Abstract
Memory T cells (TMs) have been detected in many tissues but their quantitative distribution remains largely undefined. We show that in mice there is a remarkably biased accumulation of long-term CD4 TMs into mucosal sites (mainly gut, especially Peyer patches), and CD8 TMs into lymph nodes and spleen (in particular, peripheral lymph nodes [PLNs]). This distinction correlates with their differentiated expression of PLN- and gut-homing markers. CD8 and CD4 TMs selectively require the expression of PLN-homing marker CCR7 or gut-homing marker α4β7 for maintenance. PLNs and gut supply CD8 and CD4 TMs with their individually favored homeostatic cytokine, IL-15, or IL-7. Cytokine stimulation in turn regulates the different gut-homing marker expression on CD4 and CD8 TMs. IL-15 plays a major role in vivo regulating CD8 TMs homing to PLNs. Thus, the reservoir segregation of CD4 and CD8 TMs meets their individual needs for homeostatic cytokines and is under feedback control of cytokine stimulation.
Introduction
A prominent feature of T-cell immunity is the formation of memory T cells (TMs) after exposure to infectious agents, leading to more effective immune protection on re-encountering the same stimulus.1 CD4 and CD8 T cells carry out distinct immunologic functions: while CD8 T cells are specialized for cytotoxicity,2 CD4 T cells play a more comprehensive role through helping CD8 T cells and B cells to respond and generate memory,3,4 and through constituting specialized compartments like TH17 cells and T regulatory cells.5,6 CD4 and CD8 T cells show considerable differences in their memory generation and maintenance.7-10 For example, CD8 TM differentiation appears to be efficient and follows a linear differentiation pathway, whereas CD4 TM differentiation is more complex and requires a prolonged period of stimulation.8 In the absence of antigen, TMs are maintained mainly through interactions with homeostatic cytokines, with CD8 TMs particularly depending on IL-15 while CD4 TMs primarily depend on IL-7.10
Homeostatic cytokines are mainly produced by nonhematopoietic tissue cells and considered to be supplied largely in a tissue-restricted manner in limited amount.11,12 TMs need to home to special “niches” to efficiently access and compete for these survival factors.13,14 The search for such “niches” and the associated reservoir distribution for TMs has been severely hampered by the technical difficulty of systemically tracking and quantifying TMs.9,11 Recently, bone marrow (BM) has been proposed as an important reservoir for both CD8 and CD4 TMs, which is considered to contain niches defined by IL-7–expressing stromal cells.15-17 However, the systemic distribution of TM reservoirs, especially the direct comparison between CD4 and CD8 TM reservoirs, remains largely unexamined.
In the past decade, the increasing application of molecular imaging techniques for studying the immune system has yielded valuable insights into the dynamics of the immune cells.18-20 Among them, BLI is especially suitable for studying T-cell trafficking in small animal models, because of its capacity for noninvasive measurement of T-cell dynamics in vivo, its excellent signal-to-noise ratios, and its user-friendly and relatively inexpensive instrumentation.20
Aided by the BLI technique, we have systemically analyzed and compared the reservoir distribution of long-term CD4 and CD8 TMs in mice. We find that although both TMs are found in multiple tissues, their preferences for individual tissues are quite different: CD8 TMs accumulate mainly in lymph nodes and spleen, particularly PLNs whereas CD4 TMs accumulate preferentially in mucosal sites, mainly gut and especially Peyer patches (PPs). This polarized accumulation correlates with their differing expression of PLN- and gut-homing markers. Deficiency of a gut-homing marker α4β7 or a PLN-homing marker CCR7 selectively impairs the formation and maintenance of CD4 or CD8 TMs. PLNs produce high level of IL-15, making them particularly fit as a home for CD8 TMs while gut expresses IL-7, making it appropriate for CD4 TMs. In addition, IL-7 and IL-15 stimulation sustains the differentiated expression of homing markers on the CD4 and CD8 TMs, providing an apparent feedback control to stabilize their reservoir segregation.
Methods
Mice and materials, antibodies and flow cytometry, tissue lymphocytes isolation, DC-directed lentivirus infection, CD4 effector T cells (TEs) differentiation in vitro, CD4 memory T cells (TMs) functional analysis, and IL-7 and IL-15 mRNA tissue expression are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All mouse experiments were approved by the California Institute of Technology Institutional Animal Care and Use Committee.
MFG retrovirus
Effector T cell (TE) culture, transduction, MACS sorting and adoptive transfer
To generate polyclonal TEs, SP, and LN cells harvested from B6 female mice were cultured in T-cell culture media containing 0.5 μg/mL anti–mouse CD3 and 0.5 μg/mL anti–mouse CD28 (Biolegend) for 3 days. To generate antigen-specific TEs, SP, and LN cells harvested from OT1 or OT2 Tg mice were cultured in T-cell culture medium containing either 0.1 μg/mL OVAp257-269 or 1 μg/mL OVAp323-339 for 3 days. To generate MFG-labeled TEs, on day 1 and day 2 of the culture, T cells were spin-infected with retroviral supernatant supplemented with 10 μg/mL polybrene for 90 minutes at 770g at 30°C. CD4 and CD8 TEs were purified using MACS sorting through positive-selection (Miltenyi Biotec). For adoptive transfer, purified CD4 or/and CD8 TEs (2-20 × 106/recipient), supplemented with freshly isolated BM cells (5-10 × 106/recipient), were injected intravenously into recipient mice that had received 1000 rads of total body irradiation. Postadoptive transfer, the recipient mice were maintained on the mixed antibiotic sulfmethoxazole and trimethoprim oral suspension (Hi-Tech Pharmacal) for 4 weeks.
Bioluminescence imaging
Bioluminescence imaging (BLI) was performed using an IVIS200 imaging system (Xenogen/Caliper LifeSciences). Live animal imaging was acquired 5 minutes after intraperitoneal injection of D-Luciferin (1 mg/mouse, Xenogen/Caliper Lifesciences). To image tissues, mice that received intraperitoneal injection of D-Luciferin (3 mg/mouse) were dissected 5 minutes after injection; the individual tissues were imaged within the following 15 minutes. Imaging results were analyzed using a Living Imaging 2.50 software.
In vitro and in vivo cytokine stimulation for TMs
For in vitro stimulation, CD4 or CD8 TMs were cultured in T-cell culture media supplemented with 100 ng/mL of either IL-7 or IL-15 for 3 days. For in vivo stimulation, each mouse received a single intraperitoneal injection of 10 μg of either IL-7 or IL-15 once daily for 5 sequential days.
Statistical analyses
Student t test was used for paired comparisons. Data are presented as mean ± SEM, unless otherwise indicated.
