeared (Figure 4B; supplemental Video 4). Viewed in real-time, the ongoing cellular protrusion and movement of neutrophils indicate that these neutrophils are not always completely dead before their phagocytosis by macrophages (supplemental Video 3).
Interactions between macrophages and neutrophils in vivo. (A) Interactions between macrophages (red represents photoconverted Kaede) and neutrophils (green represents EGFP) in Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) compound transgenic F1 embryos. Two examples of apoptotic neutrophils being phagocytosed by macrophages at the wound margin. The first occurs at 340 minutes by macrophage 1 (Mϕ-1) at the top of frame followed by the second at 348 minutes by Mϕ-2. Loss of green neutrophil fluorescence is evident in the Mϕ-1 after 76 minutes and within Mϕ-2 occurs in 45 minutes. Stills from supplemental Video 3. Bar represents 10 μm. (B) Demonstration that the loss of cytoplasmic neutrophil fluorescence is dependent on macrophage phagocytosis. At 568 to 571 minutes, partial phagocytosis of a segment of a neutrophil. At 572.5 to 634 minutes, loss of fluorescence in the phagocytosed fragment occurs after approximately 6 hours (dotted circle represents region of lost EGFP fluorescence). Providing an internal control for this process, an unphagocytosed still-fluorescing neutrophil fragment remains stationary throughout the first phagocytic phase, until it is subsequently engulfed by the same macrophage at 727.5 minutes, with loss of its EGFP fluorescence at 736.5 minutes. Stills from supplemental Video 4. Bar represents 10 μm. In both panels, white arrowheads and yellow arrowheads indicate unphagocyted and phagocytosed states, respectively, of neutrophil corpse. (C-D) Two examples of cytoplasmic transfer from live neutrophils (green represents EGFP) to macrophages (red represents photoconverted Kaede). (C) A neutrophil/macrophage interaction near a wound margin results in a cytoplasmic fragment from a live neutrophil transferring to within a macrophage. (Top panel) Merged images. (Bottom panel) EGFP channel only. Dashed outlines indicate position of macrophage (blue) and neutrophil/fragment. Stills extracted from supplemental Video 5. Bar represents 20 μm. (D) A neutrophil/macrophage interaction in the trunk of the embryo. Merged fluorescence and bright-field images clearly demonstrate interaction resulting in cytoplasm transfer from neutrophil to macrophage. Stills from supplemental Video 6. Bar represents 20 μm. In both examples, white arrowheads and yellow arrowheads indicate unphagocytosed and phagocytosed states, respectively, of the transferring neutrophil cytoplasmic fragment. Neutrophil viability throughout the process is demonstrated by its subsequent migration out of the field of view (direction of white arrow in subpanels iv and v).
Interactions between macrophages and neutrophils in vivo. (A) Interactions between macrophages (red represents photoconverted Kaede) and neutrophils (green represents EGFP) in Tg(mpeg1:Gal4-VP16/UAS:Kaede/mpx:EGFP) compound transgenic F1 embryos. Two examples of apoptotic neutrophils being phagocytosed by macrophages at the wound margin. The first occurs at 340 minutes by macrophage 1 (Mϕ-1) at the top of frame followed by the second at 348 minutes by Mϕ-2. Loss of green neutrophil fluorescence is evident in the Mϕ-1 after 76 minutes and within Mϕ-2 occurs in 45 minutes. Stills from supplemental Video 3. Bar represents 10 μm. (B) Demonstration that the loss of cytoplasmic neutrophil fluorescence is dependent on macrophage phagocytosis. At 568 to 571 minutes, partial phagocytosis of a segment of a neutrophil. At 572.5 to 634 minutes, loss of fluorescence in the phagocytosed fragment occurs after approximately 6 hours (dotted circle represents region of lost EGFP fluorescence). Providing an internal control for this process, an unphagocytosed still-fluorescing neutrophil fragment remains stationary throughout the first phagocytic phase, until it is subsequently engulfed by the same macrophage at 727.5 minutes, with loss of its EGFP fluorescence at 736.5 minutes. Stills from supplemental Video 4. Bar represents 10 μm. In both panels, white arrowheads and yellow arrowheads indicate unphagocyted and phagocytosed states, respectively, of neutrophil corpse. (C-D) Two examples of cytoplasmic transfer from live neutrophils (green represents EGFP) to macrophages (red represents photoconverted Kaede). (C) A neutrophil/macrophage interaction near a wound margin results in a cytoplasmic fragment from a live neutrophil transferring to within a macrophage. (Top panel) Merged images. (Bottom panel) EGFP channel only. Dashed outlines indicate position of macrophage (blue) and neutrophil/fragment. Stills extracted from supplemental Video 5. Bar represents 20 μm. (D) A neutrophil/macrophage interaction in the trunk of the embryo. Merged fluorescence and bright-field images clearly demonstrate interaction resulting in cytoplasm transfer from neutrophil to macrophage. Stills from supplemental Video 6. Bar represents 20 μm. In both examples, white arrowheads and yellow arrowheads indicate unphagocytosed and phagocytosed states, respectively, of the transferring neutrophil cytoplasmic fragment. Neutrophil viability throughout the process is demonstrated by its subsequent migration out of the field of view (direction of white arrow in subpanels iv and v).
