Abstract
We examined 807 productive IGHV-IGHD-IGHJ gene rearrangements from mantle cell lymphoma (MCL) cases, by far the largest series to date. The IGHV gene repertoire was remarkably biased, with IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23 accounting for 46.3% of the cohort. Eighty-four of 807 (10.4%) cases, mainly using the IGHV3-21 and IGHV4-34 genes, were found to bear stereotyped heavy complementarity-determining region 3 (VH CDR3) sequences and were placed in 38 clusters. Notably, the MCL stereotypes were distinct from those reported for chronic lymphocytic leukemia. Based on somatic hypermutation (SHM) status, 238/807 sequences (29.5%) carried IGHV genes with 100% germ line identity; the remainder (569/807; 70.5%) exhibited different SHM impact, ranging from minimal (in most cases) to pronounced. Shared replacement mutations across the IGHV gene were identified for certain subgroups, especially those using IGHV3-21, IGHV1-8, and IGHV3-23. Comparison with other entities, in particular CLL, revealed that several of these mutations were “MCL-biased.” In conclusion, MCL is characterized by a highly restricted immunoglobulin gene repertoire with stereotyped VH CDR3s and very precise SHM targeting, strongly implying a role for antigen-driven selection of the clonogenic progenitors. Hence, an antigen-driven origin of MCL could be envisaged, at least for subsets of cases.
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell malignancy that represents 5%-10% of non-Hodgkin lymphomas. The median overall survival is 3-5 years, and at present, conventional treatment is not curative and long-term remission is rare. However, subsets of MCL patients follow a more indolent clinical course without disease progression for a relatively long period.1
The cytogenetic hallmark of MCL is the chromosomal translocation t(11;14)(q13;q32). This aberration leads to the juxtaposition of the CCDN1 locus on chromosome 11 to the immunoglobulin heavy chain (IGH) locus on chromosome 14. As a consequence, cyclin D1 is constitutively overexpressed, causing gross cell cycle deregulation.2 In addition to the primary t(11;14), several secondary genomic aberrations can be identified in most MCL cases,2,3 supporting early findings that cyclin D1 overexpression must be accompanied by other genetic abnormalities to promote lymphomagenesis.2,4 Various gene expression changes that may cooperate with cyclin D1 deregulation also have been described in MCL, mainly involved in DNA repair pathways and the control of cell cycle and apoptosis.4,5 That notwithstanding, alterations in the local tumor microenvironment also may play an important role in regulating the growth and survival of MCL neoplastic cells.2,5,6
In B-cell malignancies, immunogenetic analysis of the clonogenic B-cell receptors (BcRs) offers valuable insight into both the ontogenetic derivation and the possible involvement of antigen selection. In particular, a biased repertoire of immunoglobulin heavy variable (IGHV) genes is generally considered as evidence for the involvement of a limited set of antigens, superantigens, or both in lymphoma development.7,8 In addition, many B-cell lymphomas exhibit somatic hypermutation (SHM) patterns in IGHV genes, typical of antigen receptors that have undergone selection by antigen.9,10 For certain lymphomas, most notably chronic lymphocytic leukemia (CLL), the involvement of antigens in lymphomagenesis is further supported by the identification of closely homologous antigen binding sites between unrelated cases (“stereotyped” BcRs).11
Studies of the immunoglobulin (IG) gene repertoire in MCL have demonstrated biases, with a preferential use of the IGHV4-34, IGHV3-21, and IGHV3-23 genes. However, their relative frequencies differed between studies, probably because of relatively small cohort sizes (the largest available study included 141 cases).12-17 Moreover, there is evidence suggesting potential associations with molecular and clinical features for cases expressing certain IGHV genes (eg, IGHV3-21 and IGHV3-23).12,13,16-18
All relevant studies consistently showed that in the majority of MCL cases the clonotypic IGHV genes were either unmutated or exhibited a low impact of SHM activity. Based on this and other observations, the cellular origin of MCL was traced to a stage of B-cell development before the transition through the germinal center.13,14 This view was adopted by the 2008 World Health Organization (WHO) classification that considers the postulated normal counterpart of MCL B cell as “a peripheral B cell of the inner mantle zone, mostly of naive pre-germinal center type.”19p232
Against this, virtually all aforementioned studies have reported the existence of a subset of MCL patients with mutated IGHV genes, variably accounting for from 16%-38% of cases.12-18 This poses a conundrum, given the prevailing views about MCL ontogeny. In all studies, assignment to the “mutated” IGHV subset followed the 2% cut-off value for deviation from the closest IGHV germ line gene, which is widely used for prognostication in CLL.20 Accordingly, IGHV-mutated status was appointed to rearrangements only when the germ line identity (GI) was < 98%, thereby giving little if any attention to cases with IGHV genes carrying a few mutations, leading to > 98% but < 100% GI. From a biologic perspective, this may be an oversimplification, in view of ample evidence from normal, autoreactive and malignant B-cell clones where even a low mutational “burden” can be functionally relevant.9,21,22
Here, we performed a detailed immunogenetic analysis of the IG receptors from 807 cases with MCL, by far the largest series to date. Our aim was to obtain a comprehensive view of the IG gene repertoire, with a special focus on SHM targeting and the configuration of the antigen-binding site. We report that MCL is characterized by a highly selective IG gene repertoire and very precise SHM targeting. We also document for the first time the existence of subsets of cases with stereotyped BcRs and disease-biased molecular features, distinct from those previously reported in other B-cell malignancies, especially CLL. On this evidence, we propose an antigen-driven origin of MCL, at least for certain subsets of cases.