Results
Visualizing the segregation of CD4 and CD8 TM reservoirs in mice
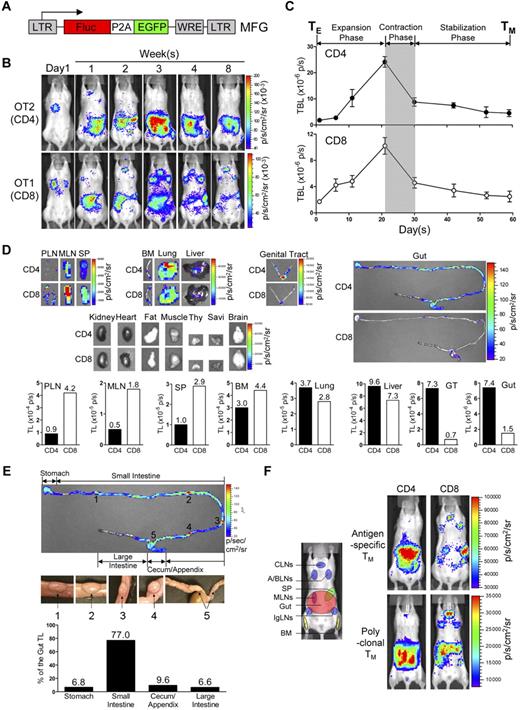
To track TMs in mice, we constructed a dual-reporter retroviral vector, MFG, coexpressing firefly luciferase (FLuc) and enhanced green fluorescence protein (EGFP; Figure 1A). OVA-specific CD4 (OT2) and CD8 (OT1) effector T cells (TEs) were generated in vitro, transduced with MFG, separately transferred into albino B6 recipient mice and tracked in vivo using BLI (supplemental Figure 1A). This approach allowed us to visualize the formation of TMs in an antigen-free host developing from a relatively homogeneous and synchronized population of TEs.21,23,24 Estimation of the pool size of the transferred cells by measuring the total body luminescence (TBL) of a recipient mouse divided TM formation into 3 phases: expansion (weeks 1-3), contraction (week 4) and stabilization (> 1 month; Figure 1B,C), which closely resembles the TM formation kinetics during an acute infection.25 FACS analysis confirmed that both OT2 and OT1 T cells at the stabilization phase have acquired the typical TM phenotype: CD25−CD69−CD62Lhi/loCD44hi (supplemental Figure 1B).
Visualizing the segregation of CD4 and CD8 TM reservoirs in mice. (A) Schematic representation of dual-reporter retrovector MFG. LTR indicates long-term repeats; Fluc, firefly luciferase; P2A, porcine teschovirus 2A sequence; EGFP, enhanced green fluorescence protein; and WRE, woodchuck responsive element. (B,C) Visualization of the CD4 and CD8 TM formation in albino B6 mice postadoptive transfer of 1 × 106 MFG-labeled OT2 (CD4) or OT1 (CD8) effector T cells (TEs) using BLI. Representative BLI images (B) and the measurements of total body luminescence (TBL, mean ± SEM; C) are shown (n = 4). Shaded area marks contraction phase (C). (D,E) Visualization of the CD4 and CD8 TMs in individual tissues using excised tissue BLI. (D) Tissues were collected from albino B6 recipient mice 2 months after transfer of 10 × 106 MFG-labeled OT2 (CD4) or OT1 (CD8) TEs. PLN indicates peripheral lymph node; MLN, mesenteric LN; SP, spleen; BM, bone marrow; Thy, thymus; GT, genital tract; and Savi, salivary gland. (E) Detailed analysis of CD4 TM homing to gut. BLI images of the gut tract (top, the “hot spots” are numbered), identification of the “hot spots” as Peyer patches (PPs; middle), and the measurements of luminescence in gut sub-regions (bottom) are shown. Data are representative of 3 independent experiments. (F) Visualization of the antigen-specific or polyclonal CD4 and CD8 TMs formed in albino B6 recipient mice each receiving 1 × 106 either MFG-labeled OT2 or OT1 TEs, or MFG-labeled B6 CD4 or CD8 TEs using BLI. Representative BLI images collected 1 month after transfer are shown (n = 4). Schematic showing the individual tissue localization in mice is provided for reference. CLNs indicate cervical LNs; A/BLNs, axillary/brachial LNs; and IgLN, inguinal LNs. PLNs refer to the combination of CLNs, A/BLNs, and IgLNs.
Visualizing the segregation of CD4 and CD8 TM reservoirs in mice. (A) Schematic representation of dual-reporter retrovector MFG. LTR indicates long-term repeats; Fluc, firefly luciferase; P2A, porcine teschovirus 2A sequence; EGFP, enhanced green fluorescence protein; and WRE, woodchuck responsive element. (B,C) Visualization of the CD4 and CD8 TM formation in albino B6 mice postadoptive transfer of 1 × 106 MFG-labeled OT2 (CD4) or OT1 (CD8) effector T cells (TEs) using BLI. Representative BLI images (B) and the measurements of total body luminescence (TBL, mean ± SEM; C) are shown (n = 4). Shaded area marks contraction phase (C). (D,E) Visualization of the CD4 and CD8 TMs in individual tissues using excised tissue BLI. (D) Tissues were collected from albino B6 recipient mice 2 months after transfer of 10 × 106 MFG-labeled OT2 (CD4) or OT1 (CD8) TEs. PLN indicates peripheral lymph node; MLN, mesenteric LN; SP, spleen; BM, bone marrow; Thy, thymus; GT, genital tract; and Savi, salivary gland. (E) Detailed analysis of CD4 TM homing to gut. BLI images of the gut tract (top, the “hot spots” are numbered), identification of the “hot spots” as Peyer patches (PPs; middle), and the measurements of luminescence in gut sub-regions (bottom) are shown. Data are representative of 3 independent experiments. (F) Visualization of the antigen-specific or polyclonal CD4 and CD8 TMs formed in albino B6 recipient mice each receiving 1 × 106 either MFG-labeled OT2 or OT1 TEs, or MFG-labeled B6 CD4 or CD8 TEs using BLI. Representative BLI images collected 1 month after transfer are shown (n = 4). Schematic showing the individual tissue localization in mice is provided for reference. CLNs indicate cervical LNs; A/BLNs, axillary/brachial LNs; and IgLN, inguinal LNs. PLNs refer to the combination of CLNs, A/BLNs, and IgLNs.
In the early expansion phase (weeks 1 and 2), OT2 and OT1 T cells exhibited similar disseminated distribution patterns: they were initially detected in the lung (as early as 14 hours after transfer), probably because lung is the first organ they enter on exiting the heart (Figure 1B and supplemental Figure 2A), followed by movement to many other lymphoid and nonlymphoid tissues, including lymph nodes (LNs), spleen (SP), bone marrow (BM), liver, pancreas (Pan), thymus (Thy), genital tract (GT) and gut (supplemental Figure 2B). Differences in distribution patterns appeared in the later expansion phase (week 3), became evident during the contraction phase (week 4), and were maintained throughout the stabilization phase (> 1 month), for as long as the TMs were detectable (up to 1 year in our experiments; Figure 1B-D). Comparison of individual tissues for OT2 and OT1 TM accumulation showed that they could be classified into 4 groups (Figure 1D): (1) OT1 TM-favored reservoirs, which included peripheral LNs (PLNs), mesenteric LNs (MLNs) and SP, showing ∼ 3-5 fold higher total luminescence (TL) for OT1 TMs than OT2 TMs; (2) common reservoirs, which included BM, lung, and liver, showing similar TL levels; (3) OT2 TM-favored reservoirs, which included mucosal sites like gut and genital tract, showing ∼ 5- to 10-fold higher TL signals for OT2 TMs than OT1 TMs; and (4) un-favored reservoirs, which included all other tissues.
Overall, the OT1 TMs preferentially accumulated in LNs and SP, particularly PLNs; whereas OT2 TMs preferred to accumulate in mucosal sites including genital tract and gut, with gut containing the most OT2 TMs because of its large size. Imaging of the isolated gut showed that OT2 TMs distributed through the entire gut tract from stomach to colon, with PPs identified as “hot spots” containing highly concentrated OT2 TMs (Figure 1E). Quantification of the TL in gut sub-regions showed that the small intestine, appendix/cecum, stomach and colon contained ∼ 80%, 10%, 5%, and 5% of the gut OT2 TMs, respectively (Figure 1E).