Fluorophore loss in neutrophils has been attributed to cell death, possibly after phagocytosis,37 although separation of these events has not previously been possible. Observations in vivo in these compound transgenic lines strongly support the notion that fluorophore loss most often represents phagocytosis by macrophages. After neutrophil spherification, loss of fluorescence occurred after a median of 70.5 minutes (range, 6.5-472 minutes; n = 9) in phagocytosed cells, whereas unphagocytosed cells were rarely observed to lose fluorescence independently. In a particularly instructive example, only a portion of a neutrophil was initially phagocytosed, with subsequent loss of fluorescence in only that portion; the remaining portion retained its fluorescence until it too was later phagocytosed (Figure 4B; supplemental Video 4). This example indicates that neutrophil fluorescence can be maintained even in immobile cell fragments and that macrophage phagocytosis results in loss of EGFP protein activity.
Macrophages interacted directly with actively motile, living neutrophils. An unexpected occasional outcome of these interactions was the transfer of portions of fluorescent cytoplasm from neutrophil to macrophage (Figure 4C-D; supplemental Videos 5-6). The direction of transfer was always from a donor neutrophil to a recipient macrophage. The viability of the donor neutrophil in such exchanges was demonstrated by its ongoing active pseudopodial activity and its subsequent migration away from the site of the interaction (Figure 4C-D; supplemental Videos 5-6). The detached neutrophil cytoplasmic fragment was not just coincidently superimposed on a macrophage because, after detachment from a neutrophil, its subsequent movement was in concert with that of the macrophage (supplemental Video 7; supplemental Video 6 examined in slow motion). After its transfer to macrophages, the cytoplasmic fragment lost fluorescence (Figure 4Cv,Dv).
Discussion
Previously, to mark zebrafish macrophages, several genes and/or their promoters have been used. However, the degree of overlap of expression of various leukocyte-expressed genes in different leukocyte subtypes has been an ongoing point of confusion in the field for some time. Some markers originally considered to be macrophage-specific later appeared less specific as the number of markers for leukocyte subtypes expanded and coexpression was examined. Expression of the csf1r gene28 has become a commonly used benchmark for defining macrophage identity, but a transgenic line using its promoter is not yet described. However, as this gene is expressed in neural crest cells as well, full recapitulation of its promoter activity for transgenic expression of a fluorophore would also be expected to mark this nonmacrophage cell type. Macrophages have also been identified by elimination: on the basis of nonoverlapping expression between pan-leukocytic markers, such as l-plastin (lcp1) and neutrophil-specific expression of mpx.20,38 Cells expressing low levels of EGFP in the Tg(mpx:EGFP)uwm1 line display macrophage characteristics20 ; in our hands, a similar population is discernable by confocal microscopy in the Tg(mpx:EGFP)i114 line (data not shown). Markers based on early fate-specification genes, such as spi1, do not have leukocyte sublineage specificity and are limited in expression timeframe.39 Descriptions of lyz-driven transgene expression in macrophages5,6 required reevaluation given the complete overlap of lyz expression with mpx,40 and in the context of our new data. Recently, mpeg1 and cxcr3.2 were reported as genes with tight macrophage-specific expression in zebrafish,23 based on static expression studies.
For the first time in zebrafish embryos, with these new mpeg1-driven transgenes, macrophage-specific transgene expression has been achieved. mpeg1-driven transgene expression is nonoverlapping with the neutrophil-specific transgenes driven from mpx and lyz promoters. This allows for clear separation of embryonic zebrafish leukocyte populations by different fluorophores. These tools provide for a wide range of experimental utility in embryonic zebrafish. The macrophage-constraining mpeg1 promoter functionality resides in a 1.86-kb DNA fragment, making its recloning and reuse highly practical. Because the fluorophore reporter constructs are in a tol-2 vector, high-level mosaic transient expression can be achieved, meaning that a subset of macrophages at least can be marked in extant compound transgenic backgrounds by microinjection alone, without complex interbreeding. Judging from its performance in transient assays, the promoter fragment is strong and not generally susceptible to off-target expression. The first mpeg1-driven stable transgenic line provides the versatility of the Gal4-VP16 transactivator; hence, any currently available or desired UAS-effector transgene functionality can now be engaged for macrophage-specific expression.
Several issues may affect the stability of the germline Tg(mpeg1:Gal4-VP16/UAS:Kaede) reporter line we have made, although they do not undermine its current utility or the usefulness of the constructs themselves for transient macrophage labeling. There is a possibility of genetic drift resulting from segregation in subsequent generations if there are multiple integration sites in the Tg(mpeg1:Gal4-VP16) founder. To date, we have not recognized any overt toxicity in macrophages resulting from mechanisms, such as squelching (ie, transcription factor sequestration).41 Although there is strong embryonic and larval transgene expression, to date we have not been able to demonstrate reporter transgene activity in adult kidney and spleen macrophages by direct fluorescence microscopy in the compound Tg(mpeg1:Gal4-VP16/UAS-Kaede) fish (Figure 2B; data not shown). Comparison with directly driven trangene constructs demonstrates that this loss of expression is unique to the Gal4-VP16/UAS based system and most probably reflects somatic transgene silencing. Embryos produced by these F0 and F1 adults exhibit full transgene expression, suggesting that silencing occurs somatically and does not affect expression in embryos/larvae of subsequent generations. Epigenetic transgene silencing has been observed in other zebrafish Gal4-UAS systems, although more related to CpG-rich concatamerized UAS sequences42 than to the Gal4 driver line. Despite this, this reporter line appears sustainably useful for studies in embryos and larvae.
Videos of side-by-side neutrophil and macrophage behaviors after wounding display, in real time and in vivo, the strikingly different dynamics of each cell type. Although some aspects of these different behaviors have previously been inferred from static studies and other models, the simplicity and elegance of these transgenic fish provide advantages. These include the internally controlled observations made in single animals and opportunity for quantification. Previous zebrafish studies of neutrophil flux after wounding sought to demonstrate the fate of apoptotic neutrophils using fixed embryos and costaining of TdT-mediated dUTP nick end labeling and neutrophil material within cells positive for the pan-leukocytic marker lcp1.37 The interaction between macrophages and senescing or dead, but still-fluorescent, Tg(mpx:EGFP) neutrophil corpses can now be examined directly in real time in vivo.
The extent of the direct, dynamic interaction between living macrophages and neutrophils revealed in these embryos was surprising, particularly the occasional exchange of substantial fragments of fluorophore-marked neutrophil cytoplasm from living neutrophils to macrophages. Several features of this exchange distinguish it from several other previously described neutrophil processes. As well as identifying the phenomenon, these studies provide tools that will enable examination of hypotheses about its function. The extent to which one cell-type's behavior may be dependent on another can now also be explored, in embryos and larvae at least, by deleting the macrophage lineage: by the combination of Tg(mpeg1:Gal4-VP16) fish and UAS:deleter-gene constructs, such as Tg(UAS:nitroreductase).43
The lack of a macrophage-specific transgenic zebrafish has constrained zebrafish-based research, particularly in the field of innate immunity. The mpeg1-driven transgenic lines allow for: specific inducible ablation of macrophages; macrophage-specific overexpression of transgenes; fluorescence-activated cell sorter and analysis of macrophages as a pure population; imaging of interactions between macrophages and other immune cells in vivo; interaction studies between macrophages and intracellular pathogens, such as Mycobacterium marinum and Candida albicans; reexamination of macrophage roles in inflammation, wound healing, and development; their interactions with other cell types (eg, vasculature, muscle) in vivo; and macrophage-focused chemical genetic screens. Trangenes using this new zebrafish promoter represent a major step forward for the study of macrophage behavior and function in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ethan Scott, Herwig Baier, Stephen Renshaw, Chris Hall, and Phil Crosier for transgenic lines; Cameron Nowell and Kelly Rogers for assistance with imaging; Harshini Weerasinghe and Sony Varma for technical assistance; Mark Greer, Kelly Turner, and Prue Chamberlain for animal care in the aquarium; Ben Kile, Joan Heath, and Ben Croker for discussions; Steve Jane, David Curtis, and the Royal Melbourne Hospital Bone Marrow Research Laboratory for support. Microscopy used instruments in the Center for Advanced Microscopy (Ludwig Institute for Cancer Research) and the Australian Cancer Research Foundation Center for Therapeutic Target Discovery.
This work was supported by the National Institutes of Health (grant R01 HL079545) and the National Health and Medical Research Council (grant 637394; G.J.L.). F.E. was supported by an Australian Postgraduate Award and Walter and Eliza Hall Institute of Medical Research Edith Moffatt Scholarship. Walter and Eliza Hall Institute of Medical Research receives infrastructure support from the Commonwealth National Health and Medical Research Council Independent Research Institutes Infrastructure Support Scheme (361646) and a Victorian State Government Operational Infrastructure Support Scheme grant.
National Institutes of Health
Authorship
Contribution: G.J.L. and F.E. conceived the project and wrote the manuscript; F.E. assessed candidate genes, built the constructs, performed microinjections, and characterized the lines; L.P. provided advice on experimental design and performed live cell imaging; J.W.H. performed some in situ hybridization experiments; A.A. provided fungal spores; and all authors discussed the project, interpreted data, and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Graham J. Lieschke, Australian Regenerative Medicine Institute, Monash University, Victoria 3800, Australia; e-mail: Lieschke@monash.edu.