Methods
Patient group
In total, 807 patients with a diagnosis of MCL from collaborating institutions in France (n = 132), Germany (n = 270), Greece (n = 56), Italy (n = 22), the Nordic countries (Denmark, Finland, Norway, and Sweden; n = 266), Spain (n = 39), and The Netherlands (n = 22) were included in the study. The diagnosis was established according to the 2008 World Health Organization classification criteria.19 The study was approved by the local ethics review committee of each institution.
PCR amplification of IGHV-IGHD-IGHJ rearrangements
PCR amplification of IGHV-IGHD-IGHJ gene rearrangements was performed on either genomic DNA (gDNA) or complementary DNA (cDNA), extracted mainly from fresh blood and bone marrow aspirates, but also from fresh and formalin-fixed, paraffin-embedded solid tissue specimens, including lymph nodes, bone marrow biopsies, spleen, orbit, and colon.
In fresh or fresh-frozen samples, PCR amplification of IGHV-IGHD-IGHJ rearrangements was performed using IGHV leader primers or consensus primers for the IGHV framework region (FR) 1 region along with appropriate IGHJ gene primers, as described previously9,12,15-17 or following the BIOMED-2 protocol.23 In formalin-fixed, paraffin-embedded material, deparaffinization was done according to standard methods, and gDNA was extracted using the QIAamp tissue kit (QIAGEN) following the manufacturer's recommendations. PCR amplification of IGHV-IGHD-IGHJ rearrangements was performed with a seminested approach, using the same IGHV FR1 or, in 61/807 cases (7.6% of the cohort), IGHV FR2 5′ consensus primers in both amplification rounds along with 2 different IGHJ 3′ consensus primers, of which that used in the second round was more internal.
Sequence analysis and interpretation
PCR amplicons were subjected to direct sequencing on both strands. Sequence data were analyzed using the ImMunoGeneTics (IMGT) databases24,25 and the IMGT/V-QUEST tool (http://www.imgt.org).26 Codons and amino acid positions are according to the IMGT unique numbering for V domain.27 To avoid misidentification of mutations when IGHV FR1 or FR2 consensus primers were used in the amplification reactions, nucleotide substitutions in the obtained sequences were evaluated from codon 27 in CDR1-IMGT or, in those sequences obtained with a VH FR2 primer, codon 56 in CDR2-IMGT, respectively. The downstream end of the analyzed V region corresponds to the 5′ end of the germ line CDR3-IMGT as defined by IMGT/JunctionAnalysis.28
Data mining
Output data from IMGT/V-QUEST for all productive IGHV-IGHD-IGHJ rearrangements were parsed, reorganized, and exported to a spreadsheet through the use of computer programming with the Perl programming language. The following information was extracted: (1) IGHV gene use, percentage of identity to the closest germ line gene and VH CDR3 length; and (2) SHM characteristics. Each nucleotide mutation in every sequence was recorded, as was the change or preservation of the corresponding amino acid, identified as replacement (R) or silent (S), respectively. Amino acids were grouped according to standardized biochemical criteria and based on physicochemical properties (eg, hydropathy, volume, chemical characteristics), as described previously.29 To account for the fact that a mutation is more likely to occur in a VH FR than a VH CDR simply because of greater length, each mutation was “weighted,” or normalized, as recently reported by our group.9 We extracted additional information on all amino acid changes codon by codon and examined whether the somatically introduced amino acid belonged to the same physicochemical class as the mutating amino acid (“conservative” change) or not (“nonconservative” change).29,30
Identification of stereotyped rearrangements based on sequence pattern discovery
To comprehensively identify possible restrictions in the VH CDR3 amino acid composition of rearrangements using the same IGHV gene, leading to an overall homology between different IGHV-IGHD-IGHJ rearrangements, we used the pattern-based method described previously in CLL.11 For the present analysis, more stringent criteria, on top of requiring at least 50% amino acid identity and 70% similarity between stereotyped sequences, were applied, including use of the same IGHV gene, identical VH CDR3 length, and identical offset of the identified pattern within VH CDR3 sequences of unrelated rearrangements.
The identification of shared patterns led to the clustering of MCL rearrangements on a first (ground) level or level 0. VH CDR3 sequences can occur in more than 1 ground level cluster, highlighting complex relationships that were used for further clustering at higher levels.
Statistical analysis
Descriptive statistics for discrete parameters included counts and frequency distributions. For quantitative variables, statistical measures included means, medians, standard deviation, and minimum–maximum values. Significance of bivariate relationships between variables was assessed with the use of χ2 and t tests. For all comparisons, a significance level of P = .05 was set, and all statistical analyses were performed with the statistical package SPSS Version 12.0 (SPSS).
Results
IG gene repertoires
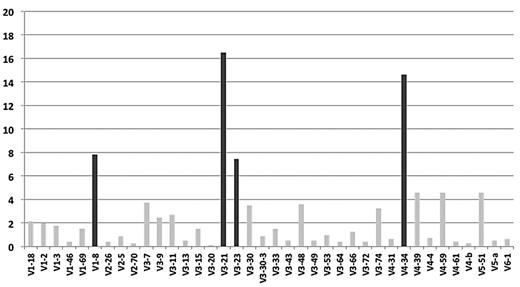
In total, 807 productive IGHV-IGHD-IGHJ rearrangements were amplified. IGHV gene repertoire analysis showed that IGHV3 was the predominant subgroup (416/807; 51.6%), followed by IGHV4 (208/807; 25.8%; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Thirty-eight functional IGHV genes were identified (supplemental Table 2), of which only 4 (IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23 in 133, 118, 63, and 60 cases, respectively) accounted for 46.3% of the series (Figure 1).
IGHV gene repertoire biases in MCL. The IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23 genes (highlighted in black) collectively account for 46% of the cohort.
IGHV gene repertoire biases in MCL. The IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23 genes (highlighted in black) collectively account for 46% of the cohort.
Significant differences were identified regarding the frequencies of individual IGHV genes comparing our MCL dataset to (1) normal B cells (a recently reported extensive dataset of IG sequences from transitional, naive, IgM memory, and switched B cells)31 or (2) the repertoires of other B-cell malignancies, namely, CLL and splenic marginal zone lymphoma (SMZL; taking advantage of the large, well-characterized CLL and SMZL IG sequence datasets previously analyzed by our group).32,33 In particular, (1) the IGHV3-21, IGHV4-34, and IGHV1-8 genes were significantly (P < .01) overrepresented in MCL versus any type of normal B cells, including naive B cells, whereas the IGHV3-23 gene was frequent in all settings; and (2) the IGHV3-21 and IGHV1-8 genes were significantly (P < .01) overrepresented in MCL versus either CLL or SMZL (supplemental Table 3A-B).
IGHD genes were identified in 803/807 junctions; IGHD3 and IGHD6 subgroup genes predominated (275 and 163 cases, respectively). Twenty-five IGHD genes were identified (supplemental Table 4), of which IGHD3-3 was the most frequent (81/803 cases; 10.1%). Regarding IGHJ gene use, the majority of cases used the IGHJ4 (353/807 cases; 43.7%) and IGHJ6 (239/807; 29.6%) genes (supplemental Table 5).
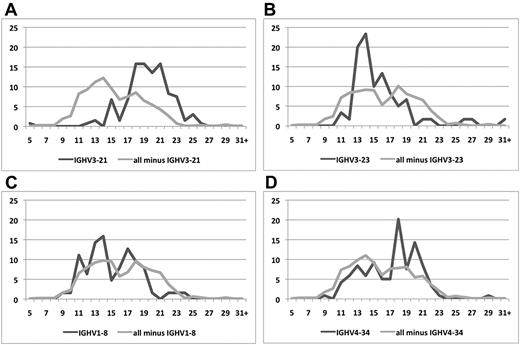
The median VH CDR3 length was 16 amino acids (range, 5-35). Focusing on rearrangements of the 4 predominant IGHV genes, IGHV3-21 and IGHV4-34 cases had significantly longer median VH CDR3 lengths (20 and 18 amino acids, respectively) than IGHV3-23 and IGHV1-8 rearrangements (15 and 14 amino acids, respectively; P < .01; supplemental Table 6; Figure 2).
Different distributions of VH CDR3 lengths in subgroups of MCL clones using different IGHV genes.IGHV3-21 (A), IGHV3-23 (B), IGHV1-8 (C), and IGHV4-34 (D) rearrangements against all other rearrangements. Skewing to longer VH CDR3s is observed among IGHV3-21 and IGHV4-34 rearrangements, contrasting IGHV3-23 and IGHV1-8 rearrangements that carry significantly shorter VH CDR3s.
Different distributions of VH CDR3 lengths in subgroups of MCL clones using different IGHV genes.IGHV3-21 (A), IGHV3-23 (B), IGHV1-8 (C), and IGHV4-34 (D) rearrangements against all other rearrangements. Skewing to longer VH CDR3s is observed among IGHV3-21 and IGHV4-34 rearrangements, contrasting IGHV3-23 and IGHV1-8 rearrangements that carry significantly shorter VH CDR3s.
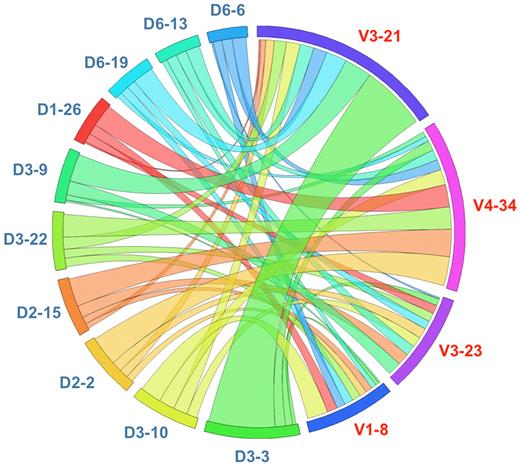
Moreover, IGHV3-21 rearrangements were strongly biased to the use of the IGHD3-3 gene (38/133 cases; 28.6%); less pronounced biases were seen for IGHV1-8 (recombined to the IGHD3-10 gene in 19% of cases) and IGHV4-34 rearrangements (recombined to either the IGHD2-2 or the IGHD2-15 gene in 14.4% and 13.6% of cases, respectively), whereas no such bias was noted for IGHV3-23 rearrangements (Figure 3).
Circular layout depicting the associations of selected IGHV and IGHD genes in MCL. The Circos software package (http://mkweb.bcgsc.ca/circos) was used to explore the combinations of the 4 predominant IGHV genes with the 10 more frequent IGHD genes. Strong biases are evident, as exemplified by the restricted pairing of IGHV3-21/IGHD3-3, IGHV1-8/IGHD3-10, and IGHV4-34 with IGHD2-2 and IGHD2-15.
Circular layout depicting the associations of selected IGHV and IGHD genes in MCL. The Circos software package (http://mkweb.bcgsc.ca/circos) was used to explore the combinations of the 4 predominant IGHV genes with the 10 more frequent IGHD genes. Strong biases are evident, as exemplified by the restricted pairing of IGHV3-21/IGHD3-3, IGHV1-8/IGHD3-10, and IGHV4-34 with IGHD2-2 and IGHD2-15.
Differential patterns of associations of certain IGHV genes with IGHJ genes also were identified. In fact, 86/133 (64.7%) IGHV3-21 rearrangements used the IGHJ6 gene, compared with only 4/60 (6.7%) IGHV3-23 rearrangements (P < .001). On the contrary, 75% of IGHV3-23 rearrangements used the IGHJ4 gene versus 16.5% of IGHV3-21 rearrangements (P < .01). Finally, IGHV4-34 rearrangements used the IGHJ4, IGHJ5, and IGHJ6 genes at roughly identical frequencies (∼ 30%; supplemental Table 7).
SHM analysis
Following the 98% GI cut-off value, which has been adopted in several MCL studies, 186/807 sequences (23%) from our series were defined as mutated, whereas the remainder (621/807 sequences; 77%) had “unmutated” IGHV genes. However, as outlined in the “Introduction,” this strategy (irrespective of the actual percentage) has questionable relevance for purposes other than prognostication in CLL.
We therefore adopted a different approach and considered sequences with no SHM separately from cases with even a single mutation. By this approach, 238/807 sequences (29.5%) could be assigned to a “truly unmutated” subgroup (100% GI), whereas the remaining sequences (569/807; 70.5%) exhibited some impact of SHM activity, ranging from minimal to pronounced. Mutated cases were further subdivided according to their mutational “load” into successive bins of 1% difference from the closest germ line gene (supplemental Table 8). For statistical comparisons, sequences with 97% to 99.9% GI were classified as “minimally/borderline mutated” (n = 458/807; 56.7%), whereas those with < 97% GI as “highly mutated” (n = 111; 13.8%). The 97% cut-off was an educated choice and although still essentially arbitrary, it does not affect our conclusions because our critical differentiation to previous approaches is the “isolation” of cases with no mutations.
The IGHV gene repertoires of the 3 subgroups described (truly unmutated, minimally/borderline mutated, and highly mutated) were significantly different (supplemental Table 9). For example, the IGHV3-21 gene was used by 4.4% of rearrangements with < 97% GI versus 22.7% of rearrangements with 100% GI (P < .01). In sharp contrast, the IGHV3-23 gene was overrepresented among rearrangements with < 97% GI (20.2%), whereas only 4.2% of truly unmutated rearrangements used this gene (P < .01).
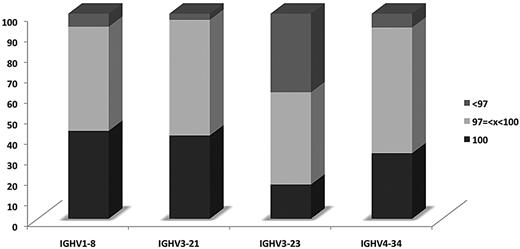
Significant differences also were identified regarding the “propensity” of certain IGHV genes to acquire SHMs when used in MCL IG receptors (supplemental Table 10). In particular, the IGHV3-23 gene had the highest proportion of highly mutated rearrangements (36.7%); in contrast, only a few (range, 3%-6.4%) IGHV1-8, IGHV3-21, and IGHV4-34 rearrangements were highly mutated (Figure 4).
Differential impact of SHM among rearrangements of different IGHV genes. Distribution of rearrangements of the 4 most frequent IGHV genes of the present series according to SHM status.
Differential impact of SHM among rearrangements of different IGHV genes. Distribution of rearrangements of the 4 most frequent IGHV genes of the present series according to SHM status.
Given the distinctive molecular features of IGHV-IGHD-IGHJ rearrangements assigned to each mutational subgroup described here, we next explored whether these features are specific to MCL. To this end, we performed comparisons to CLL and SMZL and identified that the minimally/borderline mutated status is significantly more common in MCL versus either CLL or SMZL (P < .001 for both comparisons). In addition, the IGHV gene repertoires of the mutational subgroups differed significantly between the 3 entities, indicating that the findings reported here are “MCL-specific” (supplemental Table 11A-B).
Characteristics and topology of SHM
Analysis of the molecular characteristics and distribution of SHMs was performed for all 569 IGHV-IGHD-IGHJ sequences of the present series with < 100% identity to the closest germ line gene. Overall, point mutations predominated by far, with only 5 sequence changes consistent with nucleotide duplications/insertions identified in 5 different rearrangements; all these changes occurred as multiples of 3 base pairs, therefore maintaining the original reading frame.
At cohort level, of all point mutations analyzed, transitions predominated (1545/2622 mutations; 58.9%), in keeping with a canonical SHM process.34 However, at the level of individual IGHV genes, distinctive targeting patterns were identified. For example, a significantly increased targeting of adenine (A) nucleotides was noted in IGHV3-21 rearrangements versus rearrangements using other IGHV3 subgroup genes (P < .05); results of similar comparisons for the other predominant IGHV genes of the present study are given in supplemental Table 12.
Replacement-to-silent (R/S) mutation ratios were calculated for all cases with < 100% GI; the overall R/S ratios for FRs and CDRs were 1.75 and 3.69, respectively (supplemental Table 13). In view of the IGHV gene repertoire differences between minimally/borderline mutated and highly mutated cases reported here, SHM targeting also was investigated independently within each mutational subgroup, focusing on rearrangements using the same IGHV gene (supplemental Table 14). Following this approach, significant differences were identified among minimally/borderline mutated cases. For example, IGHV3-21 rearrangements were highly targeted for R mutations in VH CDR1 and VH CDR2, whereas IGHV3-23 rearrangements exhibited a more even distribution of R and S mutations over VH FRs and VH CDRs.
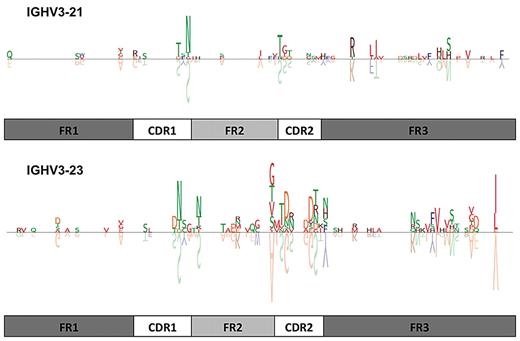
Recurrent amino acid changes (ie, the same amino acid replacement at the same position of the VH domain in different sequences) were found mainly among rearrangements using the IGHV3-21 and IGHV3-23 genes (Figure 5). In particular, in the IGHV3-21 group, the substitution of S for N at VH CDR1 codon 38 occurred in 15/78 (19.2%) cases; in the IGHV3-23 group, the amino acid changes S-to-N at VH CDR1 codon 36, A-to-G at VH FR2 codon 55, and V-to-I at VH FR3 codon 101 were detected in 12/48 (25%), 11/48 (22.9%), and 12/48 (25%) cases, respectively. A summary list of recurrent amino acid substitutions for rearrangements of the 4 predominant IGHV genes of the present cohort is given in supplemental Table 15A; the actual aligned sequences are listed in supplemental Table 15B.
Amino acid sequence logos of IGHV3-21 and IGHV3-23 rearrangements with less than 100% GI. Seventy-eight IGHV3-21 and 48 IGHV3-23 rearrangements are depicted. The letters above the line represent the amino acid changes, whereas the letters shown upside-down below the line represent the corresponding germ line amino acids of the IGHV gene. The size of the amino acid symbol represents the relative frequency of that amino acid at that position relative to all other mutations at that position in the certain IGHV group of sequences. Blank spaces represent amino acids that are unchanged in comparison to the germ line sequence. Amino acids are colored based on their similarity in terms of their physicochemical properties, as described previously. For example, in the IGHV3-21 group, the substitution of S for N at VH CDR1 codon 38 occurred in 15/78 (19.2%) cases, whereas in the IGHV3-23 group, the amino acid changes S-to-N at VH CDR1 codon 36 and V-to-I at VH FR3 codon 101 were detected in 12/48 (25%) cases each, respectively. Additional detailed information is provided in supplemental Table 12A and B.
Amino acid sequence logos of IGHV3-21 and IGHV3-23 rearrangements with less than 100% GI. Seventy-eight IGHV3-21 and 48 IGHV3-23 rearrangements are depicted. The letters above the line represent the amino acid changes, whereas the letters shown upside-down below the line represent the corresponding germ line amino acids of the IGHV gene. The size of the amino acid symbol represents the relative frequency of that amino acid at that position relative to all other mutations at that position in the certain IGHV group of sequences. Blank spaces represent amino acids that are unchanged in comparison to the germ line sequence. Amino acids are colored based on their similarity in terms of their physicochemical properties, as described previously. For example, in the IGHV3-21 group, the substitution of S for N at VH CDR1 codon 38 occurred in 15/78 (19.2%) cases, whereas in the IGHV3-23 group, the amino acid changes S-to-N at VH CDR1 codon 36 and V-to-I at VH FR3 codon 101 were detected in 12/48 (25%) cases each, respectively. Additional detailed information is provided in supplemental Table 12A and B.
Pattern discovery in VH CDR3 sequences
Based on shared VH CDR3 sequence patterns and following strict criteria as outlined in “Identification of stereotyped rearrangements based on sequence pattern discovery,” 84/807 (10.4%) sequences were placed in 38 clusters at the ground level (level 0), with 2 up to 7 sequences each (supplemental Table 16).
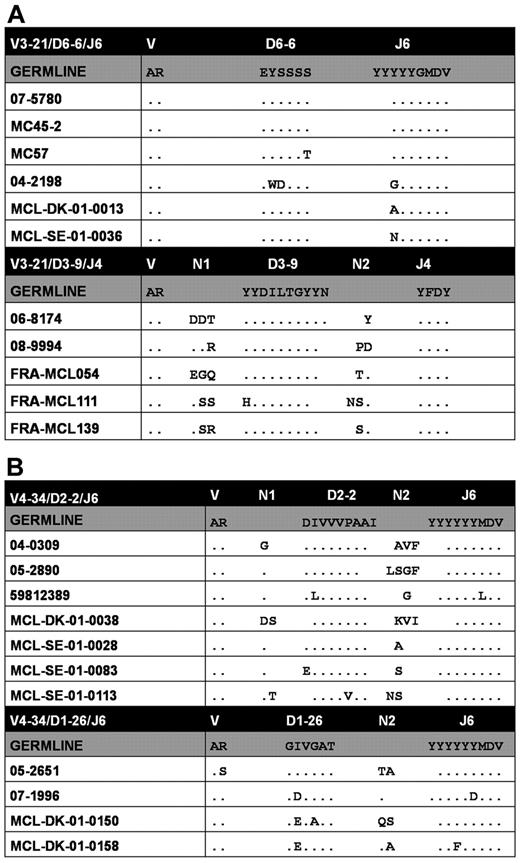
Level 0 clusters were characterized by high VH CDR3 amino acid identity and similarity stemming from the use of identical IGHD and IGHJ genes and, for some clusters, also extending to the presence of shared junctional residues (Figure 6). The common sequences between level 0 clusters led to their grouping in clusters at 2 progressively higher levels of hierarchy (levels 1-2; supplemental Table 16B); the highest level included a single cluster of 7 cases, all using the IGHV4-34 gene. Notably, analysis of the paired IG light chains in the largest level 0 cluster identified here (cluster 3; Figure 6A top panel) revealed restricted use of IGLV3-19/IGLJ2 rearrangements with stereotyped λ complementarity-determining region 3 (VL CDR3) sequences (supplemental Table 16A).
Stereotyped VH CDR3s in MCL: level 0 clusters of rearrangements with shared VH amino acid motifs. (A) Stereotyped IGHV3-21 rearrangements. (B) Stereotyped IGHV4-34 rearrangements. A detailed list of level 0 clusters is given in supplemental Table 13A. Dots represent identities.
Stereotyped VH CDR3s in MCL: level 0 clusters of rearrangements with shared VH amino acid motifs. (A) Stereotyped IGHV3-21 rearrangements. (B) Stereotyped IGHV4-34 rearrangements. A detailed list of level 0 clusters is given in supplemental Table 13A. Dots represent identities.
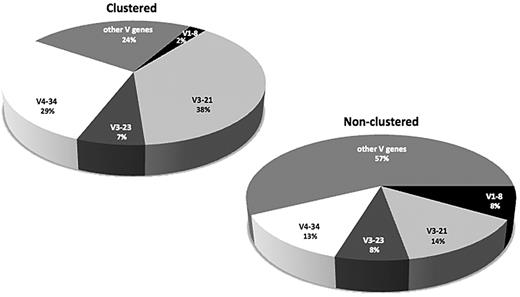
The IGHV gene repertoire differed significantly between clustered versus nonclustered cases (Figure 7). The most striking case concerned the IGHV3-21 and IGHV4-34 genes, which accounted for 67% of clustered cases versus only 27% of nonclustered cases (P < .01). Less pronounced but still statistically significant was the suppression of IGHV1-8 gene use among clustered cases (2.4% vs 8.5% in the nonclustered group; P < .01; supplemental Table 17).
In MCL, VH CDR3 stereotypy is essentially a property of IGHV3-21 and IGHV4-34 rearrangements. Differences in the IGHV gene repertoire between clustered versus nonclustered rearrangements. As the graph clearly shows, the relative frequencies of the IGHV3-21 and IGHV4-34 genes are increased considerably among clustered rearrangements; in contrast, the IGHV1-8 gene use is significantly suppressed within this group.
In MCL, VH CDR3 stereotypy is essentially a property of IGHV3-21 and IGHV4-34 rearrangements. Differences in the IGHV gene repertoire between clustered versus nonclustered rearrangements. As the graph clearly shows, the relative frequencies of the IGHV3-21 and IGHV4-34 genes are increased considerably among clustered rearrangements; in contrast, the IGHV1-8 gene use is significantly suppressed within this group.
Discussion
Despite significant recent advances, the ontogenetic puzzle of MCL is still missing many vital pieces. Here, we approached this issue from an immunogenetic perspective, on the belief (justified in other settings, most notably CLL) that the study of the clonotypic IG receptors may offer important ontogenetic clues and evidence for interactions of the malignant clones with (micro)environmental elements, implying a role for antigen in MCL development. The large size of the present series and the application of purpose-built bioinformatics tools provided new insight with important implications about the ontogenesis of MCL.
Our results confirm and significantly extend previous observations that the IGHV gene repertoire in MCL is remarkably biased,12-18 with only 4 genes (IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23) collectively accounting for almost half of the cohort, indicating the operation of selective forces shaping the IG gene repertoire. By extension, these findings also imply that a limited set of antigens, superantigens, or both may be specifically involved in MCL development. Additional evidence in support of this notion is provided by the comparison of the IG gene repertoire in MCL versus either normal B cells,31 including naive B cells, or other B-cell malignancies, namely, CLL32 and SMZL.33 This comparative assessment revealed significantly different profiles, especially regarding the use of the IGHV3-21 and IGHV1-8 genes, alluding to distinct immune pathways to lymphoma development probably through selection by different antigenic stimuli.
Immunogenetic evidence against the concept of MCL as a malignancy of exclusively naive pregerminal center B cells was also available from reports on the existence of a minor subset of MCL patients with mutated IGHV genes.12-18 In all published series, the classification of MCL cases based on SHM status was based on the 2% identity cut-off from the closest IGHV germ line gene, following the example of CLL, where this cut-off proved to be an accurate prognosticator of patient survival.20 This approach should be viewed with caution for the following reasons.
First, no solid correlations have yet been found between IGHV gene mutational status and patient survival in MCL.12,13,17,18 Although certainly of interest, we refrained from exploring potential clinical correlations given the retrospective character of our study and also taking into account that patients were not treated uniformly. Second, perhaps more importantly, on the evidence reported here, the majority of MCL cases indeed carry some level of SHM, which also seems to be distributed in a “disease-biased” manner. This important feature was overlooked in previous studies by the use of the 2% identity cut-off, which is biologically questionable and also creates an oxymoron, in that all sequences above the cut-off are considered as unmutated even when carrying one or more mutations leading to < 100% identity to the closest germ line gene. Thus, our decision to consider as mutated all cases with any type and number of IGHV gene sequence changes enabled us to obtain a far more accurate view of SHM in MCL.
In particular, a major finding of the present study is that distinct MCL subgroups of different mutational load (truly unmutated, minimally/borderline mutated, and highly mutated) display markedly different IG molecular features, indicating distinct antigen selection processes, probably responsible for even single mutations. Focusing on rearrangements expressing the 4 predominant IGHV genes in MCL (IGHV3-21, IGHV4-34, IGHV1-8, and IGHV3-23), significant differences were identified regarding their use in the 3 major mutational subgroups, strongly suggesting that the impact of SHM in MCL is related to the expression of certain VH domain specificities.
Because gene-specific patterns of SHM load have been reported in other B-cell lymphomas, and also to explore whether the findings reported here are disease-biased, we compared the IGHV-IGHD-IGHJ gene rearrangements from MCL to rearrangements of similar mutational status from CLL32 or SMZL.33 We were able to document important differences, again highlighting the unique features of the MCL IG repertoire, especially for cases assigned to the minimally/borderline mutated and truly unmutated subgroups. Prompted by these findings, we suggest that the paucity or lack of SHM in a significant proportion of MCL cases need not necessarily imply “antigen naivety” but perhaps functional selection for the preservation of the IG gene sequences in germ line or near-germ line composition, at least in a proportion of cases.
Evidence for functionally driven interactions leading to very precise and probably functionally driven SHM was obtained by the finding of recurrent amino acid changes across the entire VH domain among rearrangements using certain IGHV genes, in particular, IGHV3-21, IGHV3-23, and IGHV4-34. Recurrent amino acid changes were evident even among minimally/borderline mutated rearrangements (especially of the IGHV3-21 and IGHV4-34 genes), indicating their importance in providing malignant cells with some clonal advantage.
VH CDR3 length and amino acid composition are well-established contributors to antigen recognition.35,36 Our detailed analysis of VH CDR3 configuration in MCL supports a role of antigenic stimulation in the selection of MCL progenitors or the malignant cells themselves. We first noted different VH CDR3 length distribution patterns according to SHM status. In particular, remarkably long VH CDR3s featured on IGHV3-21 and IGHV4-34 rearrangements, which predominated in the minimally/borderline mutated and truly unmutated subgroups. In keeping with what has been reported in other settings, these length restrictions might be related to a proper positioning of critical residues under functional constraint for recognition of certain (perhaps, shared) antigenic determinants.35,37
Additional evidence for this argument is provided by our finding that a sizeable fraction of MCL cases can be assigned to clusters defined by identical IGHV gene use and restricted (stereotyped) VH CDR3 motifs. In fact, through the application of purpose-built bioinformatics tools, we were able to document for the first time stereotyped IGHV-IGHD-IGHJ gene rearrangements in MCL, collectively accounting for ∼ 10% of the cohort. The existence of striking BcR similarity in unrelated and geographically distant cases was initially identified in CLL,38,39 in which mounting evidence indicates that the clustering of cases based on shared VH CDR3 patterns may underlie common antigen reactivity and perhaps biologic behavior of the malignant clones.40,41 Importantly, detailed comparison of the amino acid motifs defining VH CDR3 stereotypes in MCL versus CLL revealed that sequence restrictions are disease-biased. Thus, the stereotyped BcRs in MCL differ significantly from those in CLL, even for cases using the same IGHV gene, alluding to distinct immune-mediated mechanisms of lymphomagenesis. Hence, the identification of stereotyped BcRs with disease-biased features in MCL, at a frequency far exceeding chance, questions the notion of MCL as a neoplasm of “mostly naive” B cells (WHO 2008 definition),19 at least for certain subsets of cases.
Despite the substantial molecular evidence for antigen selection reported here, important questions remain unresolved, especially with regard to both (1) the type of antigenic stimuli and (2) the identity of the progenitor(s) selected for transformation. Judging from other B-cell malignancies, most notably CLL, the quest for the antigens will prove long and tortuous. As for the MCL cell progenitor(s), recent studies of normal human B-cell subpopulations point to several potential candidates. In particular, Kolar et al42 identified a novel population of tonsillar B cells that seem to be an intermediate between naive and germinal center cells, express AID, bear a low impact of SHM, and have an IgM+IgD+CD27−CD23−CD5+CD10− phenotype. Noticing obvious similarities, the authors suggested that these cells might represent the progenitors of MCL neoplastic cells. Our immunogenetic findings seem to go along with this hypothesis, at least for subsets of MCL cases, without, however, excluding alternative possibilities (eg, transitional B cells that are also CD5+ and exhibit a limited number of somatic hypermutations).43-45 Admittedly, at present, these and other suggestions, including the 2008 WHO choice (ie, an inner mantle zone cell, mostly of naive pregerminal center type), are not conclusive, given differences in the molecular and immunophenotypic profiles of candidate MCL progenitors and MCL itself.
In conclusion, MCL is characterized by a highly distinctive IG gene repertoire with very precisely targeted and probably functionally driven SHM. These features, along with the presence of “receptor prototypes” characterized by biased associations of IGHV, IGHD, and IGHJ genes, specified mutational status, and restricted VH CDR3 length and amino acid composition, indicate a role for antigen(s) in MCL development and also open possibilities for future investigations into the ontogeny of MCL. Intriguingly, this notion also is supported by recent clinical evidence that targeting the BcR-dependent signaling pathway might represent a highly efficacious treatment for MCL.46,47
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Evangelia Stalika, Ariane Stuhr, Mia Thorsélius, Ulf Thunberg, and Sarah Walsh for assistance with immunoglobulin gene sequence analysis.
The analysis of Swedish cases was supported by the Swedish Cancer Society, the Swedish Research Council, the Lion's Cancer Research Foundations in Uppsala, and the Stockholm County Council. The analysis of Spanish cases was supported by the Spanish Ministry of Science and Innovation (CYCYT SAF 08/3860; E.C.). The analysis of French cases was supported by the European Community within the European MCL Network (LSHC-CT 2004-503351); the Lymphoma Research Foundation, l'Association de Recherche contre le Cancer (ARC subvention 3730); The Fondation de France Comite Leucemie (subvention 2004004029); and l'institut National du cancer (project PAIR MCL). The immunogenetic analysis was supported by Cariplo Foundation (Milan, Italy); Associazione Italiana per la Ricerca sul Cancro (AIRC–Milan, Italy); and the ENosAI project (code 09SYN-13-880), cofunded by the European Union and the Hellenic General Secretariat for Research and Technology.
Authorship
Contribution: A.H. and A.A. performed research, analyzed data, and wrote the paper; N.D. performed research and analyzed data; F.M., M.-H.D.-L., L.B.P., A.N.L., A.D., P.R., K.B., and A.K. performed research; A.T. and P.M.-T. supervised research; M.H.D., A.A., A.R., M.P., P.Gr., P.Gh., B.S., T.P., E.C., C.G., R.R., F.D., and C.P. provided samples and associated clinicopathologic data and supervised research; and K.S. designed the study, supervised research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kostas Stamatopoulos, Department of Hematology and HCT Unit, G. Papanicolaou Hospital, 57010 Exokhi, Thessaloniki, Greece; e-mail: kostas.stamatopoulos@gmail.com.
References
Author notes
A.H. and A.A. are equal first authors.
On behalf of the Nordic Lymphoma Group.