Because OT2 and OT1 T cells were specific for a model Ag, OVA, we asked whether this distinct TM homing pattern was general or unique to this Ag. When MFG-labeled polyclonal CD4 and CD8 TEs were transferred into albino B6 recipients (supplemental Figures 1C-D), the resulting CD4 and CD8 TMs showed a similar segregated accumulation (Figure 1F), suggesting that it was a general feature of long-term TMs independent of their Ag specificity. FACS analysis also confirmed that in the stabilization phase, both the MFG-labeled polyclonal CD4 and CD8 T cells displayed typical TM phenotype CD25−CD69−CD44hiCD62Lhi/lo (supplemental Figure 1D).
Various types of CD4 and CD8 TMs show a similar polarized tissue distribution
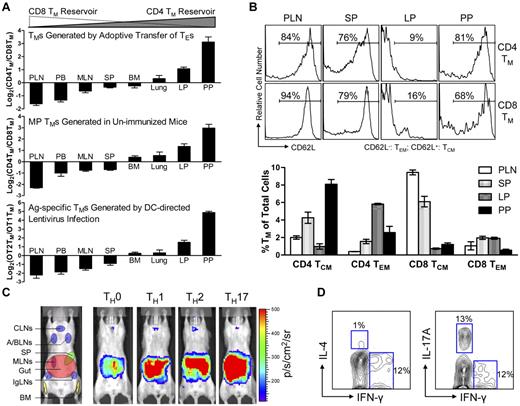
Because BLI is only a semi-quantitative method, we further analyzed the tissue distribution of polyclonal CD4 and CD8 TMs generated through adoptive transfer of effector T cells using flow cytometry. When equal numbers of B6 CD4 and CD8 TEs were cotransferred into Thy1.1 congenic recipient mice, the resulting CD4 and CD8 TMs generated in the recipients exhibited different CD4TM/CD8TM ratios in individual tissues. In agreement with the BLI study, scoring individual tissues by calculating the log2 of their CD4TM/CD8TM ratios classified them into “CD8 TM-favored reservoirs” that had negative scores (including PLN, peripheral blood, MLN and SP), “common reservoirs” that had scores close to zero (including BM and lung), and “CD4 TM-favored reservoirs” that had positive scores (including the lamina propria [LP] and PP in gut; Figure 2A top and supplemental Figure 3A). In particular, PLN and PP had the lowest (∼ −1.8) and highest (∼ 3) scores, confirming that they were the most polarized reservoirs for CD8 and CD4 TMs, respectively. Interestingly, a low score (−1.5) for peripheral blood was obtained, showing more CD8 TMs in circulation.
Various types of CD4 and CD8 TMs show a similar polarized tissue distribution. (A) Tissue distribution of CD4 and CD8 TMs of various originalities. Top: CD4 and CD8 TMs (gated as Thy1.2+CD4+ and Thy1.2+CD8+, respectively) generated in the Thy1.1 congenic mice 2 months after transfer of a mix of equal number (10 × 106) of B6 CD4 and CD8 TEs; middle: memory-phenotype (MP) CD4 TMs (gated as CD4+CD25−CD44hi, CD25 staining was included to gate off Tregs) and CD8 TMs (gated as CD8+TCRβ+CD44hi, TCRβ staining was included to gate off the CD8+ nonαβ T cells present in some tissues) spontaneously generated in 1-year-old B6 mice (similar results were observed for B6 mice aged from 3 months to 1 year); and bottom: OVA antigen-specific OT2 TMs (gated as CD4+CD25−TCRVβ5+CD44hi, CD25 staining was included to gate off Tregs) and OT1 TMs (gated as CD8+TCRVβ5+CD44hi) generated in OT2 or OT1 transgenic mice 2 months after infection with 1 × 108 TU DC-directed lentivirus expressing OVA antigen. Data are presented as mean ± SEM (n = 4). (B) Tissue distribution of CD4 and CD8 TCMs and TEMs. Various tissues were harvested from 1-year-old B6 mice and analyzed for the presence of MP CD4 and CD8 TCMs and TEMs (gated as CD4+CD25−CD44hiCD62L+, CD4+CD25−CD44hiCD62L−, CD8+TCRβ+CD44hiCD62L+, or CD8+ TCRβ+CD44hiCD62L−, respectively) using flow cytometry. Histogram plots (top) and quantification of TMs (bottom) are shown. Data are presented as mean ± SEM (n = 4). TCMs indicates central memory T cells; and TEMs, effector memory T cells. (C) Visualization of the CD4 TMs formed in abino B6 mice each receiving 1 × 106 MFG-labeled CD4 TEs that had been differentiated in vitro into TH0, TH1, TH2, or TH17 cells using BLI. Representative BLI images collected 2 months after adoptive transfer are shown (n = 4). Schematic showing the individual tissue localization in mice is provided for reference. (D) Functional analysis of MP CD4 TMs residing in PPs of 1-year-old B6 mice. PP cells (pool of 4 mice) were stimulated with PMA + Ionomycin in vitro for 6 hours and analyzed for the cytokine production of CD4 TMs (gated as CD4+CD44+) using flow cytometry. Contour plots representative of 3 independent experiments are shown.
Various types of CD4 and CD8 TMs show a similar polarized tissue distribution. (A) Tissue distribution of CD4 and CD8 TMs of various originalities. Top: CD4 and CD8 TMs (gated as Thy1.2+CD4+ and Thy1.2+CD8+, respectively) generated in the Thy1.1 congenic mice 2 months after transfer of a mix of equal number (10 × 106) of B6 CD4 and CD8 TEs; middle: memory-phenotype (MP) CD4 TMs (gated as CD4+CD25−CD44hi, CD25 staining was included to gate off Tregs) and CD8 TMs (gated as CD8+TCRβ+CD44hi, TCRβ staining was included to gate off the CD8+ nonαβ T cells present in some tissues) spontaneously generated in 1-year-old B6 mice (similar results were observed for B6 mice aged from 3 months to 1 year); and bottom: OVA antigen-specific OT2 TMs (gated as CD4+CD25−TCRVβ5+CD44hi, CD25 staining was included to gate off Tregs) and OT1 TMs (gated as CD8+TCRVβ5+CD44hi) generated in OT2 or OT1 transgenic mice 2 months after infection with 1 × 108 TU DC-directed lentivirus expressing OVA antigen. Data are presented as mean ± SEM (n = 4). (B) Tissue distribution of CD4 and CD8 TCMs and TEMs. Various tissues were harvested from 1-year-old B6 mice and analyzed for the presence of MP CD4 and CD8 TCMs and TEMs (gated as CD4+CD25−CD44hiCD62L+, CD4+CD25−CD44hiCD62L−, CD8+TCRβ+CD44hiCD62L+, or CD8+ TCRβ+CD44hiCD62L−, respectively) using flow cytometry. Histogram plots (top) and quantification of TMs (bottom) are shown. Data are presented as mean ± SEM (n = 4). TCMs indicates central memory T cells; and TEMs, effector memory T cells. (C) Visualization of the CD4 TMs formed in abino B6 mice each receiving 1 × 106 MFG-labeled CD4 TEs that had been differentiated in vitro into TH0, TH1, TH2, or TH17 cells using BLI. Representative BLI images collected 2 months after adoptive transfer are shown (n = 4). Schematic showing the individual tissue localization in mice is provided for reference. (D) Functional analysis of MP CD4 TMs residing in PPs of 1-year-old B6 mice. PP cells (pool of 4 mice) were stimulated with PMA + Ionomycin in vitro for 6 hours and analyzed for the cytokine production of CD4 TMs (gated as CD4+CD44+) using flow cytometry. Contour plots representative of 3 independent experiments are shown.
We further used this scoring method to study the tissue distribution of the CD4 and CD8 TMs developed endogenously in unperturbed mice using flow cytometry. Both memory-phenotype (MP) TMs spontaneously generated in aged mice10 (Figure 2A middle and supplemental Figure 3B) and the antigen-specific TMs induced in vivo through infecting OT1 and OT2 transgenic mice with a DC-directed lentivirus that expresses OVA antigen26 (Figure 2A bottom and supplemental Figure 3C) showed a similarly polarized tissue distribution of CD4 and CD8 TMs. Importantly, this distinct reservoir distribution is unique for memory CD4 and CD8 T cells, because a similar analysis of naive CD4 and CD8 T cells (TNs) revealed a much different distribution pattern (supplemental Figure 3E), suggesting that a unique homing propensity is acquired during memory formation.
Because TMs are heterogeneous in regard to their phenotype and functionality,7,9,27 we also studied the tissue distribution of TM subtypes. Based on their expression of CD62L and CCR7, both CD4 and CD8 TMs can be divided into TCMs and TEMs.28,29 Despite the classic definition that TCMs home to LNs while TEMs home to peripheral tissues, analysis of MP CD4 and CD8 TEMs (CD62L−) and TCMs (CD62L+) in aged mice revealed the presence of both TEMs and TCMs in all the tissues that we studied, although at various ratios (Figure 2B and supplemental Figure 3F). Of note, in addition to the overall gut-tropic accumulation for CD4 TMs and LN/SP-tropic accumulation of CD8 TMs, there are slightly differed tissue preferences for individual subsets: PPs for CD4 TCMs, LP for CD4 TEMs, PLNs for CD8 TCMs, and SP for CD8 TEMs. CD4 TMs can also be divided into various subtypes based on their differentiated functions.9 Differentiating CD4 TEs in vitro into TH0, TH1, TH2, and TH17 cells followed by adoptively transferring them into recipients resulted in CD4 TMs that all predominantly accumulated in gut (Figure 2C), implying that gut might be the common dominant reservoir for CD4 TMs of various functions.24 This notion is further supported by the observation that in aged mice, the MP CD4 TMs harvested from PPs contained subsets that exhibited cytokine production profiles featured for TH1 (IFN-γ+IL-4−), TH2 (IFN-γ−IL-4+), and TH17 (IFN-γ−IL-17A+; Figure 2D).
CD4 and CD8 TMs differ on their expression of PLN- and gut-homing markers
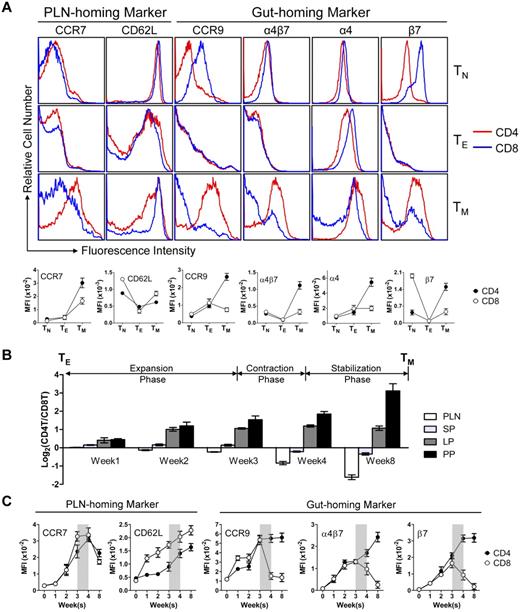
The observation of the extremely polarized gut-tropic versus PLN-tropic accumulation of CD4 and CD8 TMs raised 2 interesting questions: how do the TMs achieve this distinct distribution and what might be the rationale for them to accumulate in these 2 separate tissues? To address the first question, we examined the expression of several tissue-specific homing markers on CD4 and CD8 TMs, which have been well documented to control TM traffic to specific tissues.7,30-32 We were particularly interested in the best characterized PLN-homing markers CCR7 and CD62L,33,34 and the gut-homing markers CCR9 and α4β7.35,36 Examination of the MP TMs in the spleen of aged mice revealed that both TMs expressed similar levels of CCR7 and CD62L, but CD4 TMs expressed much more homogenous and higher levels of CCR9 and α4β7 than CD8 TMs (Figure 3A). The major difference of α4β7 expression stems from the expression of β7 (Figure 3A), its more “gut-specific” component.36 This PLN- and gut-homing marker expression correlates with their gut-tropic and PLN-tropic accumulation, and was unique for CD4 and CD8 TMs, because it was not observed for TNs and TEs (Figure 3A). Further studies of antigen-specific or polyclonal TMs generated through adoptive transfer of in vitro differentiated TEs or Ag-specific TMs generated through DC-directed lentivirus infection all confirmed a PLN- and gut-homing marker expression pattern similar to MP TMs (supplemental Figure 4A).
CD4 and CD8 TMs differ on their expression of gut-homing markers. (A) Homing marker expression on naive (TN), effector (TE), and memory (TM) CD4 and CD8 T cells analyzed by flow cytometry. CD4 and CD8 TNs (gated as CD4+CD25−CD44lo and CD8+ CD44lo, respectively) and TMs (gated as CD4+CD25−CD44hi and CD8+CD44hi, respectively) were naive or memory T cells detected in the spleen of 1-year-old B6 mice. TEs were generated by stimulating B6 spleen T cells in vitro with anti-CD3 and anti-CD28 for 3 days. Representative histogram plots (top) and measurements of mean fluorescence intensity (MFI; mean ± SEM; bottom) are shown (n = 8). (B,C) Time-course tracking of CD4 and CD8 T cells (gated as Thy1.2+CD4+ and Thy1.2+CD8+, respectively) for their tissue homing preference (B) and homing marker expression (C) during their transition from TEs to TMs in Thy1.1 congenic mice receiving adoptive transfer of a mix of equal number (10 × 106) of B6 CD4 and CD8 TEs. (B) Data are presented as mean ± SEM. (C) Representative homing marker expression on spleen T cells detected by flow cytometry, shown as mean fluorescence intensity (MFI; mean ± SEM). N = 4. Shaded area marks contraction phase.
CD4 and CD8 TMs differ on their expression of gut-homing markers. (A) Homing marker expression on naive (TN), effector (TE), and memory (TM) CD4 and CD8 T cells analyzed by flow cytometry. CD4 and CD8 TNs (gated as CD4+CD25−CD44lo and CD8+ CD44lo, respectively) and TMs (gated as CD4+CD25−CD44hi and CD8+CD44hi, respectively) were naive or memory T cells detected in the spleen of 1-year-old B6 mice. TEs were generated by stimulating B6 spleen T cells in vitro with anti-CD3 and anti-CD28 for 3 days. Representative histogram plots (top) and measurements of mean fluorescence intensity (MFI; mean ± SEM; bottom) are shown (n = 8). (B,C) Time-course tracking of CD4 and CD8 T cells (gated as Thy1.2+CD4+ and Thy1.2+CD8+, respectively) for their tissue homing preference (B) and homing marker expression (C) during their transition from TEs to TMs in Thy1.1 congenic mice receiving adoptive transfer of a mix of equal number (10 × 106) of B6 CD4 and CD8 TEs. (B) Data are presented as mean ± SEM. (C) Representative homing marker expression on spleen T cells detected by flow cytometry, shown as mean fluorescence intensity (MFI; mean ± SEM). N = 4. Shaded area marks contraction phase.
In addition, we also analyzed the MP TMs for their expression of other homing markers that have been indicated to play a role in TM trafficking, including CCR4, CCR5, CCR6, CCR10, CXCR3, and CXCR4.30 Different expressions of several markers were observed, indicating their possible roles in regulating the TM reservoir distribution as well (supplemental Figure 4B).
We then tracked the homing marker expression on CD4 and CD8 T cells during their transition from TEs to TMs in recipient mice postadoptive transfer. Both CD4 and CD8 T cells initially expressed all the LN-homing and gut-homing markers in the expansion phase (weeks 1-3; Figure 3C), correlating well with their similar and ubiquitous presence in various tissues at this stage (Figure 3B and supplemental Figure 4C). During the contraction phase (week 4), the critical “turning point” for memory formation, the CD4 T cells continued up-regulating their expression of both LN and gut-homing markers while the CD8 T cells started down-regulating their gut-homing markers (Figure 3C). This change correlated with a sharp increase of CD8 T cells in PLN, in concert with the continuous accumulation of CD4 T cells in the gut, eventually leading to the highly polarized tissue distribution of CD4 and CD8 T cells as they finished the transition into TMs and entered the stabilization phase (Figure 3B and supplemental Figure 4C). Therefore, despite their similar expression of PLN-homing markers, the biased tissue accumulation of CD4 and CD8 TMs seems to correlate with their differentiated expression of gut homing markers, which is gradually acquired as they transit from effector to memory T cells.
CD4 and CD8 TMs selectively require the expression of gut- or PLN-homing marker for their formation and maintenance
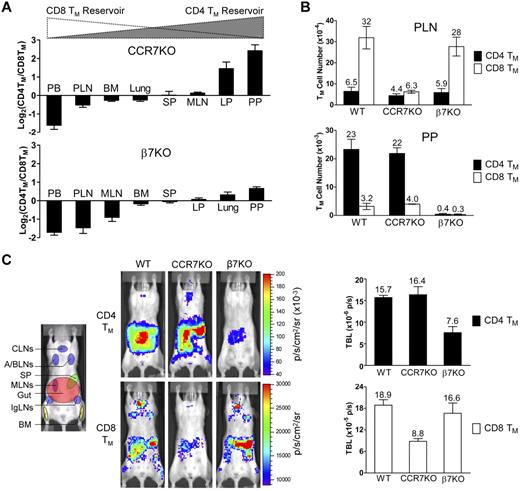
We then sought to determine whether the expression of appropriate gut- or PLN-homing markers is required for the formation and maintenance of CD4 and CD8 TMs. Analysis of mice genetically ablated for a major PLN-homing marker CCR733 revealed that compared with the age-matched wild-type (WT) mice, they had a markedly reduced MP CD8 TM population (especially in the PLNs), but a relatively normal CD4 TM population, shown in both the CD4TM/CD8TM ratio plot (Figure 4A) and the absolute TM counts (Figure 4B). On the contrary, mice deficient for a major gut-homing marker β7 (which results in a deficiency of α4β7)37 had the opposite phenotype of reduced CD4 but relatively normal CD8 TMs (Figure 4A-B). Because both CCR7KO and β7KO mice have other deficiencies in the immune system that may affect their generation of MP TMs in vivo,33,37 we extracted T cells from these mice, generated MFG-labeled CD4 and CD8 TEs in vitro, and then adoptively transferred them into WT recipients. This experimental design allowed us to confine the CCR7 or β7 deficiency to the differentiated TEs. Quantification of the TMs formed in the recipient mice confirmed the previous findings: CCR7 or β7 deficiency selectively impaired the formation of CD8 or CD4 TMs, respectively, but had little effect on the other (Figure 4C). Notably, the surviving CCR7KO CD8 TMs mainly resided in gut but not PLNs, indicating an indispensable role of CCR7 for mediating PLN-tropic accumulation of CD8 TMs. In contrast, the surviving β7KO CD4 TMs still mostly remained in gut, suggesting that gut-tropic accumulation of CD4 TMs was not solely dependent on α4β7 (Figure 4C). Thus, our data imply that CD4 and CD8 TMs selectively require the expression of gut- or PLN-homing markers, in particular α4β7 and CCR7, for their formation and maintenance.
CD4 and CD8 TMs selectively require the expression of α4β7 or CCR7 for their formation and maintenance. (A) Tissue accumulation of MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+ TCRβ+CD44hi, respectively) in 10-month-old CCR7KO or β7KO mice. Data are presented as mean ± SEM (n = 4). (B) Comparison of CD4 and CD8 TM number in PLN and PP of 10-month-old wild-type (WT), CCR7KO and β7KO mice. Data are presented as mean ± SEM (n = 4). (C) Study of the influence of CCR7 or β7 deficiency on CD4 and CD8 TM formation using BLI. Either MFG-labeled CD4 TEs (5 × 106) or CD8 TEs (1 × 106) of the indicated genotype were adoptively transferred into albino B6 recipient mice. Representative BLI images and total body luminescence (TBL) measurements of the indicated recipients (mean ± SEM, n = 4) 1 month after transfer are shown.
CD4 and CD8 TMs selectively require the expression of α4β7 or CCR7 for their formation and maintenance. (A) Tissue accumulation of MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+ TCRβ+CD44hi, respectively) in 10-month-old CCR7KO or β7KO mice. Data are presented as mean ± SEM (n = 4). (B) Comparison of CD4 and CD8 TM number in PLN and PP of 10-month-old wild-type (WT), CCR7KO and β7KO mice. Data are presented as mean ± SEM (n = 4). (C) Study of the influence of CCR7 or β7 deficiency on CD4 and CD8 TM formation using BLI. Either MFG-labeled CD4 TEs (5 × 106) or CD8 TEs (1 × 106) of the indicated genotype were adoptively transferred into albino B6 recipient mice. Representative BLI images and total body luminescence (TBL) measurements of the indicated recipients (mean ± SEM, n = 4) 1 month after transfer are shown.
CD4 and CD8 TMs accumulate in tissues that supply them with their favored homeostatic cytokines
Next, we examined the physiologic relevance of the separate CD4 and CD8 TMs accumulation in gut and PLNs. Considering the notable difference between CD4 and CD8 TMs of their dependence on the homeostatic cytokines IL-7 and IL-15,10 we hypothesized that the distinct tissue distribution of CD4 and CD8 TMs might meet their individual needs. CD4 and CD8 TMs expressed similar level of IL-7 receptor (composed of γc and IL-7Rα chains), while CD8 TMs expressed much higher and more homogenous level of IL-15 receptor (evident by a modestly higher level of IL-15Rα and a significantly higher level of IL-2/15Rβ [CD122]; Figure 5A). This distinction was TM-specific because the differences between CD4 and CD8 TNs and TEs were much smaller (supplemental Figure 5A). Tracking IL-2/15Rβ expression on CD4 and CD8 T cells during their effector to memory transition showed CD8 T cells maintaining a constant high expression of IL-2/15Rβ while CD4 T cells gradually down-regulated its expression (supplemental Figure 5B). Stimulating the purified TMs in vitro with either IL-7 or IL-15 showed that CD8 TMs generally had a proliferation advantage over CD4 TMs, especially when IL-15 was present (Figure 5B). When IL-7 or IL-15 was injected into mice harboring MFG-labeled CD4 or CD8 TMs, we found that IL-7 induced a ∼ 2.7-fold expansion of CD8 TMs and a ∼ 2-fold expansion of CD4 TMs, whereas IL-15 induced a striking ∼ 20-fold expansion of CD8 TMs and only a ∼ 2-fold expansion of CD4 TMs (Figure 5C-D). Examination of the tissue-specific expression of IL-7 and IL-15 revealed a relatively homogenous expression of IL-7 in various tissues with some preference for gut, while IL-15 was expressed at notably higher level in PLN (Figure 5E). This distribution makes PLN a particularly attractive reservoir site for the IL-15–dependent CD8 TMs. Therefore, our results supported a “homing to fitness” hypothesis: IL-15–dependent CD8 TMs tend to accumulate in PLNs, the site particularly rich in IL-15 whereas the IL-7–dependent CD4 TMs tend to accumulate in the gut where they are provided with IL-7 and do not need to compete with CD8 TMs for cytokines.
CD4 and CD8 TMs home to tissues that supply them with their favored homeostatic cytokines. (A) Cytokine receptor expression on the MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+CD44hi, respectively) in the spleen of 1-year-old B6 mice measured by flow cytometry. Representative histogram plots and mean fluorescence intensity (MFI) measurements are shown (n > 4). (B) Proliferation of CD4 and CD8 TMs in response to cytokine stimulation (100 ng/mL) for 3 days in vitro measured by cell counts. TMs were purified from 1-year-old B6 mice. Representative data are presented as mean of triplicates ± SEM (n = 4). (C,D) Proliferation of CD4 and CD8 TMs in response to cytokine stimulation in vivo measured by BLI. MFG-labeled OT2 (CD4) or OT1 (CD8) TEs (1 × 106) were adoptively transferred into each albino B6 recipient mouse. Six months later, the recipients harboring the OT2 or OT1 TMs received administration (IP) of either IL-7 or IL-15 daily (10 μg) for 5 sequential days. (C) Time-course tracking of the TBL change of the indicated recipient mice receiving either IL-7 or IL-15. Data are presented as mean ± SEM. (D) Representative BLI images of the indicated mice right before the 1st cytokine administration (day 1) and 1 day after the last cytokine administration (day 6; n = 3-4). (E) Tissue expression of IL-7 and IL-15 in 1-year-old B6 mice measured by Taqman Q-PCR. Representative data are presented as mean of triplicates ± SEM (n = 4).
CD4 and CD8 TMs home to tissues that supply them with their favored homeostatic cytokines. (A) Cytokine receptor expression on the MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+CD44hi, respectively) in the spleen of 1-year-old B6 mice measured by flow cytometry. Representative histogram plots and mean fluorescence intensity (MFI) measurements are shown (n > 4). (B) Proliferation of CD4 and CD8 TMs in response to cytokine stimulation (100 ng/mL) for 3 days in vitro measured by cell counts. TMs were purified from 1-year-old B6 mice. Representative data are presented as mean of triplicates ± SEM (n = 4). (C,D) Proliferation of CD4 and CD8 TMs in response to cytokine stimulation in vivo measured by BLI. MFG-labeled OT2 (CD4) or OT1 (CD8) TEs (1 × 106) were adoptively transferred into each albino B6 recipient mouse. Six months later, the recipients harboring the OT2 or OT1 TMs received administration (IP) of either IL-7 or IL-15 daily (10 μg) for 5 sequential days. (C) Time-course tracking of the TBL change of the indicated recipient mice receiving either IL-7 or IL-15. Data are presented as mean ± SEM. (D) Representative BLI images of the indicated mice right before the 1st cytokine administration (day 1) and 1 day after the last cytokine administration (day 6; n = 3-4). (E) Tissue expression of IL-7 and IL-15 in 1-year-old B6 mice measured by Taqman Q-PCR. Representative data are presented as mean of triplicates ± SEM (n = 4).
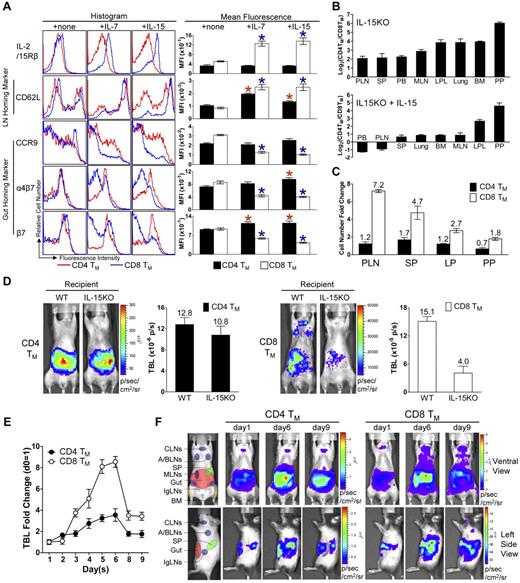
Homeostatic cytokines regulate the differentiated expression of gut-homing markers on CD4 and CD8 TMs in vitro
In addition to their role in maintaining TM homeostasis, we were interested to explore whether homeostatic cytokines might also regulate TM homing molecule expression. To this end, we purified the MP CD4 and CD8 TMs from B6 mice and cultured them in vitro in the presence or absence of IL-7 or IL-15. Without cytokines in the culture, the difference in gut-homing marker expression was almost extinguished (Figure 6A), implying that the differentiation was not fixed for TMs but was rather actively acquired through their constant interaction with external signals, such as homeostatic cytokines. Indeed, their differential marker expression was partly restored when IL-7 was added to the culture medium and, more strikingly, was almost restored to the in vivo level when IL-15 was added (Figure 6A). This regaining of the differentiation worked through reversely up-regulating or down-regulating gut homing markers on CD4 and CD8 TMs (including both CCR9 and α4β7), similarly to what was observed during the TE to TM transition (Figure 3C). This regulation was specific for gut homing markers because the PLN-homing marker CD62L was up-regulated on both CD4 and CD8 TMs in response to either IL-7 or IL-15 stimulation (Figure 6A). Notably, we observed that both IL-7 and IL-15 stimulation specifically promoted the CD8 TMs, but not CD4 TMs, to significantly up-regulate IL-2/15Rβ, thereby providing a positive-feedback control to maintain the different responsiveness to IL-15 between CD4 and CD8 TMs (Figure 6A). Thus, our results indicate that in addition to their capacity to maintain TM survival and proliferation, the homeostatic cytokines, especially IL-15, appears to also regulate the distinct homing of CD4 and CD8 TMs by maintaining their differential expression of gut-homing markers. This regulation seems to stem from an intrinsic difference between CD4 and CD8 TMs as to their response to homeostatic cytokine stimulation.
Homeostatic cytokines regulate the differentiated expression of gut-homing markers on CD4 and CD8 TMs in vitro and in vivo. (A) IL-7 and IL-15 regulation of homing marker expression on MP CD4 and CD8 TMs in vitro. MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi or CD8+CD44hi, respectively) purified from the spleen of 1-year-old B6 mice (pool of 8 mice) were cultured in triplicates in vitro for 3 days in the presence or absence of 100 ng/mL of IL-7 or IL-15. Their expression of homing markers, as well as IL-2/15Rβ (CD122), was measured by flow cytometry. Data are shown as histogram plots and measurements of mean fluorescence intensity (MFI, presented as mean of triplicates ± SEM), and are representative of at least 3 independent experiments. Red or blue *P < .05 (CD4 or CD8 TMs cultured with cytokine compared with CD4 or CD8 TMs cultured without cytokine). (B-C) Quantification of MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+TCRβ+CD44hi, respectively) in 9-month-old IL-15KO mice with or without supplementation of IL-15 (IP 10μg daily for 5 sequential days). (B) Tissue distribution of CD4 and CD8 TMs. (C) The fold changes of CD4 and CD8 TM numbers in response to IL-15 treatment in indicated tissues. Data are shown as mean ± SEM (n = 3). (D) Comparison of CD4 and CD8 TM formation in WT or IL-15KO mice using BLI. MFG-labeled B6 CD4 or CD8 TEs (1 × 106) were adoptively transferred into either WT (B6) or IL-15KO recipient mice. Representative BLI images and the measurements of TBL (shown as mean ± SEM) of the indicated recipients collected 1 month after transfer are shown (n = 3). (E,F) The impact of IL-15 supplement on the homeostasis and homing of CD4 and CD8 TMs studied using BLI. MFG-labeled WT (B6) CD4 or CD8 TEs (5 × 106) were transferred into IL-15KO recipient mice. One month later, the recipients were supplemented with IL-15 (IP 10μg daily for 5 sequential days). (E) Time-course tracking of the fold change of TBL of the indicated recipients. Data are presented as mean ± SEM. (F) Representative BLI images of the indicated recipients on day 1 (right before the 1st IL-15 administration), day 6, and day 9. N = 4. Schematics showing the individual tissue localization in mice (ventral view and left side view) are provided for reference.
Homeostatic cytokines regulate the differentiated expression of gut-homing markers on CD4 and CD8 TMs in vitro and in vivo. (A) IL-7 and IL-15 regulation of homing marker expression on MP CD4 and CD8 TMs in vitro. MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi or CD8+CD44hi, respectively) purified from the spleen of 1-year-old B6 mice (pool of 8 mice) were cultured in triplicates in vitro for 3 days in the presence or absence of 100 ng/mL of IL-7 or IL-15. Their expression of homing markers, as well as IL-2/15Rβ (CD122), was measured by flow cytometry. Data are shown as histogram plots and measurements of mean fluorescence intensity (MFI, presented as mean of triplicates ± SEM), and are representative of at least 3 independent experiments. Red or blue *P < .05 (CD4 or CD8 TMs cultured with cytokine compared with CD4 or CD8 TMs cultured without cytokine). (B-C) Quantification of MP CD4 and CD8 TMs (gated as CD4+CD25−CD44hi and CD8+TCRβ+CD44hi, respectively) in 9-month-old IL-15KO mice with or without supplementation of IL-15 (IP 10μg daily for 5 sequential days). (B) Tissue distribution of CD4 and CD8 TMs. (C) The fold changes of CD4 and CD8 TM numbers in response to IL-15 treatment in indicated tissues. Data are shown as mean ± SEM (n = 3). (D) Comparison of CD4 and CD8 TM formation in WT or IL-15KO mice using BLI. MFG-labeled B6 CD4 or CD8 TEs (1 × 106) were adoptively transferred into either WT (B6) or IL-15KO recipient mice. Representative BLI images and the measurements of TBL (shown as mean ± SEM) of the indicated recipients collected 1 month after transfer are shown (n = 3). (E,F) The impact of IL-15 supplement on the homeostasis and homing of CD4 and CD8 TMs studied using BLI. MFG-labeled WT (B6) CD4 or CD8 TEs (5 × 106) were transferred into IL-15KO recipient mice. One month later, the recipients were supplemented with IL-15 (IP 10μg daily for 5 sequential days). (E) Time-course tracking of the fold change of TBL of the indicated recipients. Data are presented as mean ± SEM. (F) Representative BLI images of the indicated recipients on day 1 (right before the 1st IL-15 administration), day 6, and day 9. N = 4. Schematics showing the individual tissue localization in mice (ventral view and left side view) are provided for reference.
IL-15 plays a major role in regulating CD8 TMs homing to PLNs in vivo
In light of the in vitro study, we further asked whether homeostatic cytokines, in particular IL-15, might regulate TM homing in vivo. First we studied the TM distribution in the IL-15KO mice. Consistent with a previous report,38 we found that compared with WT mice, IL-15KO mice have a much reduced number of MP CD8 TMs, but their CD4 TMs were relatively unaffected. Although there was a general reduction of CD8 TMs in all the tissues that we analyzed, the most significant reduction occurred in the CD8 TM-favored reservoirs, especially in PLNs, resulting in a much less dramatically polarized distribution of the CD4 and CD8 TMs. Supplementing the IL-15KO mice with IL-15 greatly expanded their CD8 TMs. In addition, there was a corrected preference for CD8 TM to accumulate in PLNs, shown as a more significant increase of CD8 TMs in PLNs compared with their increases in the other tissues, which partially restored the polarized tissue distribution of CD4 and CD8 TMs (Figures 6B-C and supplemental Figure 6B). These finding correlated with the observation that in IL-15KO mice, the CD8 TMs expressed high levels of gut-homing markers CCR9 and α4β7 similar to CD4 TMs (supplemental Figure 6A). When IL-15 was supplemented, the difference in gut-homing marker expression was largely restored between CD4 and CD8 TMs, mainly through down-regulating these markers on CD8 TMs (supplemental Figure 6A). Meanwhile, the PLN-homing marker CD62L was up-regulated on CD8 TMs, indicating that the down-regulation was specific for gut-homing markers (supplemental Figure 6A). Moreover, the difference of IL-2/15Rβ expression on CD4 and CD8 TMs was greatly reduced in IL-15KO mice, and was significantly restored on IL-15 supplementation, suggesting that IL-15 might provide a major positive feedback control in vivo to differentiate CD4 and CD8 TMs in their responsiveness to IL-15 (supplemental Figure 6A).
To further study the role of IL-15 as a tissue-restricted factor to regulate CD8 TM homing to PLN, we adoptively transferred MFG-labeled WT CD4 and CD8 TEs into either WT or IL-15KO recipients, and then follow their TM formation in vivo. The results corroborated the previous findings that mice lacking IL-15 did not effectively support CD8 TM formation, especially their homing to PLNs, whereas the maintenance and homing of CD4 TMs was mostly unaffected (Figure 6D). Supplementation with IL-15 induced a significant expansion of CD8 TMs and promoted their homing to the PLNs, while limited expansion was seen for the CD4 TMs (Figure 6E-F). Taken together, our results indicate that IL-15 supplied by the tissues plays a major role in the regulation of CD8 TM homing to PLNs in vivo. This conclusion is further supported by the finding that in WT mice, the MP CD4 and CD8 TMs harvested from PLNs where IL-15 expression is most abundant showed a more dramatic distinction of gut homing marker expression compared with those TMs harvested from SP (supplemental Figure 6C).
Discussion
Despite the detection of CD4 and CD8 TMs in many tissues,1,32,39 a dynamic and systemic study of the localization of CD4 and CD8 TMs has been lacking, probably because of the difficulty of quantifying TMs, in particular CD4 TMs, in tissues outside the dedicated lymphoid organs. Taking advantage of BLI for its capacity to visualize the TMs in a live animal and in its excised organs, and combining it with direct measurements, we have attempted to provide a global picture of CD4 and CD8 TM localization. We find a remarkably segregated tissue distribution of long-term CD4 and CD8 TMs in mice. Therefore in addition to their differences in functionality, anatomic localization is another important distinction between these 2 classes of TMs.
The accumulation of CD8 TMs mainly in the classic lymphoid organs (LNs and SP) was to be expected because they have long been recovered from these sites but the extremely specific accumulation of CD4 TMs in mucosal sites, particularly gut, and their relative paucity in LNs and SP was striking. The finding of gut as a major site for CD4 TM homing is consistent with a previous study using whole body immunohistology to study CD4 TM distribution in mice post protein antigen immunization.39 However, despite the documentation of CD4 TMs being abundant in mucosal sites in mice, nonhuman primates and humans,31,40 it has not been so evident that these sites represent specialized dominant reservoirs for CD4 TMs. What might be the physiologic rationale for the immune system to place these 2 types of TMs in almost complementary positions? Considering the different functions that CD4 and CD8 TMs serve,8,9 it is conceivable that this distribution pattern allows them to together cover the major portals for sensing pathogens that enter the body, potentially providing effective immuno-surveillance. In fact, this distribution may facilitate their specific functional activities: localization of CD8 TMs in draining LNs and SP allows them to react quickly to dendritic cells that sample the invading pathogens in tissues or in the circulation and differentiate into effector cytotoxic T cells (CTLs) that can cleanse infected tissues; localization of CD4 TMs to mucosal sites would allow them to most efficiently respond to pathogens in these sites by both defending against them directly and helping the local B cells to produce antibodies protecting the mucosal surface. It should be noted that certain pathogens take advantage of this TM distribution to aid their infection process. One example is SIV/HIV that primarily targets the CCR5-positive CD4 TMs and uses mucosal sites as its main entry route.40 Massive infection and depletion of CD4 TMs in gut precedes the infection of other tissues, and is the most profound T-cell population abnormality in AIDS.41-43 That gut is the predominant reservoir for CD4 TMs helps to explain the HIV/SIV tissue tropism, emphasizing the importance of providing mucosal protection in the design of therapeutic or vaccination strategies against HIV.
What is the physiologic significance for CD4 and CD8 TMs to separately accumulate in gut or PLNs? TMs depend on homeostatic cytokines for long-term survival, with CD8 TMs heavily reliant on IL-15 while CD4 TMs mainly respond to IL-7.10 Our study of tissue production of these cytokines revealed a relatively homogenous expression of IL-7 among the tissues (albeit slightly higher in gut) but a much higher expression of IL-15 in PLNs. Therefore, it seems that CD8 TMs do benefit from accumulating in PLNs for easy access to IL-15. Meanwhile, the CD4 TMs in gut have access to IL-7 and avoid a direct competition with CD8 TMs for cytokines. Interestingly, commensal microflora have recently been shown to promote intestinal epithelia cells to produce IL-7,44 implying that these microbes may play a role in the maintenance of CD4 TMs and their accumulation at mucosal sites. This notion is supported by our observation that in germ-free mice, in contrast to a relative constant level of memory-phenotype CD8 TMs, there is a significant reduction of CD4 TMs, especially in the gut (supplemental Figure 7). Importantly, we found that homeostatic cytokine stimulation of CD4 and CD8 TMs actively regulates their expression of homing markers, providing a potent feedback control for stabilizing their distinct homing pattern. In particular, IL-15 appears to play the dominant role by greatly down-regulating the expression of gut-homing markers on CD8 TMs. Indeed, depriving or supplementing IL-15 in vivo regulates the capacity of CD8 TMs to home to PLNs. Notably, the expression of IL-15 receptor, particularly its β subunit (IL-2/15Rβ or CD122), is quite different between CD4 and CD8 TMs and is also subjected to cytokine feedback control, strengthening their differentiated needs for IL-15.
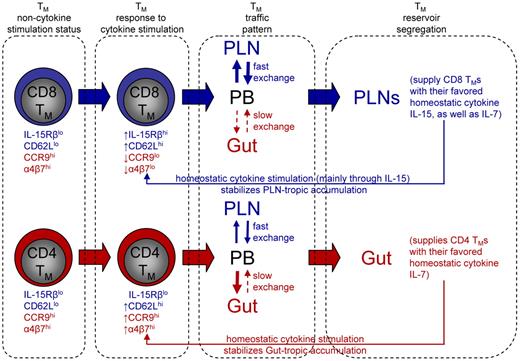
In Figure 7, we bring together all of our observations into a “Memory Compartmentalization Model.” Key to the model is that the memory development process programs CD4 and CD8 TMs into different regulatory pathways so that they respond differently to the hematopoietic cytokines. In the absence of cytokine stimulation, there is little difference between CD4 and CD8 TMs for their homing marker expression (CD62LloCCR9hiα4β7hi). The cytokines up-regulate the PLN-homing marker CD62L on both TMs, but greatly down-regulate the gut-homing markers CCR9 and α4β7 on CD8 TMs (CD62LhiCCR9loα4β7lo) while further up-regulate these markers on CD4 TMs (CD62LhiCCR9hiα4β7hi). This distinction of gut-homing marker expression leaves CD8 TMs to accumulate in PLNs by default. We postulate that CD4 TMs accumulate in gut because of their relative free exchange between PLNs and circulation but a more restricted exchange back to circulation once they enter the gut, as suggested by previous report.45 It is relevant to point out that expression of certain tissue homing marker on TMs allow them to access the corresponding tissue and stay in that tissue for certain time, but by no means making them the permanent residents of that tissue. Instead, all TMs retain their capacity to survey the body through blood and lymphoid circulation. In the PLNs, CD8 TMs find a high level of their favored cytokine IL-15, while in the gut, CD4 TMs find an environment rich in their favored cytokine IL-7 which they can access without competing with CD8 TMs. The cytokine stimulation in turn stabilizes the PLN-tropic and gut-tropic accumulation of CD8 and CD4 TMs through down-regulating or up-regulating their gut-homing markers, respectively. In addition, the cytokine stimulation also maintains the different expression of IL-2/15Rβ on CD4 and CD8 TMs, strengthening their different reliance on IL-15. We should point out that this model mainly addresses TMs that heavily depend on IL-7 and IL-15 for maintenance. It is possible that TMs that are sustained through other forms of regulation may exhibit other homing patterns. For instance, it has been reported that TMs generated from a localized infection in a peripheral tissue, such as skin, gut and lung, tend to be “imprinted” with the capacity to home back to that particular tissue.31,32,46 It will be interesting for future study to characterize the homeostatic regulation of these TMs and their whole-body reservoir distribution.
Memory compartmentalization model. This schematic model illustrates the segregation of CD4 and CD8 TM reservoirs in mice, and the possible molecular controls and physiologic relevance of this phenomenon (for details, see main text).
Memory compartmentalization model. This schematic model illustrates the segregation of CD4 and CD8 TM reservoirs in mice, and the possible molecular controls and physiologic relevance of this phenomenon (for details, see main text).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sarkis Mazmanian and June L. Round for technical advice, and Pin Wang, Dan Kahn, Xin Luo and Shengli Hao for critical reading of this manuscript and insightful discussions.
This work was supported by the Skirball Foundation and National Institutes of Health P01 CA132681A.
National Institutes of Health
Authorship
Contribution: L.Y. designed, performed and analyzed experiments, and wrote the paper; Y.Y., M.K., and T.-W.J.T. performed and analyzed experiments; and D.B. analyzed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.-W.J.T. is Boehringer Ingelheim Pharmaceuticals Inc.
Correspondence: Lili Yang, Division of Biology, M/C 147-75, California Institute of Technology, 1200 E California Blvd, Pasadena, CA, 91125; e-mail: liyang@caltech.edu; or David Baltimore, Division of Biology, M/C 147-75, California Institute of Technology, 1200 E California Blvd, Pasadena, CA 91125; e-mail: baltimo@caltech.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal