Abstract
Dendritic cells (DCs) are a heterogeneous group of professional antigen-presenting cells functioning as sentinels of the immune system and playing a key role in the initiation and amplification of innate and adaptive immune responses. DC development and functions are acquired during a complex differentiation and maturation process influenced by several factors present in the local milieu. A common feature at pathologic sites is represented by hypoxia, a condition of low pO2, which creates a unique microenvironment affecting cell phenotype and behavior. Little is known about the impact of hypoxia on the generation of mature DCs (mDCs). In this study, we identified by gene expression profiling a significant cluster of genes coding for immune-related cell surface receptors strongly up-regulated by hypoxia in monocyte-derived mDCs and characterized one of such receptors, TREM-1, as a new hypoxia-inducible gene in mDCs. TREM-1 associated with DAP12 in hypoxic mDCs, and its engagement elicited DAP12-linked signaling, resulting in ERK-1, Akt, and IκBα phosphorylation and proinflammatory cytokine and chemokine secretion. Finally, we provided the first evidence that TREM-1 is expressed on mDCs infiltrating the inflamed hypoxic joints of children affected by juvenile idiopathic arthritis, representing a new in vivo marker of hypoxic mDCs endowed with proinflammatory properties.
Introduction
Dendritic cells (DCs) are a heterogeneous group of professional antigen-presenting cells involved in the initiation and amplification of innate and adaptive immunity, which develop through different hematopoietic pathways.1 Myeloid DC immunostimulatory properties are acquired during a complex differentiation and maturation process. Their precursors extravasate from the bloodstream to nonlymphoid peripheral tissues, where they reside in an “immature” stage (iDCs) at sites of potential pathogen entry and are an important component of the leukocyte infiltrate in inflammatory tissues.1,2 iDCs are specialized for antigen capture and processing, functioning as sentinels of the immune system.3 Antigen uptake and activation by endogenous factors, such as proinflammatory cytokines and tissue damage-associated molecular patterns, or exogenous factors, such as pathogen-associated molecular patterns, induce iDCs to undergo phenotypic and functional changes that culminate in their maturation into mature DCs (mDCs), which have a higher capacity for antigen presentation.1 mDCs switch their chemokine receptor repertoire down-regulating inflammatory receptors and up-regulating those required for homing to secondary lymphoid organs, where they prime naive T cells triggering specific immune responses.2-4
The local microenvironment contributes to the regulation of DC development and functions.2,5,6 A common feature of inflamed tissues is represented by hypoxia, a condition of low partial oxygen pressure (pO2, 0-20 mmHg), which arises as a result of dysfunctional vascular network and diminished O2 supply and affects the phenotype and functions of every cell exposed to it.6-9 Activation of gene transcription is the primary mechanism by which mammalian cells respond to decreased pO2, and the underlying molecular pathways have been elucidated in detail and extensively reviewed. Briefly, transcriptional activation is mediated primarily by the hypoxia-inducible factor-1 (HIF-1), a heterodimer of the constitutive HIF-1β subunit and an oxygen-sensitive α-subunit (HIF-1α/-2α). α-Subunits are post-translationally stabilized under hypoxia and translocate to the nucleus where they dimerize with HIF-1β transactivating the hypoxia responsive element (HRE) present in the promoter of many O2-sensitive genes.7,8,10 HIF-1 expression and activity are tightly regulated by various cofactors and transcription factors, and HIF-independent pathways mediating gene induction by hypoxia have also been described.6,10,11
Recent studies have investigated hypoxia effects on DC differentiation, maturation, and functions. We reported that monocyte differentiation into iDCs under chronic hypoxia promotes the onset of a unique migratory phenotype by differentially modulating the expression profile of chemokines/receptors and genes involved in cell adhesion and tissue remodeling.12,13 Other studies indicated that acute iDC exposure to low pO2 may either impair14 or promote5,15 their maturation, by affecting expression of chemokine receptors, costimulatory molecules, and T cell-priming ability. However, the impact of chronic hypoxia on mDC development and functional behavior in inflammatory states is still largely unknown.
In this study, we demonstrate that the gene expression pattern of mDCs generated from human monocytes under chronic hypoxia (H-mDCs) is distinct from that of mDCs developing under normal O2 levels and characterized by up-regulation of genes coding for surface costimulatory and adhesion molecules, immunoregulatory and pattern recognition receptors. Among them, we identify the triggering receptor expressed on myeloid cells (TREM)–1, a member of the Ig superfamily of immunoreceptors and a strong amplifier of the immune responses,16,17 as a new hypoxia-inducible gene in mDCs. We provide evidence that this receptor is biologically active and expressed in vivo on mDCs recruited to the hypoxic joints of juvenile idiopatic arthritis (JIA) patients, pointing to a pathogenetic role for this molecule in the disease.
Methods
DC generation and culture
Blood monocytes were isolated from healthy volunteers at a purity of more than 93%, plated into 6-well culture plates (BD Biosciences Discovery Labware) in RPMI 1640 (Euroclone) supplemented with 10% heat-inactivated fetal calf serum (HyClone), and incubated for 4 days under normoxic (20% O2) or hypoxic (1% O2) conditions in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) (both 100 ng/mL), as detailed.12,13 A cocktail of proinflammatory mediators containing tumor necrosis factor-α (TNF-α; 50 ng/mL), IL-1β (50 ng/mL), IL-6 (10 ng/mL), and prostaglandin E2 (1mM) was added for the last 48 hours to induce DC maturation. Hypoxic conditions were obtained by culturing cells in an anaerobic work-station incubator (CARLI Biotec) flushed with a mixture of 1% O2/5% CO2/94% N2. Medium was allowed to equilibrate in the hypoxic incubator for 2 hours before use, and pO2 was monitored using a portable oxygen analyzer (Oxi 315i/set, WTW).
SFMC isolation
Synovial fluid (SF) samples were obtained at the time of therapeutic knee arthrocentesis from 8 children affected by oligoarticular JIA18 and collected into sodium-heparin tubes under vacuum. pO2 levels in SF samples were monitored to confirm hypoxic conditions. Paired peripheral blood samples and peripheral blood from 5 age-matched control subjects undergoing venipuncture for minor orthopedic procedures were obtained on the occasion of routine venipuncture and collected as for SF. Informed consent was obtained according to the procedure approved by the Gaslini's Ethical Committee. Specimens were centrifuged to prepare cell-free SF and plasma and separated by Ficoll to isolate mononuclear cells (SF mononuclear cells [SFMCs] and peripheral blood mononuclear cells [PBMCs]). SF-derived samples were handled in the anaerobic incubator to prevent cell reoxygenation, as detailed.19,20
Cytokines and antibodies
Human recombinant GM-CSF, IL-4, TNF-α, IL-1β, and IL-6 were from PeproTech; prostaglandin E2 was from Sigma-Aldrich.
Monoclonal antibodies (mAbs) used for fluorescence-activated cell sorter include: anti-CD83-fluorescein isothiocyanate (FITC) or phycoerythrin (PE)-Cy5 and anti-CD86-FITC (BD Biosciences PharMingen), anti-TREM-1-PE, anti-CXCR4-FITC, and anti-CCR7-FITC (R&D Systems), anti-CD1a-FITC or -allophycocyanin (Serotec), anti-CD1c-FITC and anti-CD141-allophycocyanin (Miltenyi-Biotec). Proper isotype-matched control Abs (Dako North America) were used.
Abs used for Western blot include: mouse anti–human TREM-1 (R&D Systems), mouse anti–human HIF-1α (BD-Biosciences), rabbit anti–human phospho (p)-ERK)-1, p-Akt, and p-IκBα (Cell Signaling Technology), mouse anti–human DAP12, rabbit anti–human ERK-1, Akt, IkBα, and β-actin (Santa Cruz Biotechnology),
Flow cytometry
Flow cytometry was performed as described.12,13 Cells resuspended with phosphate-buffered saline supplemented with 0.2% bovine serum albumin, 0.01% NaN3 were incubated with fluorochrome-conjugated mAbs for 30 minutes at 4°C, after blocking nonspecific sites with rabbit IgG (Sigma-Aldrich). Fluorescence was quantitated on a FACSCalibur flow cytometer equipped with CellQuest software Version 2002 (BD Biosciences). Cells were gated according to their light-scatter properties to exclude cell debris.
RNA isolation and GeneChip hybridization
Total RNA was purified from different donor-derived mDCs using the QIAGEN RNeasy MiniKit and reverse-transcribed into double-stranded cDNA on a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems) using the one-cycle cDNA synthesis kit (Affymetrix). cDNA derived from 3 donors was purified and biotin labeled with the IVT-expressed kit (Affymetrix), as described.21 Fragmented cRNA was hybridized to Affymetrix HG-U133 plus 2.0 arrays (Genopolis Corporation) containing 54 000 probe sets coding for 38 500 genes; chips were stained with streptavidin-phycoerythrin (Invitrogen) and scanned using an Affymetrix GeneChip Scanner 3000, as described.21 Data capturing was conducted with standard Affymetrix analysis software algorithms (Microarray Suite 5.0). Comparative analysis of hypoxic relative to normoxic expression profiles was carried out with GeneSpring Expression Analysis Software Gx9.0 (Silicon Genetics), and expression data were normalized using “per chip normalization” and “per gene normalization” algorithms. Fold change was calculated as the ratio between the average expression level under hypoxia and normoxia. We selected a modulated gene list of 2-fold induction/inhibition with a false discovery rate of 0%. The significance of gene expression differences between the 2 experimental conditions was calculated using the Mann-Whitney U test. Only genes that passed the test at a confidence level of 95% (P < .05) were considered significant. A complete dataset for each microarray experiment was logged in the Gene Expression Omnibus public repository at National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo; accession no. GSE22282). Gene Ontology (GO) data mining for biologic process at level 1 was conducted online using the Database for Annotation, Visualization and Integrated Discovery (DAVID) software Version 2008 (www.david.niaid.nih.gov).
HRE consensus elements consisting of a 4-nt core (CGTG) flanked by degenerated sequences ((T G C)(A G)(CGTG)(C G A)(G C T)(G T C)(C T G)) were mapped in the promoter regions of genes represented in the chip, as detailed.12
Real-time RT-PCR
Real-time quantitative reverse-transcribed polymerase chain reaction (RT-PCR) was performed on a 7500 Real Time PCR System (Applied), using SYBR Green PCR Master Mix and sense/antisense oligonucleotide primers (TIBMolbiol) (listed in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Expression data were normalized on the values obtained in parallel for 3 reference genes (indicated in supplemental Table 1) selected among those not affected by hypoxia in the Affymetrix analysis using the Bestkeeper software Version 2004, and relative expression values were calculated using Q-gene software Version 2003, as detailed.21
Cross-linking of TREM-1+ cells
Twelve-well flat-bottom tissue culture plates (Corning Life Sciences) precoated with 10 μg/mL of agonist anti-TREM-1 mAb or control IgG1 were incubated overnight at 37°C before seeding 8 × 105 H-mDCs/well/mL of RPMI 1640 without cytokines. Plates were briefly spun at 130g to engage TREM-1. After 24-hour stimulation under hypoxia, supernatants were harvested and tested for cytokine/chemokine content by enzyme-linked immunosorbent assay (ELISA). In a set of experiments, cells were plated in medium without fetal calf serum, and plates were centrifuged and incubated for 20 minutes at 37°C under hypoxic conditions. ERK1, Akt, and IκBα phosphorylation was then assessed by Western blot.
Western blot analysis and immunoprecipitation
Total protein extracts were prepared as described,13 subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride membranes (Millipore), and probed with specified Abs. In a few experiments, lysates were subjected to immunoprecipitation with 10 μg/mL anti-TREM-1 mAb or control IgG1 ON at 4°C, and protein G-Sepharose 4B (GE-Healthcare) for 45 minutes at 4°C. Precipitates were separated by SDS-PAGE and immunoblotted with anti-DAP12 mAb. Chemiluminescence detection was carried out with peroxidase-conjugated goat antirabbit and antimouse Abs using an ECL kit (Pierce Chemical).
ELISA
Conditioned medium (CM) was replaced on day 3 or 4 of mDC generation with fresh medium supplemented with cytokines for 24 hours and tested for soluble TREM-1 (sTREM-1) content by ELISA (R&D Systems) after an additional 24-hour culture. sTREM-1 was also quantified in SF and plasma samples. TNF-α, IL-6, IL-12p70, IL-10, IL-8, CCL4, and CCL5 were measured in CM from mDCs triggered with anti-TREM-1 mAb or control mAb by specific ELISA (R&D Systems). Data were analyzed with the GraphPad Prism-5 Software.
Statistical analysis
Data are the mean plus or minus SE of 3 independent experiments, unless differently specified. The Student t test was used to determine significance of results (P < .05). sTREM-1 concentrations in SF and plasma specimens were evaluated by the Wilcoxon rank test (P < .05 statistically significant).
Results
Gene expression profile of hypoxic mDCs
mDCs were generated by culturing human monocytes under normoxic and hypoxic conditions in the presence of GM-CSF/IL-4 for 4 days and a cocktail of proinflammatory stimuli for the last 48 hours. As determined by flow cytometry (Figure 1), both mDCs and H-mDCs displayed the mature phenotype, characterized by high surface expression of CD1a differentiation marker, CD83 maturation marker, and CCR7 chemokine receptor, undetectable in fresh monocytes, and by up-regulation of CD86 costimulatory molecule and CXCR4 chemokine receptor, in agreement with previous observations.13
Phenotype of monocyte-derived mDCs generated under hypoxic conditions. Human monocytes were cultured for 48 hours with IL-4 and GM-CSF followed by incubation with the proinflammatory mediators, TNF-α, IL-1β, IL-6, and prostaglandin E2 for an additional 48 hours under 20% O2 (mDCs) or 1% O2 (H-mDCs) conditions, and expression of the indicated cell surface molecules was analyzed by flow cytometry after 4-day culture, as described in “Flow cytometry.” The expression profile of unstimulated monocytes is shown for comparison. Cells were gated according to their light scatter properties to exclude cell debris. Solid histograms represent the fluorescent profile of cells stained with specific FITC-conjugated Abs; and open histograms, the fluorescent profile of cells stained with isotype-matched controls. Data are plotted as fluorescence intensity on a log scale versus the number of positive cells. In each histogram, the percentage of positive cells is indicated. Results are from one representative experiment of 10 performed with cells from different donors.
Phenotype of monocyte-derived mDCs generated under hypoxic conditions. Human monocytes were cultured for 48 hours with IL-4 and GM-CSF followed by incubation with the proinflammatory mediators, TNF-α, IL-1β, IL-6, and prostaglandin E2 for an additional 48 hours under 20% O2 (mDCs) or 1% O2 (H-mDCs) conditions, and expression of the indicated cell surface molecules was analyzed by flow cytometry after 4-day culture, as described in “Flow cytometry.” The expression profile of unstimulated monocytes is shown for comparison. Cells were gated according to their light scatter properties to exclude cell debris. Solid histograms represent the fluorescent profile of cells stained with specific FITC-conjugated Abs; and open histograms, the fluorescent profile of cells stained with isotype-matched controls. Data are plotted as fluorescence intensity on a log scale versus the number of positive cells. In each histogram, the percentage of positive cells is indicated. Results are from one representative experiment of 10 performed with cells from different donors.
H-mDC transcriptional profile was then assessed by microarray analysis. Pairwise comparison between datasets from normoxic and hypoxic samples revealed differential modulation of a large number of transcripts. The majority of differentially expressed genes were identified as unique and named in the GenBank, whereas the remaining transcripts were either expressed sequence tags (ESTs) or hypothetical. After restricting the profile to sequences exhibiting more than or equal to 2-fold expression differences, we identified 563 up- and 402 down-regulated genes by hypoxia (Figure 2), which were selected for further analysis. These results provided the first indication that mDCs and H-mDCs had a different gene expression signature.
Functional classification of hypoxia-responsive genes by GO data mining. The gene expression profile of H-mDCs versus mDCs was analyzed using high-density oligonucleotide arrays, as described in “RNA isolation and GeneChip hybridization.” Unique genes showing at least 2-fold change in expression levels between cells generated under hypoxic and normoxic conditions were selected and clustered in different KEGG pathways according to GO data mining for biologic process at level 1 using the DAVID software Version 2008. Based on this classification scheme, genes can be placed in more than one category if more than one function of the encoded protein was established. Bars on the right of the y-axis represent up-regulated genes; and bars on the left of the y-axis, down-regulated genes. Immune-related pathways are highlighted in boldface.
Functional classification of hypoxia-responsive genes by GO data mining. The gene expression profile of H-mDCs versus mDCs was analyzed using high-density oligonucleotide arrays, as described in “RNA isolation and GeneChip hybridization.” Unique genes showing at least 2-fold change in expression levels between cells generated under hypoxic and normoxic conditions were selected and clustered in different KEGG pathways according to GO data mining for biologic process at level 1 using the DAVID software Version 2008. Based on this classification scheme, genes can be placed in more than one category if more than one function of the encoded protein was established. Bars on the right of the y-axis represent up-regulated genes; and bars on the left of the y-axis, down-regulated genes. Immune-related pathways are highlighted in boldface.
To gain insights into the nature of hypoxia-induced changes, selected genes were clustered into various functional categories according to GO data mining. We highlighted 33 interrelated functional pathways, containing a statistically significant portion of hypoxia-modulated genes (Figure 2). H-mDC transcriptional profile was mainly associated with cell growth, differentiation, and/or maturation, and angiogenesis regulation. Several up-regulated genes fell into pathways implicated in signal transduction, metabolic processes, and apoptosis. Interestingly, a prominent set of up-regulated genes had immunologic relevance, coding for proteins involved in immune regulation, inflammatory responses, cell migration, and adhesion.
Characterization of hypoxia-inducible immune-related genes
Among immune-related genes, profound differences were observed in the expression of a large cluster (63) of cell surface receptor-encoding genes (Table 1). These include several pattern recognition receptors critical to host defense, such as CD180, various complement receptor components, Toll-like receptors 1 and 2, C-type lectin receptors CLEC-2D, -2B, -7A, and macrophage receptor with collagenous structure. Of interest is also the up-regulation of scavenger receptors implicated in the regulation of fatty acid and/or cholesterol uptake/transport, such as thrombospondin receptor (CD36) and apolipoprotein B48 receptor. A set of genes coding for costimulatory and adhesion/homing molecules was also up-regulated, such as several integrin family members, L1 cell adhesion molecule (CD171), neuropilin-1, ADAM metallopeptidase 8 (CD156), platelet/endothelial cell adhesion molecule (CD31), and lymphocyte adhesion molecule-1. Other hypoxia-inducible genes coded for immunoregulatory receptors, the most relevant of which are: Ig-Fc receptors, TREM-1, SLAM family member-9, blood dendritic cell antigen (CD141), leukocyte immunoglobulin-like receptors A1 and A2, and semaphorins 4B and 4D. These data suggest that hypoxia can exert a profound modulatory effect on mDC innate and adaptive immune functions.
Relative expression of genes encoding immune-related cell surface receptors in H-mDCs versus mDCs*
| GenBank accession no. . | Gene symbol . | Full name . | Main function(s) of gene product . | Fold increase‡ . | HRE§ . |
|---|---|---|---|---|---|
| Pattern recognition/scavenger receptors | |||||
| NM_005582 | CD180 (RP105)† | CD180 antigen | Belongs to the TLR family; negative regulator of TLR4 signaling on myeloid and B cells and of innate immune responses to pathogens and danger; NF-κB activator | 13.5 | − |
| NM_012072 | CD93 (C1qR1)† | Complement component 1q, receptor 1 | Defense collagen receptor for C1q, mannose-binding lectin, and surfactant protein A; mainly expressed on monocytes, neutrophils, and endothelial cells; modulates phagocytosis and clearance of apoptotic cells; mediates leukocyte-endothelial cell interactions | 8.6 | − |
| NM_003264 | TLR2† | Toll-like receptor 2 | Member of the TLR family expressed on cells of the innate immune system; plays an essential role in PAMP recognition leading to the generation of immune responses; implicated in the pathogenesis of autoimmune and inflammatory diseases | 7.7 | + |
| AW299226 | CD36† | Thrombospondin receptor | Multifunctional class B scavenger receptor; involved in the uptake of oxidized LDL, collagen, thrombospondin, anionic phospholipids, and apoptotic cells; adhesion molecule | 5 | + |
| BC005254 | CLEC2B† | C-type lectin domain family 2, member B | Member of the NK cell C-type lectin-like receptor family; platelet activation receptor; involved in tumor cell growth/metastatization | 4.8 | − |
| BE676623 | CD71† | Transferrin receptor | Regulator of cellular uptake of iron and iron metabolism; IgA receptor expressed on DCs | 4.5 | − |
| NM_001877 | CD21 (CR2)† | Complement component receptor 2 | EBV receptor overexpressed on B cells and follicular DCs; required for generation of immune responses to T cell antigens | 4.4 | − |
| AF285089 | CLEC2D† | C-type lectin domain family 2, member D | Member of the NK cell C-type lectin receptor family; pattern recognition receptor expressed on myeloid cells; mediates phagocytosis and proinflammatory cytokine production; regulates NK and APC cross-talk | 4.1 | + |
| NM_006770 | MARCO† | Macrophage receptor with collagenous structure | Member of the class A scavenger receptor family; pattern recognition receptor up-regulated on phagocytes in response to TLR agonists and Gram+/− bacteria; implicated in phagocytic activity and proinflammatory cytokine production; plays a role in host defense to pathogens, cell-cell recognition, and modified LDL scavenging | 3.7 | − |
| AL050262 | TLR1† | Toll-like receptor 1 | Member of the TLR family expressed on cells of the innate immune system; plays a fundamental role in PAMP recognition; mediates production of cytokines for the activation of innate immunity | 3.6 | + |
| AF313468 | CLEC7A | C-type lectin domain family 7, member A | Member of the C-type lectin-like receptor family; pattern recognition receptor for fungal β-glucan expressed on myeloid cells including DCs; activates phagocytosis, ROS and cytokine production, DC maturation, and T-cell costimulatory activity | 2.9 | + |
| NM_018690 | APOB48R | Apolipoprotein B48 receptor | Binds to apolipoprotein B48 on reticuloendothelial cells and regulates lipid transport into the cell | 2.9 | + |
| NM_000573 | CD35 (C3b/4bR1) | Complement component 3b/4b, receptor 1 | Member of the complement activation receptor (RCA) family expressed on follicular DCs and other leukocytes; mediates binding of complement-bound opsonins and immune complexes; enhances adaptive immune responses | 2.6 | − |
| U81501 | FPRL1 | Formyl peptide receptor-like 1 | High affinity receptor for bacteria-derived and host-derived N-formyl-methionyl peptides; key player in innate immunity and host defense against pathogen infection; mediates phagocyte chemotactic migration and IL-1 production | 2.6 | + |
| M74921 | EDNRB | Endothelin receptor type B | G protein-coupled receptor for the vasoactive peptide, endothelin; functions as a clearance receptor contributing to the regulation of vascular homeostasis | 2.2 | + |
| Costimulatory and adhesion/homing molecules | |||||
| 201666_at | TIMP1 | TIMP metallopeptidase inhibitor 1 | Matrix metalloproteinase inhibitor; promotes proliferation of several cell types and has an antiapoptotic function | 15.1 | + |
| NM_000889 | ITGB7 | Integrin, β 7 | Receptor for VCAM1 and fibronectin; interacts with MADCAM1 adhesion molecule expressed on vascular endothelium; plays a role in cell adhesive interactions | 8.3 | − |
| AK025016 | CD9† | CD9 antigen | Member of the tetraspanin family; facilitates the organization of multimolecular membrane complexes, including integrins; plays a role in leukocyte activation, differentiation, and migration | 8 | + |
| N95414 | ITGA2 (CD49B) | Integrin, α 2 | Receptor for laminin, collagen, fibronectin, and E-cadherin; involved in cell adhesion, cell surface-mediated signaling, extracellular matrix generation, and organization | 6.9 | + |
| AI653981 | L1CAM (CD171) | L1 cell adhesion molecule | Costimulatory/adhesion molecule belonging to the Ig superfamily; regulates DC transendothelial migration and trafficking; stimulates cancer cell motility and spreading | 6.7 | + |
| AF280547 | NRP1 | Neuropilin 1 | Coreceptor of tyrosine kinase receptors for vascular endothelial growth factor, class 3 semaphorins, and placenta growth factor; involved in the regulation of cell growth and migration, angiogenesis, DC and T-cell interaction, and in the initiation of primary immune responses | 5.8 | + |
| NM_001109 | ADAM8 (CD156) | ADAM metallopeptidase domain 8 | Membrane-anchored protein implicated in cell-cell and cell-matrix interactions | 4.5 | + |
| AF213460 | EPHA3 | Ephrin receptor A3 | Receptor for members of the ephrin-A family of receptor tyrosine kinases; mediates cell-cell contact, cellular adhesion, angiogenesis, and developmental remodeling | 4 | − |
| X74039 | CD87 | Plasminogen activator, urokinase receptor | Receptor for urokinase plasminogen activator; regulates cell-surface plasminogen activation; mediates cell adhesion by binding to vitronectin; modulates the proliferative, adhesive, and migratory cell phenotype | 3.8 | − |
| NM_013447 | CD312 (MR2) | EGF-like domain containing hormone receptor-like 2 | Member of the GPCR family of adhesion molecules differentially expressed during myeloid cell differentiation, maturation, and activation; plays a role in migration and adhesion | 3.8 | − |
| NM_000560 | CD53 | CD53 antigen | Member of the tetraspanin family; complexes with integrins and plays a role in the regulation of leukocyte activation, growth, and motility; contributes to the transduction of CD2-generated signals in T and NK cells | 3.6 | − |
| NM_002205 | ITGA5 (CD49) | Integrin, α 5 | Receptor for fibronectin and fibrinogen; involved in cell adhesion to fibronectin and cell surface mediated signaling | 3.6 | + |
| NM_002414 | CD99 | CD99 antigen | Mediates leukocyte adhesion amd transendothelial migration | 3.4 | + |
| BE870509 | MET (HGFR)† | Met proto-oncogene (hepatocyte growth factor receptor) | Widely expressed proto-oncogene encoding the tyrosine-kinase receptor for hepatocyte growth factor and scatter factor; plays a role in cell growth and motility in many physiologic and pathologic processes | 3.4 | + |
| M37780 | PECAM-1 (CD31)† | Platelet/endothelial cell adhesion molecule | Cell adhesion molecule predominantly expressed on endothelial cells | 3 | + |
| AW131039 | ITGB8 | Integrin, β 8 | Receptor for fibronectin; mediates cell-cell and cell-extracellular matrix interaction | 3 | + |
| NM_000655 | LECAM1 | Lymphocyte adhesion molecule 1 (selectin L) | Member of a family of adhesion/homing receptors required for leukocyte binding and rolling on endothelial cells; facilitates leukocyte migration into secondary lymphoid organs and inflammatory sites | 3 | − |
| M80436 | PAFR | Platelet-activating factor receptor | G protein-coupled receptor for platelet-activating factor with pleiotropic functions, including regulation of cell motility, smooth muscle contraction, cytokine synthesis, and release; implicated in allergy, asthma, septic shock, arterial thrombosis, and inflammatory processes | 2.8 | − |
| NM_001778 | CD48† | B-cell membrane protein | Activation-associated glycoprotein, member of the Ig superfamily; binds CD2 and regulates NK and T-cell activation | 2.7 | + |
| U94903 | CD44† | CD44 antigen | Receptor for hyaluronic acid, osteopontin, collagens, and matrix metalloproteinases; involved in cell-cell interaction, leukocyte adhesion, recirculation and homing, hematopoiesis, and tumor spreading | 2.7 | − |
| U87967 | CD39 | Ectonucleoside triphosphate diphosphohydrolase | Surface enzyme involved in the reduction of inflammatory cell adhesion and trafficking and tissue injury; contributes to vascular homeostasis regulation | 2.7 | − |
| D37781 | CD148† | Protein tyrosine phosphatase, receptor type, J | Receptor PTP induced in macrophages by proinflammatory stimuli; regulates cell spreading, cytoskeletal rearrangements, and chemotaxis | 2.3 | + |
| AJ277151 | CD134† | Tumor necrosis factor receptor superfamily, member 4 | Regulates T-cell expansion and survival, NK cell functions, and cross-talk with APCs; NF-κB activator | 2.1 | − |
| S78505 | PRLR | Prolactin receptor | Widespread receptor, major mediator of prolactin effects on cell proliferation, survival, cytoskeletal modification and differentiation | 2.1 | + |
| NM_002207 | ITGA9 | Integrin, α 9 | Receptor for VCAM1, tenascin, and osteopontin; participates in cell adhesion and motility | 2 | − |
| Immunoregulatory receptors | |||||
| U43677 | FCAR (CD89) | Fc fragment of IgA, receptor | ITAM-bearing receptor of the Fc region of IgA expressed on myeloid cells; mediates both pro- and anti-inflammatory functions of IgA | 55 | + |
| NM_018643 | TREM-1† | Triggering receptor expressed on myeloid cells 1 | Ig superfamily immunoregulatory receptor expressed on monocytes/macrophages and neutrophils; stimulates their proinflammatory functions and acts as an inflammatory amplifier via functional interaction with TLRs; involved in inflammatory diseases and septic shock | 39.8 | + |
| NM_001432 | EREG† | Epiregulin | Member of the epidermal growth factor (EGF) family; binds EGFR and members of ErbB tyrosine-kinase receptor family; mediates cell proliferation and proinflammatory cytokine production; promotes wound healing | 20.8 | + |
| NM_033438 | SLAMF9† | SLAM family member 9 | Member of the signaling leukocyte activation Ig subfamily; involved in innate and acquired immune responses through the regulation of multiple hematopoietic cell functions, including DC maturation | 12.2 | + |
| AW119113 | BDCA3 (CD141)† | Blood dendritic cell Ag (thrombomodulin) | Expressed on DCs; associated with TH2-skewed immune responses; plays a pathogenetic role in atopy and asthma | 11.4 | − |
| U90940 | FCGR2C (CD32) | Fc fragment of IgG, low affinity IIc, receptor | Involved in myeloid cell effector and regulatory functions, such as phagocytosis of immune complexes and modulation of antibody production by B cells | 11 | − |
| AI246590 | IRAK-2 | Interleukin-1 receptor-associated kinase 2 | IRAK kinase family member; key component of IL-1R and TLR signal transduction pathways; involved in the regulation of intracellular signaling networks controlling inflammation | 9.7 | + |
| NM_000570 | FCGR3A (CD16) | Fc fragment of IgG, low affinity IIIa, receptor | Type 1 transmembrane glycoprotein of the Ig superfamily expressed on several immune cells, including phagocytes; activating receptor involved in phagocytosis, inflammatory mediator release, ADCC, and immune complex clearance | 9.6 | + |
| AJ130712 | SIGLEC7 | Sialic acid binding Ig-like lectin 7 | ITIM-containing member of the CD33-related Siglec receptor family, weakly expressed on monocytic cells; acts as an inhibitory receptor | 8 | − |
| NM_006866 | LILRA2 (ILT1)† | Leukocyte immunoglobulin-like receptor A2 | Member of the leukocyte Ig-like immunoreceptor family, selectively expressed on myeloid cells, including DCs, and inflammatory tissues; acts as an activating receptor pairing with FcγR for signaling; promotes inflammatory responses | 6.4 | + |
| 1557444_at | TREML3† | Triggering receptor expressed on myeloid cells-like 3 | Ig superfamily immunoregulatory receptor expressed on myeloid cells, structurally related to the TREM gene family | 5.9 | − |
| AW241983 | CD307 | Fc receptor-like 5 | Differentiation antigen of B-lineage cells; plays an immunoregulatory role | 5 | − |
| NM_001772 | CD33 | CD33 antigen | Marker of myeloid cell differentiation belonging to the sialic acid-binding Ig-like lectin (Siglec) family; negative regulator of myeloid cell activation and DC generation | 4.7 | − |
| AK026133 | SEMA4B† | Semaphorin 4B | Integral membrane protein with an Ig extracellular domain, widely expressed throughout the nervous system; provides axonal guidance cues during neuronal development | 4.1 | − |
| NM_000956 | PTGER2 | Prostaglandin E receptor 2 | Receptor for prostaglandin E2, expressed on macrophages and mDCs; activates the expression of multiple genes implicated in the regulation of immunity and inflammation; involved in CCR7-mediated mDC migration | 3.7 | − |
| NM_004106 | FCER1G | Fc fragment of IgE, high affinity I, receptor | ITAM-containing subunit of the high-affinity IgE receptor and of other Fc receptors; expressed on mast cells and APCs; involved in allergic reactions mediated by IgE and in the regulation of immune responses | 3.6 | + |
| AF025529 | LILRA1 (LIR6) | Leukocyte immunoglobulin-like receptor A1 | Member of the leukocyte Ig-like immunoreceptor family expressed predominantly on monocytes/macrophages and B cells and at lower levels on DCs; associates with FcγR and acts as receptor for MHCI antigens | 3.5 | + |
| NM_006378 | SEMA4D (CD100)† | Semaphorin 4D | Integral membrane protein with an Ig extracellular domain; controls proliferation, survival, and migration of nervous cells of the system; positive regulator of angiogenesis; plays a role in the activation of both humoral and cellular immunity, including induction of DC maturation, functions, and interaction with T cells | 3.1 | + |
| NM_020377 | CYSLT2R | Cysteinyl leukotriene receptor 2 | Member of the superfamily of G protein-coupled receptors; mediates cell trafficking and innate immune responses; involved in the pathogenesis of chronic inflammatory diseases and allergic reactions | 2.9 | − |
| AI681260 | CD85 (LILRB1)† | Leukocyte immunoglobulin-like receptor B1 | ITIM-containing inhibitory leukocyte Ig-like receptor expressed on myeloid lineage cells and up-regulated during DC differentiation; binds MHCI; inhibitor of immune and inflammatory responses | 2.6 | + |
| NM_002346 | LY6 | Lymphocyte antigen 6 complex, locus E | Glycosylphosphatidylinositol-anchored molecule that belongs to the Ly-6 family, also known as stem cell antigen 2 (SCA2); type I interferon-inducible gene | 2.5 | − |
| M34986 | EPOR | Erythropoietin receptor | Member of the cytokine receptor family expressed on various hematopoietic cells with immunostimulatory activities | 2.1 | + |
| BE908995 | MYADM (SB135) | Myeloid-associated differentiation marker | Differentiation marker selectively expressed in myeloid cells, including DCs | 2.1 | + |
| GenBank accession no. . | Gene symbol . | Full name . | Main function(s) of gene product . | Fold increase‡ . | HRE§ . |
|---|---|---|---|---|---|
| Pattern recognition/scavenger receptors | |||||
| NM_005582 | CD180 (RP105)† | CD180 antigen | Belongs to the TLR family; negative regulator of TLR4 signaling on myeloid and B cells and of innate immune responses to pathogens and danger; NF-κB activator | 13.5 | − |
| NM_012072 | CD93 (C1qR1)† | Complement component 1q, receptor 1 | Defense collagen receptor for C1q, mannose-binding lectin, and surfactant protein A; mainly expressed on monocytes, neutrophils, and endothelial cells; modulates phagocytosis and clearance of apoptotic cells; mediates leukocyte-endothelial cell interactions | 8.6 | − |
| NM_003264 | TLR2† | Toll-like receptor 2 | Member of the TLR family expressed on cells of the innate immune system; plays an essential role in PAMP recognition leading to the generation of immune responses; implicated in the pathogenesis of autoimmune and inflammatory diseases | 7.7 | + |
| AW299226 | CD36† | Thrombospondin receptor | Multifunctional class B scavenger receptor; involved in the uptake of oxidized LDL, collagen, thrombospondin, anionic phospholipids, and apoptotic cells; adhesion molecule | 5 | + |
| BC005254 | CLEC2B† | C-type lectin domain family 2, member B | Member of the NK cell C-type lectin-like receptor family; platelet activation receptor; involved in tumor cell growth/metastatization | 4.8 | − |
| BE676623 | CD71† | Transferrin receptor | Regulator of cellular uptake of iron and iron metabolism; IgA receptor expressed on DCs | 4.5 | − |
| NM_001877 | CD21 (CR2)† | Complement component receptor 2 | EBV receptor overexpressed on B cells and follicular DCs; required for generation of immune responses to T cell antigens | 4.4 | − |
| AF285089 | CLEC2D† | C-type lectin domain family 2, member D | Member of the NK cell C-type lectin receptor family; pattern recognition receptor expressed on myeloid cells; mediates phagocytosis and proinflammatory cytokine production; regulates NK and APC cross-talk | 4.1 | + |
| NM_006770 | MARCO† | Macrophage receptor with collagenous structure | Member of the class A scavenger receptor family; pattern recognition receptor up-regulated on phagocytes in response to TLR agonists and Gram+/− bacteria; implicated in phagocytic activity and proinflammatory cytokine production; plays a role in host defense to pathogens, cell-cell recognition, and modified LDL scavenging | 3.7 | − |
| AL050262 | TLR1† | Toll-like receptor 1 | Member of the TLR family expressed on cells of the innate immune system; plays a fundamental role in PAMP recognition; mediates production of cytokines for the activation of innate immunity | 3.6 | + |
| AF313468 | CLEC7A | C-type lectin domain family 7, member A | Member of the C-type lectin-like receptor family; pattern recognition receptor for fungal β-glucan expressed on myeloid cells including DCs; activates phagocytosis, ROS and cytokine production, DC maturation, and T-cell costimulatory activity | 2.9 | + |
| NM_018690 | APOB48R | Apolipoprotein B48 receptor | Binds to apolipoprotein B48 on reticuloendothelial cells and regulates lipid transport into the cell | 2.9 | + |
| NM_000573 | CD35 (C3b/4bR1) | Complement component 3b/4b, receptor 1 | Member of the complement activation receptor (RCA) family expressed on follicular DCs and other leukocytes; mediates binding of complement-bound opsonins and immune complexes; enhances adaptive immune responses | 2.6 | − |
| U81501 | FPRL1 | Formyl peptide receptor-like 1 | High affinity receptor for bacteria-derived and host-derived N-formyl-methionyl peptides; key player in innate immunity and host defense against pathogen infection; mediates phagocyte chemotactic migration and IL-1 production | 2.6 | + |
| M74921 | EDNRB | Endothelin receptor type B | G protein-coupled receptor for the vasoactive peptide, endothelin; functions as a clearance receptor contributing to the regulation of vascular homeostasis | 2.2 | + |
| Costimulatory and adhesion/homing molecules | |||||
| 201666_at | TIMP1 | TIMP metallopeptidase inhibitor 1 | Matrix metalloproteinase inhibitor; promotes proliferation of several cell types and has an antiapoptotic function | 15.1 | + |
| NM_000889 | ITGB7 | Integrin, β 7 | Receptor for VCAM1 and fibronectin; interacts with MADCAM1 adhesion molecule expressed on vascular endothelium; plays a role in cell adhesive interactions | 8.3 | − |
| AK025016 | CD9† | CD9 antigen | Member of the tetraspanin family; facilitates the organization of multimolecular membrane complexes, including integrins; plays a role in leukocyte activation, differentiation, and migration | 8 | + |
| N95414 | ITGA2 (CD49B) | Integrin, α 2 | Receptor for laminin, collagen, fibronectin, and E-cadherin; involved in cell adhesion, cell surface-mediated signaling, extracellular matrix generation, and organization | 6.9 | + |
| AI653981 | L1CAM (CD171) | L1 cell adhesion molecule | Costimulatory/adhesion molecule belonging to the Ig superfamily; regulates DC transendothelial migration and trafficking; stimulates cancer cell motility and spreading | 6.7 | + |
| AF280547 | NRP1 | Neuropilin 1 | Coreceptor of tyrosine kinase receptors for vascular endothelial growth factor, class 3 semaphorins, and placenta growth factor; involved in the regulation of cell growth and migration, angiogenesis, DC and T-cell interaction, and in the initiation of primary immune responses | 5.8 | + |
| NM_001109 | ADAM8 (CD156) | ADAM metallopeptidase domain 8 | Membrane-anchored protein implicated in cell-cell and cell-matrix interactions | 4.5 | + |
| AF213460 | EPHA3 | Ephrin receptor A3 | Receptor for members of the ephrin-A family of receptor tyrosine kinases; mediates cell-cell contact, cellular adhesion, angiogenesis, and developmental remodeling | 4 | − |
| X74039 | CD87 | Plasminogen activator, urokinase receptor | Receptor for urokinase plasminogen activator; regulates cell-surface plasminogen activation; mediates cell adhesion by binding to vitronectin; modulates the proliferative, adhesive, and migratory cell phenotype | 3.8 | − |
| NM_013447 | CD312 (MR2) | EGF-like domain containing hormone receptor-like 2 | Member of the GPCR family of adhesion molecules differentially expressed during myeloid cell differentiation, maturation, and activation; plays a role in migration and adhesion | 3.8 | − |
| NM_000560 | CD53 | CD53 antigen | Member of the tetraspanin family; complexes with integrins and plays a role in the regulation of leukocyte activation, growth, and motility; contributes to the transduction of CD2-generated signals in T and NK cells | 3.6 | − |
| NM_002205 | ITGA5 (CD49) | Integrin, α 5 | Receptor for fibronectin and fibrinogen; involved in cell adhesion to fibronectin and cell surface mediated signaling | 3.6 | + |
| NM_002414 | CD99 | CD99 antigen | Mediates leukocyte adhesion amd transendothelial migration | 3.4 | + |
| BE870509 | MET (HGFR)† | Met proto-oncogene (hepatocyte growth factor receptor) | Widely expressed proto-oncogene encoding the tyrosine-kinase receptor for hepatocyte growth factor and scatter factor; plays a role in cell growth and motility in many physiologic and pathologic processes | 3.4 | + |
| M37780 | PECAM-1 (CD31)† | Platelet/endothelial cell adhesion molecule | Cell adhesion molecule predominantly expressed on endothelial cells | 3 | + |
| AW131039 | ITGB8 | Integrin, β 8 | Receptor for fibronectin; mediates cell-cell and cell-extracellular matrix interaction | 3 | + |
| NM_000655 | LECAM1 | Lymphocyte adhesion molecule 1 (selectin L) | Member of a family of adhesion/homing receptors required for leukocyte binding and rolling on endothelial cells; facilitates leukocyte migration into secondary lymphoid organs and inflammatory sites | 3 | − |
| M80436 | PAFR | Platelet-activating factor receptor | G protein-coupled receptor for platelet-activating factor with pleiotropic functions, including regulation of cell motility, smooth muscle contraction, cytokine synthesis, and release; implicated in allergy, asthma, septic shock, arterial thrombosis, and inflammatory processes | 2.8 | − |
| NM_001778 | CD48† | B-cell membrane protein | Activation-associated glycoprotein, member of the Ig superfamily; binds CD2 and regulates NK and T-cell activation | 2.7 | + |
| U94903 | CD44† | CD44 antigen | Receptor for hyaluronic acid, osteopontin, collagens, and matrix metalloproteinases; involved in cell-cell interaction, leukocyte adhesion, recirculation and homing, hematopoiesis, and tumor spreading | 2.7 | − |
| U87967 | CD39 | Ectonucleoside triphosphate diphosphohydrolase | Surface enzyme involved in the reduction of inflammatory cell adhesion and trafficking and tissue injury; contributes to vascular homeostasis regulation | 2.7 | − |
| D37781 | CD148† | Protein tyrosine phosphatase, receptor type, J | Receptor PTP induced in macrophages by proinflammatory stimuli; regulates cell spreading, cytoskeletal rearrangements, and chemotaxis | 2.3 | + |
| AJ277151 | CD134† | Tumor necrosis factor receptor superfamily, member 4 | Regulates T-cell expansion and survival, NK cell functions, and cross-talk with APCs; NF-κB activator | 2.1 | − |
| S78505 | PRLR | Prolactin receptor | Widespread receptor, major mediator of prolactin effects on cell proliferation, survival, cytoskeletal modification and differentiation | 2.1 | + |
| NM_002207 | ITGA9 | Integrin, α 9 | Receptor for VCAM1, tenascin, and osteopontin; participates in cell adhesion and motility | 2 | − |
| Immunoregulatory receptors | |||||
| U43677 | FCAR (CD89) | Fc fragment of IgA, receptor | ITAM-bearing receptor of the Fc region of IgA expressed on myeloid cells; mediates both pro- and anti-inflammatory functions of IgA | 55 | + |
| NM_018643 | TREM-1† | Triggering receptor expressed on myeloid cells 1 | Ig superfamily immunoregulatory receptor expressed on monocytes/macrophages and neutrophils; stimulates their proinflammatory functions and acts as an inflammatory amplifier via functional interaction with TLRs; involved in inflammatory diseases and septic shock | 39.8 | + |
| NM_001432 | EREG† | Epiregulin | Member of the epidermal growth factor (EGF) family; binds EGFR and members of ErbB tyrosine-kinase receptor family; mediates cell proliferation and proinflammatory cytokine production; promotes wound healing | 20.8 | + |
| NM_033438 | SLAMF9† | SLAM family member 9 | Member of the signaling leukocyte activation Ig subfamily; involved in innate and acquired immune responses through the regulation of multiple hematopoietic cell functions, including DC maturation | 12.2 | + |
| AW119113 | BDCA3 (CD141)† | Blood dendritic cell Ag (thrombomodulin) | Expressed on DCs; associated with TH2-skewed immune responses; plays a pathogenetic role in atopy and asthma | 11.4 | − |
| U90940 | FCGR2C (CD32) | Fc fragment of IgG, low affinity IIc, receptor | Involved in myeloid cell effector and regulatory functions, such as phagocytosis of immune complexes and modulation of antibody production by B cells | 11 | − |
| AI246590 | IRAK-2 | Interleukin-1 receptor-associated kinase 2 | IRAK kinase family member; key component of IL-1R and TLR signal transduction pathways; involved in the regulation of intracellular signaling networks controlling inflammation | 9.7 | + |
| NM_000570 | FCGR3A (CD16) | Fc fragment of IgG, low affinity IIIa, receptor | Type 1 transmembrane glycoprotein of the Ig superfamily expressed on several immune cells, including phagocytes; activating receptor involved in phagocytosis, inflammatory mediator release, ADCC, and immune complex clearance | 9.6 | + |
| AJ130712 | SIGLEC7 | Sialic acid binding Ig-like lectin 7 | ITIM-containing member of the CD33-related Siglec receptor family, weakly expressed on monocytic cells; acts as an inhibitory receptor | 8 | − |
| NM_006866 | LILRA2 (ILT1)† | Leukocyte immunoglobulin-like receptor A2 | Member of the leukocyte Ig-like immunoreceptor family, selectively expressed on myeloid cells, including DCs, and inflammatory tissues; acts as an activating receptor pairing with FcγR for signaling; promotes inflammatory responses | 6.4 | + |
| 1557444_at | TREML3† | Triggering receptor expressed on myeloid cells-like 3 | Ig superfamily immunoregulatory receptor expressed on myeloid cells, structurally related to the TREM gene family | 5.9 | − |
| AW241983 | CD307 | Fc receptor-like 5 | Differentiation antigen of B-lineage cells; plays an immunoregulatory role | 5 | − |
| NM_001772 | CD33 | CD33 antigen | Marker of myeloid cell differentiation belonging to the sialic acid-binding Ig-like lectin (Siglec) family; negative regulator of myeloid cell activation and DC generation | 4.7 | − |
| AK026133 | SEMA4B† | Semaphorin 4B | Integral membrane protein with an Ig extracellular domain, widely expressed throughout the nervous system; provides axonal guidance cues during neuronal development | 4.1 | − |
| NM_000956 | PTGER2 | Prostaglandin E receptor 2 | Receptor for prostaglandin E2, expressed on macrophages and mDCs; activates the expression of multiple genes implicated in the regulation of immunity and inflammation; involved in CCR7-mediated mDC migration | 3.7 | − |
| NM_004106 | FCER1G | Fc fragment of IgE, high affinity I, receptor | ITAM-containing subunit of the high-affinity IgE receptor and of other Fc receptors; expressed on mast cells and APCs; involved in allergic reactions mediated by IgE and in the regulation of immune responses | 3.6 | + |
| AF025529 | LILRA1 (LIR6) | Leukocyte immunoglobulin-like receptor A1 | Member of the leukocyte Ig-like immunoreceptor family expressed predominantly on monocytes/macrophages and B cells and at lower levels on DCs; associates with FcγR and acts as receptor for MHCI antigens | 3.5 | + |
| NM_006378 | SEMA4D (CD100)† | Semaphorin 4D | Integral membrane protein with an Ig extracellular domain; controls proliferation, survival, and migration of nervous cells of the system; positive regulator of angiogenesis; plays a role in the activation of both humoral and cellular immunity, including induction of DC maturation, functions, and interaction with T cells | 3.1 | + |
| NM_020377 | CYSLT2R | Cysteinyl leukotriene receptor 2 | Member of the superfamily of G protein-coupled receptors; mediates cell trafficking and innate immune responses; involved in the pathogenesis of chronic inflammatory diseases and allergic reactions | 2.9 | − |
| AI681260 | CD85 (LILRB1)† | Leukocyte immunoglobulin-like receptor B1 | ITIM-containing inhibitory leukocyte Ig-like receptor expressed on myeloid lineage cells and up-regulated during DC differentiation; binds MHCI; inhibitor of immune and inflammatory responses | 2.6 | + |
| NM_002346 | LY6 | Lymphocyte antigen 6 complex, locus E | Glycosylphosphatidylinositol-anchored molecule that belongs to the Ly-6 family, also known as stem cell antigen 2 (SCA2); type I interferon-inducible gene | 2.5 | − |
| M34986 | EPOR | Erythropoietin receptor | Member of the cytokine receptor family expressed on various hematopoietic cells with immunostimulatory activities | 2.1 | + |
| BE908995 | MYADM (SB135) | Myeloid-associated differentiation marker | Differentiation marker selectively expressed in myeloid cells, including DCs | 2.1 | + |
Gene expression profiling was carried out independently on the RNA purified from 3 independent mDC and H-mDC preparations, and comparative analysis of gene expression differences between the two experimental conditions was conducted as described in “Methods.” A GenBank accession number, a common gene symbol, a full name, a brief description of the gene product main functions, and the fold increase value are specified for each gene. Genes are ordered in each group by fold change.
Genes validated by quantitative RT-PCR.
The indicated values are calculated as the ratio of hypoxic/normoxic signals (mean of expression level of 3 experiments). Genes increased by ≥ 2-fold are shown.
The + sign indicates genes whose promoter contain members of the HRE family.
The possible relationship between gene inducibility by hypoxia and HRE presence in the promoter was then investigated by mapping HRE sequences in the first 2000 bases upstream the transcription initiation site. Interestingly, we found that approximately 56% of up-regulated genes contained at least one member of the HRE family in the promoter, whereas the others were HRE− (Table 1), indicating that a remarkable portion of the hypoxic transcriptome does not require HIF-1 binding.
To validate microarray results, mRNA levels for a subset of genes selected among those listed in Table 1 were quantified by quantitative RT-PCR in pooled RNA from mDCs and H-mDCs generated from different donors (supplemental Figure 1). A few known hypoxia target genes were analyzed in parallel as positive controls (indicated in supplemental Figure 1). We found an approximately 90% concordance between quantitative RT-PCR and Affymetrix data with respect to the direction of the expression changes, although differences in the extent of modulation were observed for some genes. CD85, CD9, and CLEC2B up-regulation by hypoxia was not confirmed by quantitative RT-PCR.
TREM-1 is expressed in hypoxic mDCs
Among validated genes, TREM-1 was that displaying the highest expression difference in H-mDCs versus mDCs (supplemental Figure 1). TREM-1 was previously reported to be selectively expressed in blood neutrophils and a subset of blood monocytes and tissue macrophages and to be completely down-regulated during monocyte differentiation into DCs.16,17 Hence, we were interested in further investigating TREM-1 regulation and functional significance in H-mDCs.
To address the issue of donor-to-donor variability, we evaluated TREM-1 mRNA expression in H-mDCs generated from 5 donors by quantitative RT-PCR (Figure 3A). Expression of CAXII metalloenzyme was assessed in parallel as an index of response to hypoxia.22 TREM-1 transcript levels were significantly higher in H-mDCs than in mDCs from all tested samples, paralleling those of CAXII, with differences ranging from 23- to 516-fold among individual donors. In keeping with the mRNA data, marked TREM-1 immunoreactivity was observed by Western blot analysis in H-mDC lysates, whereas TREM-1 was almost undetectable in mDCs (Figure 3B), suggesting that its expression is restricted to cells generated under hypoxia.
TREM-1 expression in H-mDCs. mDCs and H-mDCs were generated from different donors, CM was replaced on day 3 of generation with fresh medium supplemented with cytokines, and TREM-1 was analyzed at day 4 of culture. (A) TREM-1 mRNA expression. Total RNA was reverse-transcribed and tested for TREM-1 expression by quantitative RT-PCR. CAXII mRNA levels were assayed in parallel as positive control. Expression changes were evaluated as detailed in “Real-time RT-PCR.” Data are expressed as mean normalized gene expression values, calculated on the basis of triplicate measurements for each experiment/donor, relative to the values obtained for the reference genes. (B) TREM-1 protein expression. Total cell lysates were prepared from mDCs generated from 3 of the donors shown in panel A under normoxic (−) or hypoxic (+) conditions. Proteins (100 μg) were resolved on 10% SDS-PAGE, and the blots were hybridized with Abs directed to TREM-1 and β-actin as a loading control. A representative immunoblot is shown. Strong bands are seen at approximately 30 kDa, the expected TREM-1 molecular weight. (C) TREM-1 surface expression. mDCs and H-mDCs were double-stained with anti-CD83-FITC and anti-TREM-1-PE Abs and analyzed by flow cytometry on a FACScan, as specified in “Flow cytometry.” Cells were electronically gated according to their light scatter properties to exclude cell debris. Left panel: Results from one of 8 independent experiments are shown as dot plots. The percentage of single- and double-positive cells is indicated: TREM-1/CD83 double-positive cells are contained in the top right quadrant, whereas CD83 and TREM-1 single-positive cells are contained in the bottom right and top left quadrants, respectively. Cells stained with control Abs were contained the bottom left quadrants. Right panel: Data are expressed as percentage of TREM-1+ cells within CD83+ mDCs and H-mDCs generated from 8 individual donors (dots). Horizontal lines represent median values for each group. P value by the Student t test is indicated. (D) sTREM-1 secretion. Cell-free supernatants were harvested and assayed for sTREM-1 content by ELISA. Data were obtained from the same preparations analyzed in panel C and are expressed as picograms/1 × 106 cells/mL (dots). Horizontal lines represent median values for each group. P value by the Student t test is indicated.
TREM-1 expression in H-mDCs. mDCs and H-mDCs were generated from different donors, CM was replaced on day 3 of generation with fresh medium supplemented with cytokines, and TREM-1 was analyzed at day 4 of culture. (A) TREM-1 mRNA expression. Total RNA was reverse-transcribed and tested for TREM-1 expression by quantitative RT-PCR. CAXII mRNA levels were assayed in parallel as positive control. Expression changes were evaluated as detailed in “Real-time RT-PCR.” Data are expressed as mean normalized gene expression values, calculated on the basis of triplicate measurements for each experiment/donor, relative to the values obtained for the reference genes. (B) TREM-1 protein expression. Total cell lysates were prepared from mDCs generated from 3 of the donors shown in panel A under normoxic (−) or hypoxic (+) conditions. Proteins (100 μg) were resolved on 10% SDS-PAGE, and the blots were hybridized with Abs directed to TREM-1 and β-actin as a loading control. A representative immunoblot is shown. Strong bands are seen at approximately 30 kDa, the expected TREM-1 molecular weight. (C) TREM-1 surface expression. mDCs and H-mDCs were double-stained with anti-CD83-FITC and anti-TREM-1-PE Abs and analyzed by flow cytometry on a FACScan, as specified in “Flow cytometry.” Cells were electronically gated according to their light scatter properties to exclude cell debris. Left panel: Results from one of 8 independent experiments are shown as dot plots. The percentage of single- and double-positive cells is indicated: TREM-1/CD83 double-positive cells are contained in the top right quadrant, whereas CD83 and TREM-1 single-positive cells are contained in the bottom right and top left quadrants, respectively. Cells stained with control Abs were contained the bottom left quadrants. Right panel: Data are expressed as percentage of TREM-1+ cells within CD83+ mDCs and H-mDCs generated from 8 individual donors (dots). Horizontal lines represent median values for each group. P value by the Student t test is indicated. (D) sTREM-1 secretion. Cell-free supernatants were harvested and assayed for sTREM-1 content by ELISA. Data were obtained from the same preparations analyzed in panel C and are expressed as picograms/1 × 106 cells/mL (dots). Horizontal lines represent median values for each group. P value by the Student t test is indicated.
TREM-1 surface expression was then measured by cytofluorimetry. Figure 3C (left panel) shows the CD83/TREM-1 staining pattern of mDCs and H-mDCs from a representative donor, and Figure 3C (right panel) shows the percentage of TREM-1+ cells in the CD83-gated population from 8 individual samples at day 4 of culture. A total of 50% to 86% of CD83+ H-mDCs expressed TREM-1. Conversely, no TREM-1+ mDCs were detectable in any of the donors examined, confirming previous evidence.17 Immunophenotypic analysis of TREM-1+ and TREM-1− cells demonstrated a different expression profile of CD1c (BDCA-1) and CD141 (BDCA-3), 2 surface markers recently identified in functionally distinct subgroups of myeloid DCs (MDC1 and MDC2) in vivo,23 with a significantly (P < .05) higher percentage of TREM-1+ H-mDCs displaying CD141 and a higher CD1c expression detectable on TREM-1− cells (supplemental Table 2), suggesting a correlation between TREM-1 and CD141 expression. sTREM-1 was described in biologic fluids during inflammation.16,24 We evaluated sTREM-1 content in CM from mDCs and H-mDCs. sTREM-1 was released by H-mDCs but not by mDCs, ranging from 79 to 207 pg/106 cells/mL in 8 different donors (Figure 3D), consistent with the expression pattern of the membrane-bound form.
H-mDC reoxygenation by exposure to normoxic conditions (Reox) for 24 hours resulted in a pronounced down-regulation of TREM-1 mRNA levels compared with cells maintained under hypoxia for the same length of time (Figure 4A). Accordingly, reduction of TREM-1 surface expression was measured by fluorescence-activated cell sorter both in terms of mean fluorescence intensity and percentage of positive cells (Figure 4B) and was paralleled by a comparable decrease of sTREM-1 release into the supernatant (Figure 4C), suggesting that hypoxia-stimulatory effects on TREM-1 were reversible. The role of low pO2 as a stimulus for TREM-1 in mDCs was further supported by data showing that TREM-1 mRNA (Figure 5A) and surface protein (Figure 5B) were induced in mDCs exposed to acute (24 hours) hypoxia, although at levels and in a percentage of cells lower than those detectable in H-mDCs.
Effects of reoxygenation on TREM-1 expression. Four-day H-mDCs were generated from different donors, CM was replaced with fresh medium supplemented with cytokines, and TREM-1 was analyzed after additional 24-hour culture under 20% O2 (Reox). (A) TREM-1 and CAXII mRNA expression was assessed by quantitative RT-PCR. Data from one representative experiment of 3 performed are expressed as in the legend to Figure 3A. (B) TREM-1 surface expression was evaluated by flow cytometry. Solid histograms represent the fluorescent profile of TREM-1-expressing cells; and open histograms, the fluorescent profile of cells stained with the isotype-matched control Ab. The percentage of TREM-1+ cells and the mean fluorescent intensity (in parentheses) are indicated. Results from one of 3 independently tested donors are shown. (C) sTREM-1 release was measured by ELISA. Data from one representative experiment of 3 performed are expressed as picograms/1 × 106 cells/mL.
Effects of reoxygenation on TREM-1 expression. Four-day H-mDCs were generated from different donors, CM was replaced with fresh medium supplemented with cytokines, and TREM-1 was analyzed after additional 24-hour culture under 20% O2 (Reox). (A) TREM-1 and CAXII mRNA expression was assessed by quantitative RT-PCR. Data from one representative experiment of 3 performed are expressed as in the legend to Figure 3A. (B) TREM-1 surface expression was evaluated by flow cytometry. Solid histograms represent the fluorescent profile of TREM-1-expressing cells; and open histograms, the fluorescent profile of cells stained with the isotype-matched control Ab. The percentage of TREM-1+ cells and the mean fluorescent intensity (in parentheses) are indicated. Results from one of 3 independently tested donors are shown. (C) sTREM-1 release was measured by ELISA. Data from one representative experiment of 3 performed are expressed as picograms/1 × 106 cells/mL.
Effects of acute hypoxia on TREM-1 expression. Four-day mDCs were exposed to hypoxia for 24 hours, and (A) TREM-1 and CAXII mRNA expression was assessed by quantitative RT-PCR. Data from one representative experiment of 3 performed are expressed as in the legend to Figure 3A. (B) TREM-1 surface expression was evaluated by flow cytometry. Solid histograms represent the fluorescent profile of TREM-1-expressing cells; and open histograms, the fluorescent profile of cells stained with the isotype-matched control Ab. The percentage of TREM-1+ cells and the mean fluorescent intensity are indicated. Results from one of 3 independently tested donors are shown.
Effects of acute hypoxia on TREM-1 expression. Four-day mDCs were exposed to hypoxia for 24 hours, and (A) TREM-1 and CAXII mRNA expression was assessed by quantitative RT-PCR. Data from one representative experiment of 3 performed are expressed as in the legend to Figure 3A. (B) TREM-1 surface expression was evaluated by flow cytometry. Solid histograms represent the fluorescent profile of TREM-1-expressing cells; and open histograms, the fluorescent profile of cells stained with the isotype-matched control Ab. The percentage of TREM-1+ cells and the mean fluorescent intensity are indicated. Results from one of 3 independently tested donors are shown.
DC exposure to hypoxia was previously shown to be associated with HIF-1α protein accumulation and target gene induction.13-15 Given the presence of HRE in the gene promoter (Table 1), we investigated the potential involvement of HIF-1α in TREM-1 hypoxic induction by RNA interference. As shown in supplemental Figure 2, high levels of HIF-1α were present in mDCs cultured for 24 hours under hypoxia and were associated with TREM-1 expression. HIF-1α silencing decreased TREM-1 protein levels, whereas it did not modify the expression of β-actin used as an internal control. These data suggest that TREM-1 inducibility by hypoxia in mDCs is mediated at least in part by HIF-1α.
TREM-1 cross-linking on H-mDCs promotes DAP12-signaling activation and proinflammatory cytokine and chemokine release
TREM-1 is a transmembrane receptor that lacks signaling motifs in the short cytoplasmic tail and noncovalently associates with the adapter molecule, DAP12, for signal transduction in human monocytes and neutrophils.17,24,25 To confirm TREM-1 association with DAP12, 4-day H-mDCs were subjected to immunoprecipitation with anti-TREM-1 mAb, and the precipitates were analyzed by anti-DAP12 immunoblotting. As shown in Figure 6A, TREM-1 paired with DAP12, consistent with a role for this protein in mediating TREM-1 signaling in H-mDCs. To determine whether TREM-1 was functionally competent, H-mDCs were plated on a plastic surface coated with an agonist mAb or control IgG for 20 minutes under hypoxia, and activation of DAP12-signaling pathway was assessed. TREM-1 cross-linking resulted in increased phosphorylation of DAP12-linked molecules, ERK-1, Akt, and IκBα, indicating that TREM-1 engagement can transduce activating signals in H-mDCs (Figure 6B).
Activation of H-mDCs by TREM-1 cross-linking. (A) mDCs were generated under hypoxic conditions from 2 different donors, and total cell lysates were subjected to immunoprecipitation with anti-TREM-1 mAb or control IgG1. The precipitates were resolved on a 12% SDS-PAGE and immunoblotted with anti-DAP12 mAb, as described in “Western blot analysis and immunoprecipitation.” DAP12 and Ig light chain (IgL) are indicated by arrows. Molecular weight markers are given at the side. (B-C) H-mDCs were seeded onto plates precoated with agonist anti-TREM-1 mAb or control IgG1 at 10 μg/mL, after extensive washing to remove cytokines, and cultured for 20 minutes (B) or 24 hours (C) under hypoxic conditions. (B) Protein phosphorylation. Cell lysates (40 μg) were resolved on 10% SDS-PAGE and immunoblotted with Abs antiphospho (p)-ERK, Akt, and IκBα. Abs against the nonphosphorylated forms (tot) were used as loading controls. Representative experiments performed with cells from 2 different donors are shown. T0 indicates H-mDCs from one of the donors not subjected to Ab cross-linking, used as a negative control of phosphorylation. The figure was arranged by combining the lanes containing not-crosslinked samples (T0) with those containing samples subjected to cross-linking from different parts of the same gel. (C) Cytokine production. CM was assayed for TNF-α, IL-6, IL-12p70, IL-10, CXCL8, CCL4, and CCL5 content by specific ELISA. Results are expressed as picograms or nanograms/8 × 105 cells/mL and are represented as the mean ± SE of 3 different experiments. Values significantly different from those of H-mDCs cross-linked with IgG1: * P < .05; **P < .001; ***P < .0001.
Activation of H-mDCs by TREM-1 cross-linking. (A) mDCs were generated under hypoxic conditions from 2 different donors, and total cell lysates were subjected to immunoprecipitation with anti-TREM-1 mAb or control IgG1. The precipitates were resolved on a 12% SDS-PAGE and immunoblotted with anti-DAP12 mAb, as described in “Western blot analysis and immunoprecipitation.” DAP12 and Ig light chain (IgL) are indicated by arrows. Molecular weight markers are given at the side. (B-C) H-mDCs were seeded onto plates precoated with agonist anti-TREM-1 mAb or control IgG1 at 10 μg/mL, after extensive washing to remove cytokines, and cultured for 20 minutes (B) or 24 hours (C) under hypoxic conditions. (B) Protein phosphorylation. Cell lysates (40 μg) were resolved on 10% SDS-PAGE and immunoblotted with Abs antiphospho (p)-ERK, Akt, and IκBα. Abs against the nonphosphorylated forms (tot) were used as loading controls. Representative experiments performed with cells from 2 different donors are shown. T0 indicates H-mDCs from one of the donors not subjected to Ab cross-linking, used as a negative control of phosphorylation. The figure was arranged by combining the lanes containing not-crosslinked samples (T0) with those containing samples subjected to cross-linking from different parts of the same gel. (C) Cytokine production. CM was assayed for TNF-α, IL-6, IL-12p70, IL-10, CXCL8, CCL4, and CCL5 content by specific ELISA. Results are expressed as picograms or nanograms/8 × 105 cells/mL and are represented as the mean ± SE of 3 different experiments. Values significantly different from those of H-mDCs cross-linked with IgG1: * P < .05; **P < .001; ***P < .0001.
TREM-1 is an amplifier of the inflammatory response.17,24 Hence, we investigated whether its activation triggered proinflammatory cytokine and chemokine production by H-mDCs. Supernatants from H-mDCs were collected 24 hours after stimulation with agonist anti-TREM-1 or control IgG1 under hypoxic conditions and analyzed for cytokine and chemokine content by ELISA (Figure 6C). TREM-1 cross-linking significantly enhanced TNF-α, IL-6, IL-12p70, CXCL8, CCL4, and CCL5 secretion compared with Ig, whereas production of the anti-inflammatory cytokine IL-10 was not affected. No changes in cytokine secretion were observed on triggering with an isotype-matched anti-MHC-I mAb (data not shown), confirming that H-mDC activation by anti-TREM-1 mAb was not the result of a specific Fc receptor ligation. Overall, these data suggest that TREM-1 is a functional receptor selectively expressed by H-mDCs and not by the normoxic counterpart whose engagement can drive production of cytokines and chemokines involved in innate and adaptive immunity, raising the question of the existence of TREM-1+ mDCs in vivo.
TREM-1 is expressed in vivo on H-mDCs recruited to the synovial fluid of JIA patients
TREM-1 has been recently implicated in the pathogenesis of noninfectious inflammatory disorders, including rheumatoid arthritis.26,27 Hypoxia is a common feature of the inflamed rheumatoid synovium,8,28 and evidence suggests that DCs are enriched in arthritic joints playing a role in the initiation and perpetuation of the inflammatory process.23,29 To evaluate TREM-1+ mDC occurrence in vivo, we investigated whether TREM-1 was expressed in synovial mDCs from children affected by JIA. We first analyzed plasma and SF samples obtained from JIA patients for sTREM-1 content by ELISA. As shown in Figure 7A, plasma sTREM concentrations in JIA patients (61-394 pg/mL) were significantly higher (P < .01) than those in control subjects (12-16 pg/mL) and were further increased (P < .001) in matched SF samples (1120-3800 pg/mL). These data are in line with previous observations in other forms of inflammatory arthritis, such as rheumatoid arthritis,26,27 indicating disease activity.
TREM-1 expression in vivo in hypoxic mDCs from JIA-SF. SF was collected from children affected by JIA, and SFMCs were purified as detailed in “SFMC isolation.” (A) sTREM-1 concentrations were quantified by ELISA in paired plasma and SF from 8 JIA patients and plasma from 5 age-matched control subjects. Individual samples were run in triplicate. Boxes represent the values falling between the 25th and 75th percentiles; whiskers, the highest and lowest values for each subgroup. Bold horizontal lines represent median values. P value by the Wilcoxon rank test is indicated: *P < .01 relative to plasma controls; **P < .001 relative to plasma from JIA patients. (B) HIF-1α expression was assessed by Western blot analysis on whole protein extracts from 3 pairs of fresh PBMCs and SFMCs purified from a subset of the JIA patients shown in panel A. Protein (100 μg) was resolved on 8% SDS-PAGE, and the blots were hybridized with anti-HIF-1α mAb. β-actin was evaluated as a loading control. (C) Flow cytometric analysis of TREM-1 expression in SF-mDCs. SFMCs from the 3 JIA patients shown in panel B were stained with anti-CD1a-allophycocyanin, CD83-FITC, and-TREM-1-PE Abs and analyzed on a FACScan. Cells were electronically gated according to their light scatter properties to exclude cell debris. The gated population was analyzed for CD1a positivity (left panels, region R1), and CD1a+ cells were then examined for CD83 and TREM-1 expression (right panels). Nonspecific staining was corrected using isotype-matched Abs. Numbers represent the percentage of single- and double-positive cells within the CD1a-gated population and are indicated for each patient: TREM-1/CD83 double-positive cells are contained in the top right quadrant, whereas CD83 and TREM-1 single-positive cells are contained in the bottom right and top left quadrants, respectively. Cells stained with control Abs are in the bottom left quadrant. The percentage of TREM-1+ cells relative to the total number of CD83+ cells (representing TREM-1-expressing mDCs) was 88%, 60%, and 80% in patients 1, 3, and 6, respectively, whereas the percentage of TREM-1+ cells relative to the total number of CD83− cells (representing TREM-1-expressing iDCs) was 74%, 80%, and 80% in patients 1, 3, and 6, respectively.
TREM-1 expression in vivo in hypoxic mDCs from JIA-SF. SF was collected from children affected by JIA, and SFMCs were purified as detailed in “SFMC isolation.” (A) sTREM-1 concentrations were quantified by ELISA in paired plasma and SF from 8 JIA patients and plasma from 5 age-matched control subjects. Individual samples were run in triplicate. Boxes represent the values falling between the 25th and 75th percentiles; whiskers, the highest and lowest values for each subgroup. Bold horizontal lines represent median values. P value by the Wilcoxon rank test is indicated: *P < .01 relative to plasma controls; **P < .001 relative to plasma from JIA patients. (B) HIF-1α expression was assessed by Western blot analysis on whole protein extracts from 3 pairs of fresh PBMCs and SFMCs purified from a subset of the JIA patients shown in panel A. Protein (100 μg) was resolved on 8% SDS-PAGE, and the blots were hybridized with anti-HIF-1α mAb. β-actin was evaluated as a loading control. (C) Flow cytometric analysis of TREM-1 expression in SF-mDCs. SFMCs from the 3 JIA patients shown in panel B were stained with anti-CD1a-allophycocyanin, CD83-FITC, and-TREM-1-PE Abs and analyzed on a FACScan. Cells were electronically gated according to their light scatter properties to exclude cell debris. The gated population was analyzed for CD1a positivity (left panels, region R1), and CD1a+ cells were then examined for CD83 and TREM-1 expression (right panels). Nonspecific staining was corrected using isotype-matched Abs. Numbers represent the percentage of single- and double-positive cells within the CD1a-gated population and are indicated for each patient: TREM-1/CD83 double-positive cells are contained in the top right quadrant, whereas CD83 and TREM-1 single-positive cells are contained in the bottom right and top left quadrants, respectively. Cells stained with control Abs are in the bottom left quadrant. The percentage of TREM-1+ cells relative to the total number of CD83+ cells (representing TREM-1-expressing mDCs) was 88%, 60%, and 80% in patients 1, 3, and 6, respectively, whereas the percentage of TREM-1+ cells relative to the total number of CD83− cells (representing TREM-1-expressing iDCs) was 74%, 80%, and 80% in patients 1, 3, and 6, respectively.
SFMCs were then isolated from a subset of patients and analyzed for TREM-1 surface expression. HIF-1α protein was also evaluated in parallel by Western blotting to confirm cell adaptation to the hypoxic synovial environment. SFMCs, but not paired PBMCs, constitutively accumulated high levels of HIF-1α (Figure 7B), confirming recent observations.19,20 The presence of TREM-1+ mDCs in SFMCs was then determined by 3-color flow cytometric analysis with mAbs to CD1a, CD83, and TREM-1. As shown in Figure 7C, a subset of SFMCs, ranging from 4.6% to 6.6% in 3 different patients, expressed CD1a (left panels), confirming DC enrichment in JIA SF. A high proportion of DCs in JIA SF expressed CD83 (Figure 7C, right panels), composing 42%, 25%, and 20% of the total CD1a-gated cells in patients 1, 3, and 6, respectively (top and bottom right quadrants). It is noteworthy that 88%, 60%, and 80% of cells within the CD83+ subset expressed TREM-1, accounting for 37%, 15%, and 16% of the total CD1a-gated population (top right quadrants). These findings provide the first evidence of the existence in a human hypoxic tissue in vivo of a population of CD83+ mDCs expressing TREM-1, correspondent to mDCs generated in vitro under low pO2, suggesting the potential relevance of this molecule as an in vivo marker of hypoxic mDCs.
Discussion
mDC development from monocytic precursors recruited at sites of inflammation and infection occurs in the setting of low pO2.5,6 The impact of the hypoxic microenvironment on DC maturation process is still controversial. This study characterizes the transcriptional profile of mDCs generated from human monocytes under chronic hypoxic conditions similar to those present in diseased tissues, demonstrating that H-mDCs are functionally reprogrammed through the differential expression of a large number of genes involved in innate and adaptive immunity. Furthermore, we define a new subpopulation of hypoxic mDCs characterized by the CD1a+/CD83+/TREM-1+ phenotype.
Divergent effects of hypoxia on DC maturation were reported in previous studies based on the expression of maturation markers, costimulatory molecules, chemokine receptors, and T cell-priming ability. Mancino et al showed that acute hypoxia impaired iDC phenotypic and functional maturation in response to lipopolysaccharide,14 whereas Rama et al5 and Jantsch et al15 demonstrated that it promoted their maturation both by itself and in combination with lipopolysaccharide. Two recent reports addressing the effects of chronic hypoxia suggested inhibition of monocyte-derived DC maturation.30,31 In contrast, our results indicate that chronic exposure to low pO2 does not substantially affect mDC phenotypic maturation because comparable CD1a, CD83, CD86, CCR7, and CXCR4 surface expression was displayed by mDCs and H-mDCs, confirming and extending previous data by our group13 and by Zhao et al.32 A possible explanation for these conflicting results could be the different experimental approaches used with regard to source and purity of DC precursors, differentiation/maturation protocols, degree and duration of the hypoxic stimulus, and difference of species (human vs mouse).
Clustering of hypoxia-modulated genes according to GO data mining identified 33 interrelated functional categories, containing a statistically significant portion of genes controlling cell metabolism, differentiation/maturation, and functional properties. Interestingly, a large cluster of genes belonging to immune-related pathways was up-regulated in H-mDCs with respect to mDCs. These findings extend to mDCs the characteristic trend of response to hypoxia of other types of mononuclear phagocytes, including primary monocytes, monocyte-derived macrophages, and iDCs,12,21,33 emphasizing the critical contribution of reduced oxygenation to the control of immune responses.
mDCs are critical for the induction of protective immunity to microbial invasion and the maintenance of self-tolerance.34 Their functions are tightly regulated by a complex network of inhibitory and activating signals transduced by a defined repertoire of cell surface receptors,4,24,35,36 and dysregulated expression of these molecules may result in an aberrant response characterized by amplification of inflammation and loss of tolerance.2,4,34 DC maturation under chronic hypoxia resulted in the up-regulation of genes encoding various members of these receptor families. Profound changes were observed in genes coding for pattern recognition receptors critical to host defense, which are endowed with the capability to recognize specific pathogen-associated molecular patterns on infectious agents and trigger proinflammatory cytokine production,36-38 suggesting enhanced inflammatory functions of mDCs generated in hypoxic than in normoxic areas. Up-regulation of mRNA for the scavenger receptors, CD36 and APOB48R, is also of note and consistent with the view that hypoxia may exert a pathogenetic role in atherosclerosis7 because these molecules function as endocytic transporters for lipoproteins and phospholipids, contributing to lipid-loaded foam cell formation.39,40
mDC migration is known to be highly sensitive to microenvironmental changes.2-4 We observed higher expression of mRNA for several adhesion/homing receptors and costimulatory molecules in H-mDCs compared with mDCs. Given the role of the encoded proteins in cell-cell and cell-matrix interactions, transendothelial migration and trafficking, and T cell-costimulatory activity,41,42 it is probable that H-mDCs have an increased capacity to migrate to secondary lymphoid organs and activate adaptive immune responses than the normoxic counterparts.
Hypoxia also affected the expression pattern of genes coding for immunoregulatory Ig-like receptor family members involved in the regulation of myeloid cell functions in innate and adaptive immunity. Of relevance is the increased mRNA for Ig-Fc receptors. These are activating receptors characterized by immunoreceptor tyrosine-based activating motifs in their cytoplasmic domain (FcγRIIC, FcαR, FcϵRI) or pairing with an immunoreceptor tyrosine-based activating motif-containing transmembrane adaptor proteins for signaling (FcγRIIIA), which trigger phagocytosis and immune complex clearance, inflammatory cytokine production, antibody-dependent cellular cytotoxicity, and respiratory burst.43 Hypoxic up-regulation of other genes encoding Ig family members, such as BDCA3, LILRA2, and SEMA4D,35,44,45 was also observed. These molecules modulate DC maturation, immunogenicity, cooperation with T cells, and proinflammatory cytokine production. Our results suggest that hypoxia can affect mDC functional behavior in diseased tissues and potentially contribute to the pathogenesis of allergic inflammatory disorders and/or autoimmune diseases, which are associated with overexpression of these receptors.44-46 The challenge of future studies will be to validate these data in vivo.
An intriguing finding of this study is the identification of TREM-1 as a hypoxia-inducible gene in mDCs. Previous reports have shown that TREM-1 is selectively expressed on neutrophils and on a subset of monocytes and tissue macrophages, where it can be markedly up-regulated by various stimuli, such as Toll-like receptor ligands and proinflammatory cytokines, whereas it is completely down-regulated during monocyte differentiation/maturation into DCs.16,17,24,25 Our results provide the first evidence that TREM-1 can be expressed in mDCs generated under chronic hypoxia. TREM-1 mRNA was consistently detected in H-mDCs from different donors and paralleled by expression of the membrane-bound receptor and release of its soluble form. Interestingly, TREM-1 mRNA and protein expression was also inducible on mDC exposure to acute hypoxia, although at levels lower than those induced by chronic hypoxia, and was reversible, because cell reoxygenation resulted in TREM-1 down-regulation. Consistent with these findings, the HIF/HRE system appeared to be involved at least in part in TREM-1 hypoxic induction. These results suggest that TREM-1 expression in mDCs in vivo may vary dynamically with the degree of local oxygenation, which is quite heterogeneous and rapidly fluctuating in diseased tissues,9 giving rise to distinct mDC subsets potentially endowed with different functional properties.
TREM-1 expressed on H-mDC paired with DAP12 adapter protein and was biologically active because its engagement resulted in the activation of DAP12-linked signaling, as shown by increased ERK-1 and Akt kinase phosphorylation. These data are in agreement with previous observations in primary monocytes and neutrophils.17,24,25 TREM-1 cross-linking on H-mDCs triggered secretion of TNF-α, IL-6, CXCL8, CCL4, and CCL5, which are important proinflammatory and chemotactic factors,4,13,21,29 and of IL-12p70, a Th1-inducing cytokine essential for adaptive immune responses against intracellular pathogens and tumors.4,34 These data support and extend previous findings implicating TREM-1 as an amplifier of inflammation17,24 and suggest that the hypoxic environment enhances mDC ability to regulate leukocyte recruitment at inflammatory sites and Th1 priming by stimulating TREM-1 expression. Increased nuclear levels of nuclear factor-κB (NF-κB) were observed in response to TREM-1 cross-linking in monocytes and neutrophils.25 NF-κB proteins are sequestered in the cytoplasm by inhibitory proteins (IκBα), which become phosphorylated and subsequently degraded after cell activation, allowing NF-κB nuclear translocation and activity.47 Our data showing IκBα phosphorylation in response to TREM-1 cross-linking support a role for the NF-κB pathway in TREM-1-mediated proinflammatory responses also in H-mDCs.
Increased TREM-1 expression and sTREM-1 release were observed in several infectious and noninfectious inflammatory disorders, suggesting a pathogenetic role for this molecule.16,17,24,26,27,48 Our data add to the growing list of diseases associated with TREM-1 overexpression, providing the first evidence of elevated sTREM-1 levels in JIA. JIA is the most common chronic pediatric rheumatic disease,18 and sTREM-1 presence in JIA-SF is an index of the underlying inflammatory process whose persistence and amplification are dictated by the functional cooperation among several inflammatory cell populations recruited to the synovium of affected joints,28 including TREM-1+ monocytes/macrophages and neutrophils.26,27,48 We have recently documented the presence of reduced pO2 in the JIA synovium and the contribution of the hypoxic environment to the persistence of inflammation by triggering leukocyte recruitment and activation.19,20,28 The demonstration that SFMCs express HIF-1α confirms the adaptation of synovial inflammatory cells to local hypoxia. Previous studies have demonstrated accumulation of both DC precursors and myeloid DC subsets in the SF and synovial tissue from children affected by JIA.23,28 Consistent with these findings, our immunophenotypical data demonstrated enrichment of cells expressing the DC marker CD1a in JIA synovial effusions, which composed up to 6.6% of the total SFMC population. Although CD1a+ DCs had a predominantly immature phenotype (CD83−), a high percentage of mature (CD83+) DCs was also present in hypoxic JIA-SF, and the majority of cells in the CD83+ subset expressed TREM-1. Taken together with in vitro findings, the demonstration of TREM-1 expression in vivo on mDCs present in hypoxic JIA-SF strongly suggests the potential relevance of this molecule as a marker of mDCs generated under hypoxic conditions. Furthermore, the observation that also the majority of cells belonging to the CD83− subset in JIA-SF was TREM-1+ indicates that TREM-1 expression is independent of the DC maturation stage, representing a common in vivo marker of hypoxic DCs. Sustained TREM-1 induction on synovial DCs by intra-articular hypoxia may be of pathologic relevance, representing a potential mechanism of amplification of synovial inflammation, and future studies will be aimed at assessing the role of TREM-1 putative ligand(s)49 present in the inflamed arthritic synovium50 in triggering DC proinflammatory cytokine and chemokine production.
In conclusion, this study provides novel insights into the mechanisms linking low pO2 to the regulation of immune and inflammatory responses, leading to new perspectives of the role of hypoxia in programming mDC functions within pathologic tissues.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Italian Health Ministry, Associazione Italiana Ricerca sul Cancro, Associazione Italiana Glicogenosi, Regione Piemonte: Progetti di Ricerca Sanitaria Finalizzata e Applicata, Progetti strategici su tematiche di interesse regionale o sovraregionale, and Fondazione CRT (Progetto Alfieri). P.C. was supported by a fellowship from the Fondazione Italiana Ricerca sul Cancro.
Authorship
Contribution: M.C.B. designed and supervised research, contributed to data analysis, and wrote the manuscript; D.P. and F.B. contributed to study design, carried out most of the research, and analyzed flow cytometric and microarray data; F.R. contributed to DC culture and gene expression analysis; C.V. performed Western blot analysis; M.G. provided synovial and blood samples from JIA patients and discussion; A.E. provided critical discussion; F.N. contributed to data analysis; P.C. analyzed cytokine expression profile; and M.G. and L.V. conceived and coordinated research, contributed to data analysis, and assisted with manuscript preparation and revision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria Carla Bosco, Laboratorio di Biologia Molecolare, Istituto Giannina Gaslini, Padiglione 2, L.go G.Gaslini 5, 16147 Genova Quarto, Italy; e-mail: mariacarlabosco@ospedale-gaslini.ge.it.
References
Author notes
M.C.B., D.P., and F.B. contributed equally to this study.
M.G. and L.V. share senior authorship.

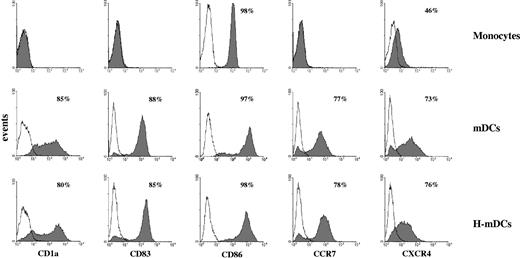
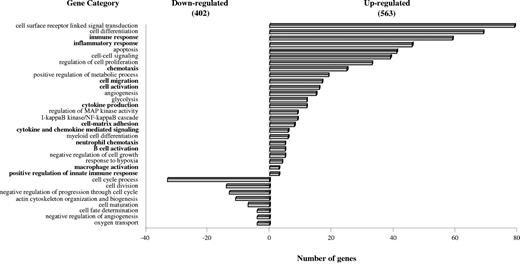
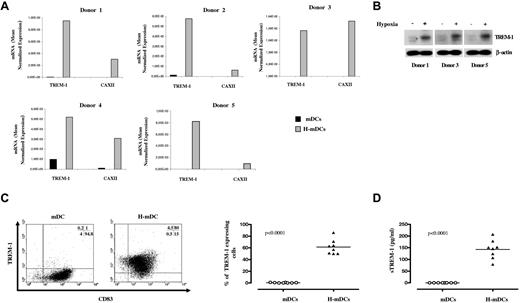
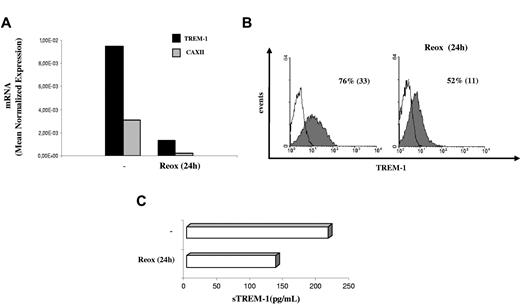
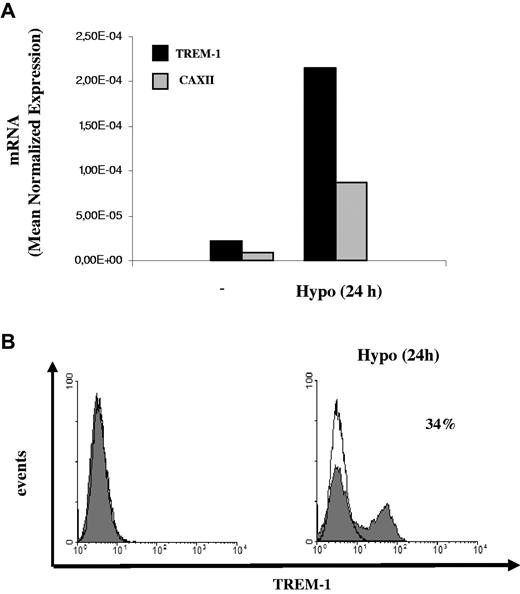
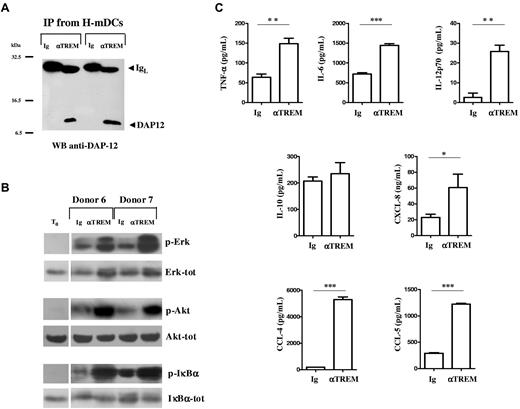
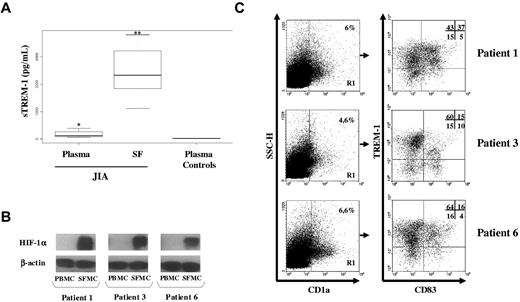
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal