Abstract
Single-nucleotide polymorphisms in genes that affect warfarin metabolism (cytochrome P450 2C9 gene, CYP2C9) and response (vitamin K epoxide reductase complex 1 gene, VKORC1) have an important influence on warfarin therapy, particularly during initiation; however, there is a lack of consensus regarding the optimal pharmacogenetics-based initiation strategy. We conducted a prospective cohort study in which patients requiring warfarin therapy for atrial fibrillation or venous thromboembolism were initiated with a novel pharmacogenetics-initiation protocol (WRAPID, Warfarin Regimen using A Pharmacogenetics-guided Initiation Dosing) that incorporated loading and maintenance doses based on genetics, clinical variables, and response (n = 167, followed up for 90 days), to assess the influence of genetic variations on anticoagulation responses. Application of the WRAPID algorithm resulted in a negligible influence of genetic variation in VKORC1 or CYP2C9 on time to achievement of first therapeutic response (P = .52, P = .28) and risk of overanticoagulation (P = .64, P = .96). After adjustment for covariates, time to stable anticoagulation was not influenced by VKORC1 or CYP2C9 genotype. Importantly, time spent within or above the therapeutic range did not differ among VKORC1 and CYP2C9 genotype groups. Moreover, the overall time course of the anticoagulation response among the genotype groups was similar and predictable. We demonstrate the clinical utility of genetics-guided warfarin initiation with the WRAPID protocol to provide safe and optimal anticoagulation therapy for patients with atrial fibrillation or venous thromboembolism.
Introduction
The vitamin K antagonist warfarin is an oral anticoagulant commonly prescribed in North America to treat venous thromboembolism (VTE) and decrease the risk of stroke in atrial fibrillation (AF).1 Warfarin therapy is challenging because of marked and often unpredictable interindividual dosing variation to reach and maintain adequate anticoagulation. For most indications, optimal warfarin therapy is achieved by maintaining the international normalized ratio (INR) within a narrow therapeutic range of 2.0 to 3.0. An insufficient warfarin dose leads to a lack of antithrombotic effect, whereas overanticoagulation is associated with elevated bleeding risk.2
Although warfarin has been in use for the past 60 years, genetic control of warfarin response has only recently been appreciated.3 In this regard, among the most studied genetic determinants are the single-nucleotide polymorphisms (SNPs) in genes that encode cytochrome P450 2C9 (CYP2C9)4 and the vitamin K epoxide reductase complex subunit 1 (VKORC1).5,6 CYP2C9 is the primary enzyme responsible for metabolism of the active S-enantiomer of warfarin, and its polymorphisms contribute significantly to variability in warfarin response.7 Possession of the common CYP2C9*2 (c.430C > T, rs1799853) and *3 (c.1075A > C, rs1057910) variant alleles results in lower dose requirement, increased time to stability, and a higher risk of overanticoagulation.8 VKORC1 is the target of warfarin that recycles oxidized vitamin K to the reduced form, an essential cofactor for activation of clotting factors II, VII, IX, and X, through γ-glutamyl carboxylation.9 Harboring common genetic variants of VKORC1, such as the functional promoter SNP −1639G > A (rs9923231), results in enhanced warfarin sensitivity, whereas rare mutations have been linked to warfarin resistance.5,6,10 In addition to VKORC1 and CYP2C9 polymorphisms, several studies have reported recently that a functional SNP in CYP4F2 (c.1297G > A, rs2108622), the metabolizing enzyme for vitamin K,11 also determines warfarin dose requirement.12
Since the Food and Drug Administration revised the label for warfarin to note the importance of VKORC1 and CYP2C9 polymorphisms,13 several groups have proposed genotype-guided maintenance dose algorithms that incorporate both genetics and demographic parameters, such as age, weight, and body surface area.14,15 However, there is a paucity of information with respect to dosing during warfarin initiation, arguably the most clinically challenging therapeutic phase, during which the risk of hemorrhage and recurrent thromboembolism is greatest.16,17 Standardized loading dose nomograms developed to date have not considered genetics and other patient-specific characteristics and have not been applied to indications other than VTE, such as AF.18-20 We and others have recently shown that VKORC1 and CYP2C9 genetic variations modulate early and stable response to warfarin during initiation when dosing by traditional means is used.21-24 Thus, in the present study, a novel and practical VKORC1- and CYP2C9-based loading and maintenance dose algorithm (WRAPID, Warfarin Regimen using A Pharmacogenetics-guided Initiation Dosing protocol) was developed and evaluated in AF and VTE patients with the aim of providing a safe, rapid, and uniform anticoagulation response.
Methods
Study sample and eligibility
This was a prospective cohort study of outpatients conducted at the London Health Sciences Center (LHSC) and The Ottawa Hospital (TOH). The study was approved by research ethics boards at both institutions. Patients requiring initiation of warfarin therapy were screened for eligibility. The requirement for warfarin therapy was determined on the basis of current American College of Chest Physicians guidelines.1 Patients who met the eligibility criteria were enrolled on provision of written informed consent in accordance with the Declaration of Helsinki. Table 1 summarizes patient characteristics.
Patient characteristics (n = 167)
| Demographics | |
| Age, y | 60 ± 17 |
| Sex, male/female, n | 96/74 |
| Weight, kg | 84 ± 19 |
| Height, cm | 171 ± 10 |
| Ethnicity | |
| White | 159 (95.2) |
| Black | 4 (2.4) |
| Asian | 3 (1.8) |
| Other | 1 (0.6) |
| Prescribed medication | |
| Amiodarone | 2 (1.2) |
| Statins | 45 (26.9) |
| Antiplatelet drugs | 55 (32.9) |
| Antibiotics | 6 (3.6) |
| Antifungal drugs | 1 (0.6) |
| NSAIDs | 12 (7.2) |
| Indication for warfarin | |
| Atrial fibrillation | 61 (36.6) |
| Deep vein thrombosis | 77 (46.1) |
| Pulmonary embolism | 21 (12.6) |
| Other | 8 (4.7) |
| Prescribed warfarin dose | |
| Mean maintenance dose, mg/d | 5.54 |
| CYP2C9 | |
| *1/*1 | 119 (71.3) |
| *1/*2 | 29 (17.4) |
| *1/*3 | 14 (8.4) |
| *2/*2 | 3 (1.7) |
| *2/*3 | 2 (1.2) |
| *3/*3 | 0 |
| VKORC1 −1639 | |
| G/G | 66 (39.5) |
| G/A | 75 (44.9) |
| A/A | 26 (15.6) |
| CYP4F2 c.1297 | |
| C/C | 80 (48.6) |
| C/T | 68 (40.7) |
| T/T | 19 (11.3) |
| Demographics | |
| Age, y | 60 ± 17 |
| Sex, male/female, n | 96/74 |
| Weight, kg | 84 ± 19 |
| Height, cm | 171 ± 10 |
| Ethnicity | |
| White | 159 (95.2) |
| Black | 4 (2.4) |
| Asian | 3 (1.8) |
| Other | 1 (0.6) |
| Prescribed medication | |
| Amiodarone | 2 (1.2) |
| Statins | 45 (26.9) |
| Antiplatelet drugs | 55 (32.9) |
| Antibiotics | 6 (3.6) |
| Antifungal drugs | 1 (0.6) |
| NSAIDs | 12 (7.2) |
| Indication for warfarin | |
| Atrial fibrillation | 61 (36.6) |
| Deep vein thrombosis | 77 (46.1) |
| Pulmonary embolism | 21 (12.6) |
| Other | 8 (4.7) |
| Prescribed warfarin dose | |
| Mean maintenance dose, mg/d | 5.54 |
| CYP2C9 | |
| *1/*1 | 119 (71.3) |
| *1/*2 | 29 (17.4) |
| *1/*3 | 14 (8.4) |
| *2/*2 | 3 (1.7) |
| *2/*3 | 2 (1.2) |
| *3/*3 | 0 |
| VKORC1 −1639 | |
| G/G | 66 (39.5) |
| G/A | 75 (44.9) |
| A/A | 26 (15.6) |
| CYP4F2 c.1297 | |
| C/C | 80 (48.6) |
| C/T | 68 (40.7) |
| T/T | 19 (11.3) |
Values are mean ± SD or n (%), unless otherwise indicated.
NSAIDs indicates nonsteroidal anti-inflammatory drugs.
Study eligibility was determined by the following inclusion criteria: (1) at least 18 years of age and (2) indication for new warfarin therapy for at least 3 months with a target INR range of 2.0 to 3.0. Exclusion criteria were diagnosis of cancer other than nonmelanoma skin cancer, alcohol or drug abuse, baseline INR > 1.4, known warfarin allergy/intolerance, terminal disease, prior use of warfarin therapy or vitamin K within 2 weeks before study enrollment, and known or suspected pregnancy.
Clinical data collection and follow-up
Demographic information was obtained at the time of enrollment. Therapy-related information was collected by patient interview and review of medical records. A baseline venous blood sample was obtained for DNA extraction and assessment of INR. Subsequent INR measurements, dose adjustments, adverse events, and therapy-related interventions were recorded at both study sites. The study period was September 2008 to August 2010.
Genotyping
Genomic DNA was isolated with Gentra Puregene or DNA Blood Midi extraction kit according to the manufacturer's protocol (QIAGEN). Genotype analysis included CYP2C9*2, CYP2C9*3, VKORC1 −1639G > A and CYP4F2 c.1297G > A. At LHSC, genotypes were determined by allelic discrimination with TaqMan drug metabolism genotyping assays using the 7500 RT-PCR system (assay IDs: C__25625805_10, C__27104892_10, C__1329189_10, C__16179493_40; Applied Biosystems). At TOH Research Institute, genotypes were determined with the Luminex 200 system (Luminex Corp). Briefly, forward and reverse primers for SNPs of interest were designed to amplify regions surrounding each SNP by standard multiplex PCR protocols. The PCR products were then hybridized with appropriate xMAP carboxylated microspheres at 52°C, followed by analysis with the Luminex 200. Genotyping was generally performed within 24 hours of receiving the baseline blood sample and used prospectively to determine individualized initiation doses before warfarin commencement for all study subjects.
Mathematical foundation for a novel pharmacogenetics-based initiation protocol
Using historical datasets compiled from Vanderbilt University (n = 297)24 and TOH (n = 63),25 we developed a pharmacogenetics-based initiation protocol aimed at providing a uniform anticoagulation response among all patients. The dosing regimen comprised both a loading and maintenance dose algorithm. Development of the loading and maintenance dose algorithms required the integration of pharmacokinetic (PK) and pharmacodynamic (PD) factors to predict the time course of warfarin plasma levels and response.
In this mechanistic model, the plasma PK of warfarin was described with a simple 1-compartment model for warfarin distribution of a set volume (V). The time course of estimated plasma S-warfarin concentration (Cplasma) arose from the interplay between drug absorption in the gut (ka) and drug elimination via CYP2C9 metabolism (ke). Values for kinetic parameters for S-warfarin were obtained from the literature.26-29 Warfarin metabolism capacity (ke) is mainly dependent on CYP2C9 genotype; thus, the values for ke were adjusted based on reported clearance reductions in heterozygous and homozygous variant allele carriers of either CYP2C9*2 or *3.29 The pooling of genotypes into 3 subgroups was performed because of the lack of confidence in the accuracy of limited available clearance rates reported in literature for the CYP2C9*2/*2, *2/*3, and *3/*3 genotype groups as a result of low allelic frequencies.29
Warfarin PD was described by an indirect response model that incorporates the known delay and magnitude of anticoagulation effects after achieving the required plasma concentration.30 In this model, the degree of suppression of vitamin K–dependent clotting factor production is related to the effectiveness of warfarin concentrations to inhibit vitamin K epoxide reductase. Here, the rate of change in INR is modeled with zero-order input (K) and first-order output (ko) variables. Plasma warfarin levels dictate the inhibition of output response according to classic competitive enzyme inhibition kinetics described by the parameters Imax (enzyme content and intrinsic activity) and IC50 (drug affinity).31 Imax values corresponding to VKORC1 genotypes were determined on simulation of maintenance drug administration to stable therapeutic coagulation. Depiction and parameters of the PK-PD model are shown in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The PK-PD model and corresponding parameter values presented here are preliminary and served only to guide the establishment of a practical WRAPID dosing protocol for various VKORC1 and CYP2C9 genotype combinations. A finalized version of the PK-PD model with data-derived parameter values based on formal modeling of R/S-warfarin concentrations and INR measurements obtained in this patient cohort, along with identification of key nongenetic and genetic determinants of warfarin kinetics and responses, will be published elsewhere.
Loading doses
Practical (5, 7.5, and 10 mg) daily loading doses were prescribed for 2 days and were dependent on VKORC1 and CYP2C9 genotypes (Table 2). These doses were used to obtain sufficient warfarin plasma concentrations to reach and maintain optimal anticoagulation response (INR 2.0-3.0) with similar rapid initial time course for all genotype groups (supplemental Figure 2).
Pharmacogenetics-based loading dose grid according to VKORC1 and CYP2C9 genotype
| VKORC1 . | CYP2C9 . | ||
|---|---|---|---|
| *1/*1 . | *1/*2 or *1/*3 . | *2/*2, *2/*3, *3/*3 . | |
| G/G | 10† | 10† | 7.5‡ |
| G/A | 10† | 7.5‡ | 5‡ |
| A/A | 5‡ | 5‡ | 5‡ |
| VKORC1 . | CYP2C9 . | ||
|---|---|---|---|
| *1/*1 . | *1/*2 or *1/*3 . | *2/*2, *2/*3, *3/*3 . | |
| G/G | 10† | 10† | 7.5‡ |
| G/A | 10† | 7.5‡ | 5‡ |
| A/A | 5‡ | 5‡ | 5‡ |
Loading doses are in milligrams.
Loading dose was adjusted to 7.5 mg for patients with weight < 60 kg.
Loading dose was decreased by 2.5 mg for patients with weight < 45 kg.
Maintenance doses
To obtain the maintenance dose, key patient clinical parameters that are known to influence warfarin dose requirement along with genetics were combined in a generalized linear regression model (Table 3). Briefly, warfarin dose was the dependent variable, the VKORC1 and CYP2C9 genetics-based dose was a constant variable (Table 4), and coefficients of independent variables were varied according to the least squares linear regression method.
Final multiple linear regression for estimation of maintenance dose
| Predictor variable . | B . | Standard error . | P in final model . |
|---|---|---|---|
| Intercept | −1.46 | 1.23 | .235 |
| Weight (kg) | 0.06 | 0.01 | < .0001 |
| Genetics-based dose grid (Table 4) | 1 | < .0001 | |
| Age, y | −0.05 | 0.01 | < .0001 |
| Sex, female | −0.90 | 0.34 | < .01 |
| Amiodarone use, yes | −1.97 | 1.1 | .07 |
| CYP4F2 c.1297G > A, per allele | 0.33 | 0.25 | .199 |
| Predictor variable . | B . | Standard error . | P in final model . |
|---|---|---|---|
| Intercept | −1.46 | 1.23 | .235 |
| Weight (kg) | 0.06 | 0.01 | < .0001 |
| Genetics-based dose grid (Table 4) | 1 | < .0001 | |
| Age, y | −0.05 | 0.01 | < .0001 |
| Sex, female | −0.90 | 0.34 | < .01 |
| Amiodarone use, yes | −1.97 | 1.1 | .07 |
| CYP4F2 c.1297G > A, per allele | 0.33 | 0.25 | .199 |
Genetics-dependent dose grid for maintenance dose regression
| VKORC1 . | CYP2C9 . | ||
|---|---|---|---|
| *1/*1 . | *1/*2 or *1/*3 . | *2/*2, *2/*3, *3/*3 . | |
| G/G | 7 | 5 | 3.5 |
| G/A | 5 | 4 | 2.5 |
| A/A | 3.5 | 2.5 | 1.5 |
| VKORC1 . | CYP2C9 . | ||
|---|---|---|---|
| *1/*1 . | *1/*2 or *1/*3 . | *2/*2, *2/*3, *3/*3 . | |
| G/G | 7 | 5 | 3.5 |
| G/A | 5 | 4 | 2.5 |
| A/A | 3.5 | 2.5 | 1.5 |
Doses are in milligrams.
After the 2-day loading dose, patients were prescribed the calculated maintenance dose to begin on day 3. On 3 occasions within the first 9 days of therapy (initiation), INR measurements were obtained (typically days 3, 5/6, and 7/8/9), because this frequency of INR measurements has been shown to be practical and efficacious for anticoagulation management.19 When the INR response at each measurement did not conform to the predicted trajectory based on the mathematical model, the daily maintenance dose was further adjusted according to a treatment day–specific dose-adjustment nomogram (Table 5). Adjusted doses were rounded to either the nearest whole number or 0.5.
Dose-adjustment nomogram during initiation
| . | INR . | Warfarin dose adjustment . |
|---|---|---|
| Day 3 | < 1.3 | ↑ 10% |
| 1.3-1.5 | No change | |
| 1.6-1.8 | ↓ 10% | |
| 1.9-2.1 | ↓ 20% | |
| 2.2-2.5 | ↓ 50% | |
| > 2.5 | Hold dose for 1 day, then ↓ 50% | |
| Day 5/6 | < 1.3 | ↑ 50% |
| 1.4-1.7 | ↑ 20% | |
| 1.8-2.5 | No change | |
| 2.6-3.0 | ↓ 20% | |
| 3.1-3.9 | ↓ 50% | |
| ≥ 4.0 | Hold dose for 1 day, then ↓ 50% | |
| Day 7/8/9 | < 1.5 | ↑ 20% |
| 1.5-1.9 | ↑ 10% | |
| 2.0-2.8 | No change | |
| 2.9-3.5 | ↓ 10% | |
| 3.6-4.0 | Hold dose for 1 day, then ↓ 15% | |
| ≥ 4.0 | Hold dose, test INR daily until in range (2.0-3.0), then ↓ 25% |
| . | INR . | Warfarin dose adjustment . |
|---|---|---|
| Day 3 | < 1.3 | ↑ 10% |
| 1.3-1.5 | No change | |
| 1.6-1.8 | ↓ 10% | |
| 1.9-2.1 | ↓ 20% | |
| 2.2-2.5 | ↓ 50% | |
| > 2.5 | Hold dose for 1 day, then ↓ 50% | |
| Day 5/6 | < 1.3 | ↑ 50% |
| 1.4-1.7 | ↑ 20% | |
| 1.8-2.5 | No change | |
| 2.6-3.0 | ↓ 20% | |
| 3.1-3.9 | ↓ 50% | |
| ≥ 4.0 | Hold dose for 1 day, then ↓ 50% | |
| Day 7/8/9 | < 1.5 | ↑ 20% |
| 1.5-1.9 | ↑ 10% | |
| 2.0-2.8 | No change | |
| 2.9-3.5 | ↓ 10% | |
| 3.6-4.0 | Hold dose for 1 day, then ↓ 15% | |
| ≥ 4.0 | Hold dose, test INR daily until in range (2.0-3.0), then ↓ 25% |
↑indicates increase; and ↓, decrease.
To make dosing practical, we devised an automated dose calculator for the initiation phase (supplemental Figure 3 and supplemental Excel file). After initiation, dosing was adjusted by pharmacists at both centers on the basis of a standardized postinitiation nomogram. Once patients had obtained 2 therapeutic INRs, dosing was assisted by Dawn AC anticoagulation software (4-S Information Systems Ltd).
Refinement of loading and maintenance dose algorithm
It was our objective to refine the loading and maintenance dose algorithm after monitoring an initial cohort of patients (n = 87) for application in a final cohort. After the initial cohort, we observed a disproportionate number of out-of-range INR responses in those patients with a high loading dose–to-weight ratio. Thus, we modified the loading dose algorithm to consider weight after the first cohort. For patients who weighed < 60 kg who had been given a 10-mg load according to the original loading algorithm, we decreased the dose to 7.5 mg. For patients who weighed < 45 kg, all loading doses were decreased by 2.5 mg (7.5-5 mg and 5-2.5 mg).
Planned optimization of the maintenance dose regression was performed by slight modification of the contribution of clinical parameters to dose, whereas the impact of VKORC1 and CYP2C9 remained unchanged. In univariate analysis, we observed a significant relationship between CYP4F2 c.1297G > A genotype and dose, in which A/A carriers required a 1-mg higher warfarin dose than the wild-type group (P < .05). Thus, we included CYP4F2 genotype in the final regression model to determine maintenance doses of the final cohort.
Sample size
The WRAPID study was powered to assess the effect of VKORC1 −1639G > A genotype on anticoagulation response, after pharmacogenetics-guided initiation. A study size of 150 patients was estimated to have 80% power to detect a response hazard of 2 at a 2-sided significance level of 0.05, which allowed for a dropout rate of 10%. A hazard ratio (HR) of 2 was chosen on the basis of previously published VKORC1 carrier status hazard risk for primary outcomes.24 Power analysis was performed with SAS Version 9.2 (SAS Institute).
Primary and secondary outcomes
The primary outcomes of the present study were time to first therapeutic INR and time to first overanticoagulation (INR ≥ 4). We choose these primary outcomes because they are critical markers of anticoagulation pace and quality of control. Therapeutic INR was defined as 2.0-3.0 for all patients.
Secondary outcomes were time to first stable anticoagulation, time spent in therapeutic range, and time spent above therapeutic range during the first 30 days (prestabilization phase) and after 30 days (stabilization phase). Stable anticoagulation was defined as 2 consecutive in-range INRs, at least 7 days apart, with no dose adjustments. For estimation of time spent in or out of range, we adopted the Rosendaal linear interpolation method to calculate the percentage of time each patient spent within and out of the therapeutic range.32 The difference between one INR value and the subsequent INR value was divided by the number of days elapsed between the 2 measurements to produce the average daily increment or decrement in INR.
Additional secondary outcomes included percentage of patients within range during initiation, percentage of patients with an INR ≥ 5, percentage of time spent above INR ≥ 4, percentage of time spent in therapeutic range, and extended therapeutic range (1.8-3.2), as well as average maintenance dose.
The outcomes were selected to assess the influence of VKORC1 genotype on anticoagulation response. Although the present study was not powered to evaluate the individual effects of CYP2C9*2 and *3 genotypes on response, analysis was conducted to compare wild-type and any CYP2C9 variant carrier status.
Statistical analysis
Patients were divided into 3 groups for VKORC1 and 2 groups for CYP2C9: VKORC1 wild-type, heterozygous, and homozygous carriers of −1639G > A; CYP2C9 wild-type (*1/*1); and a CYP2C9 variant group that included 1 or 2 variant allele carriers (*1/*2, *1/*3; *2/*2, *3/*3, or *2/*3). Hardy-Weinberg equilibrium was assessed for each genotype with the χ2 goodness-of-fit test.
The influence of VKORC1 and CYP2C9 genotype on the primary outcomes was evaluated with survival analysis techniques. Kaplan-Meier plots were used to depict the proportion of subjects without events over time. Comparison between survival curves was conducted by the log-rank test. Unadjusted HR and its 95% confidence interval (CI) between genotype groups were computed. The Cox proportional hazard model was adopted to adjust for potential confounding effects of age, sex, weight, warfarin dose, amiodarone use, indication, patient cohort, and VKORC1, CYP2C9, and CYP4F2 genotype to obtain adjusted HRs and their 95% CIs. For comparison of differences in outcomes between patients with various VKORC1 statuses, G/G genotype was considered as the reference group, because it is the most warfarin-resistant group. Statistical analysis of Schoenfeld residuals and visual inspection of log-minus-log plots revealed no significant variation from the proportional hazards assumption.
Percentages of time spent in therapeutic range and time spent above the therapeutic range were compared among VKORC1 and CYP2C9 genotype groups with the use of Kruskal-Wallis test followed by Tukey-Kramer posttest or Mann-Whitney test, as appropriate.
All enrolled patients (initial and second cohort) were included for outcome analysis to obtain at least 80% power to detect the association of VKORC1 genotype and response. This was acceptable because there were no statistical differences between the 2 cohorts with respect to primary outcomes of time to first therapeutic INR and time to first INR ≥ 4. In addition, potential variations between cohorts because of dosing-regimen modifications were accounted for as a confounding variable in the Cox regression analysis of primary survival data and should not interfere with assessment of genetic variation effects on rate of INR anticoagulation responses. Because no significant differences were observed between the initial and second cohorts for secondary outcomes, both patient groups were combined for secondary analysis.
To assess and compare the predictability of our dosing model, we determined the association between maintenance dose and model-derived dose. The proportion of variance explained was calculated as the R2 statistic. In addition, we determined the mean absolute error of each model for the same purpose.
A 2-tailed P value < .05 was considered significant for all analyses. Statistical analysis was performed with the use of GraphPad Prism Version 5.0 or SPSS Version 17.
Results
Population characteristics
Of the 196 patients enrolled, 29 were excluded from analysis for the following reasons: 3 because of entry error, 4 because they self-administered the wrong dose, 1 for failure to comply with INR measurements, 14 dropouts, 6 because of incomplete follow-up, and 1 because of death (cause not attributed to study participation). Of those included for outcome analysis, 61 and 96 patients were enrolled at the LHSC and TOH, respectively.
The allelic frequencies for VKORC1 −1639G > A and CYP4F2 c.1297G > A were 38.0% and 31.7%, respectively. The CYP2C9*2 and *3 allelic frequencies were 11.1% and 4.8%, respectively. There were no deviations from Hardy-Weinberg equilibrium.
Time to first therapeutic INR (2.0-3.0) and overanticoagulation (INR ≥ 4)
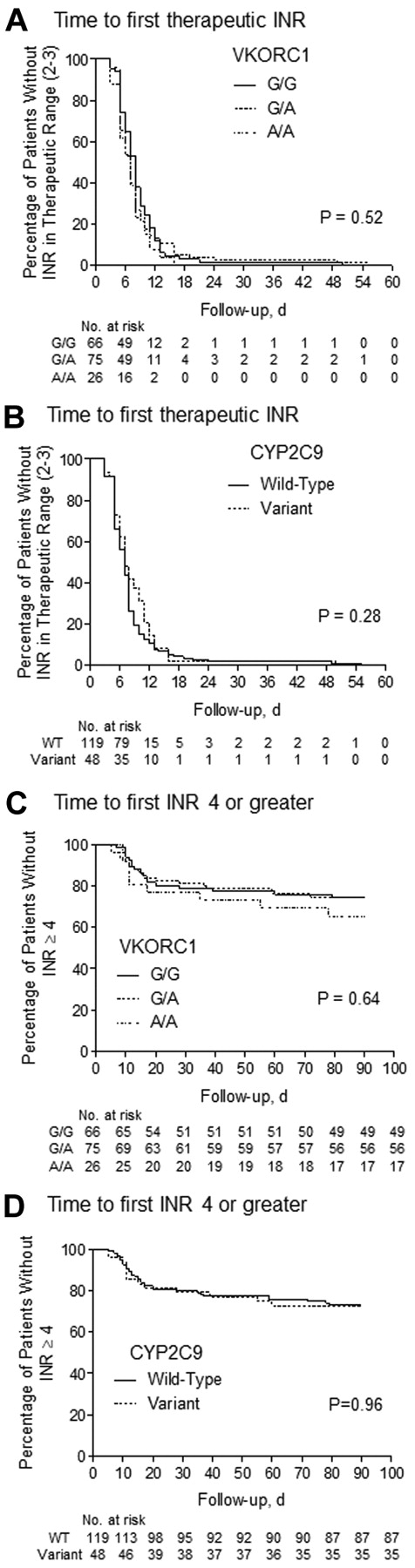
The primary outcomes were compared in terms of VKORC1 and CYP2C9 genotypes. VKORC1 genotype had no significant effect on time required to reach the first INR within the therapeutic range (P = .52) or time required to obtain an INR ≥ 4, according to log-rank test (P = .64; Figure 1A,C). Similarly, there was no significant difference between CYP2C9 wild-type and variant genotype for either of these outcomes (P = .28 for first INR, P = .96 for first INR ≥ 4; Figure 1B,D). Concordant with these findings, HR estimates for the VKORC1 and CYP2C9 genotype groups were not significantly different from unity before or after adjustment for covariates by Cox regression analysis (Table 6). Because outcomes during the first 30 days would be most sensitive to the initiation protocol, we compared the time to first INR ≥ 4 during the first month of therapy among genotype groups. VKORC1 and CYP2C9 polymorphisms were without influence on this outcome (data not shown). Importantly, these outcomes were not associated with genotype when we considered the first and second cohorts of patients independently (data not shown).
The effect of pharmacogenetics-guided dosing on time to primary events. Kaplan-Meier plots represent the lack of association for attainment of first INR within therapeutic range (2.0-3.0) and first above-range INR (INR ≥ 4) among VKORC1 (A,C) and CYP2C9 (B,D) genotype groups after initiation with WRAPID nomogram. The statistic in each panel represents the log-rank P value for testing the equality of survival functions. WT indicates wild type.
The effect of pharmacogenetics-guided dosing on time to primary events. Kaplan-Meier plots represent the lack of association for attainment of first INR within therapeutic range (2.0-3.0) and first above-range INR (INR ≥ 4) among VKORC1 (A,C) and CYP2C9 (B,D) genotype groups after initiation with WRAPID nomogram. The statistic in each panel represents the log-rank P value for testing the equality of survival functions. WT indicates wild type.
Unadjusted and adjusted HRs for anticoagulation outcomes in patients with VKORC1 G/A or A/A and CYP2C9 variant genotype
| Genotype and outcome . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| VKORC1 G/A genotype* | ||||
| Time to first therapeutic INR | 0.79 (0.50-1.25) | .32 | 0.62 (0.27-1.39) | .24 |
| Time to first above-range INR | 0.71 (0.32-1.59) | .40 | 0.58 (0.11-3.19) | .53 |
| Time to stable anticoagulation | 1.03 (0.65-1.63) | .91 | 1.11 (0.64-1.90) | .72 |
| VKORC1 A/A genotype* | ||||
| Time to first therapeutic INR | 0.87 (0.55-1.37) | .54 | 0.76 (0.34-1.57) | .43 |
| Time to first above-range INR | 0.70 (0.32-1.56) | .39 | 0.56 (0.14-3.15) | .60 |
| Time to stable anticoagulation | 0.69 (0.43-1.10) | .12 | 0.8 (0.47-1.37) | .41 |
| CYP2C9 variant genotype† | ||||
| Time to first therapeutic INR | 0.85 (0.61-1.19) | .34 | 1.04 (0.69-1.57) | .86 |
| Time to first above-range INR | 1.02 (0.53-1.94) | .96 | 0.91 (0.43-1.92) | .81 |
| Time to stable anticoagulation | 1.17 (0.83-1.65) | .38 | 0.88 (0.60-1.29) | .51 |
| Genotype and outcome . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| VKORC1 G/A genotype* | ||||
| Time to first therapeutic INR | 0.79 (0.50-1.25) | .32 | 0.62 (0.27-1.39) | .24 |
| Time to first above-range INR | 0.71 (0.32-1.59) | .40 | 0.58 (0.11-3.19) | .53 |
| Time to stable anticoagulation | 1.03 (0.65-1.63) | .91 | 1.11 (0.64-1.90) | .72 |
| VKORC1 A/A genotype* | ||||
| Time to first therapeutic INR | 0.87 (0.55-1.37) | .54 | 0.76 (0.34-1.57) | .43 |
| Time to first above-range INR | 0.70 (0.32-1.56) | .39 | 0.56 (0.14-3.15) | .60 |
| Time to stable anticoagulation | 0.69 (0.43-1.10) | .12 | 0.8 (0.47-1.37) | .41 |
| CYP2C9 variant genotype† | ||||
| Time to first therapeutic INR | 0.85 (0.61-1.19) | .34 | 1.04 (0.69-1.57) | .86 |
| Time to first above-range INR | 1.02 (0.53-1.94) | .96 | 0.91 (0.43-1.92) | .81 |
| Time to stable anticoagulation | 1.17 (0.83-1.65) | .38 | 0.88 (0.60-1.29) | .51 |
All Cox regression models were adjusted for age, sex, weight, warfarin dose, indication for therapy, cohort, and VKORC1, CYP2C9, and CYP4F2 genotype.
Survival function was compared with VKORC1 wild-type G/G genotype group.
Time to stable anticoagulation
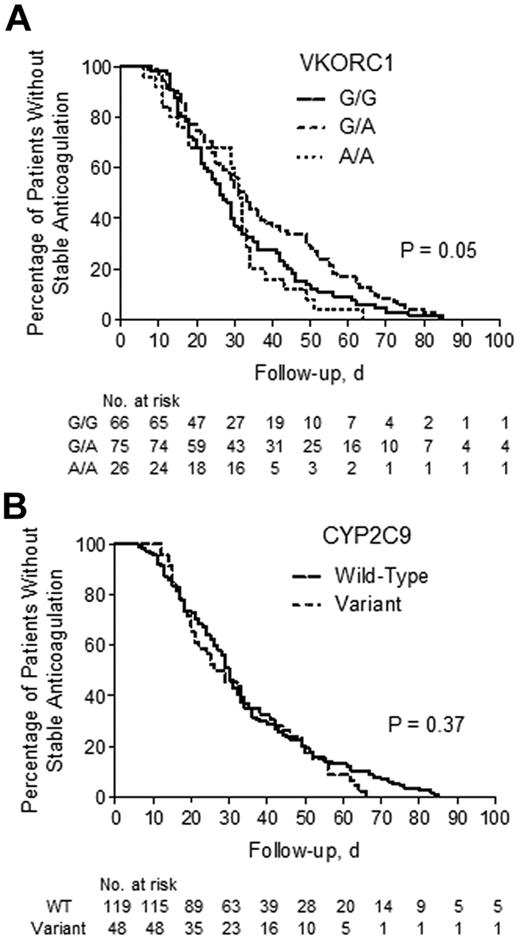
The time to first stable anticoagulation was significantly different between VKORC1 (P < .05) genotype groups, whereas there were no differences between CYP2C9 groups (P = .37; Figure 2). However, when adjusted for confounding covariates, neither VKORC1 nor CYP2C9 showed a significant influence on time to stability (Table 6).
The effect of pharmacogenetics-guided dosing on time to stability. Kaplan-Meier plots represent the time to stable anticoagulation among VKORC1 (A) and CYP2C9 (B) genotype groups. The statistic in each panel represents the log-rank P value for testing the equality of survival functions. WT indicates wild type.
The effect of pharmacogenetics-guided dosing on time to stability. Kaplan-Meier plots represent the time to stable anticoagulation among VKORC1 (A) and CYP2C9 (B) genotype groups. The statistic in each panel represents the log-rank P value for testing the equality of survival functions. WT indicates wild type.
Time spent within therapeutic range (INR 2.0-3.0) and above therapeutic range (INR > 3)
To separate the initiation and stabilization phases of therapy, we considered prestabilization as days 1 to 30 and the stabilization phase as day 31 to the end of the study period. We chose 30 days because the median time to stability was 29 days. There was no significant influence of VKORC1 or CYP2C9 genotype on time spent in therapeutic range or above range during the prestabilization or stabilization phase (supplemental Table 1). The present study was not powered to detect secondary outcomes.
INR response time course during first 3 weeks of therapy
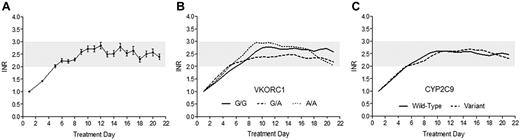
With the PK-PD model, the response profiles for various genotype groups were predicted to be similar during the attainment of therapeutic INR. Our algorithm was developed to enable patients to reach the first therapeutic response in a steady and safe manner, with a goal of reaching optimal anticoagulation by the end of initiation. Figure 3A illustrates the average INR time course of patients in the present study up to treatment week 3. The time course observed was similar to the model-predicted response profile, particularly during the critical first week. Concordant with our primary outcomes, average INR during initiation rose to the target range in a similar fashion among VKORC1 and CYP2C9 genotype groups, and importantly, anticoagulation stability, as represented by maintenance of INR within range after initiation, was comparable (Figure 3B-C).
The effect of genotype-guided dosing on response time course during the first 3 weeks of warfarin therapy. (A) The average response observed in patients dosed by the WRAPID nomogram, represented as mean with 95% CI of the SE, is similar to the PK-PD model–predicted anticoagulation response time course. The observed INR time courses among VKORC1 (A) and CYP2C9 (B) genotype groups, presented as LOWESS (locally weighted scatterplot smoothing regression) smoothed plots, rise and are maintained within the therapeutic range in a parallel and similar manner.
The effect of genotype-guided dosing on response time course during the first 3 weeks of warfarin therapy. (A) The average response observed in patients dosed by the WRAPID nomogram, represented as mean with 95% CI of the SE, is similar to the PK-PD model–predicted anticoagulation response time course. The observed INR time courses among VKORC1 (A) and CYP2C9 (B) genotype groups, presented as LOWESS (locally weighted scatterplot smoothing regression) smoothed plots, rise and are maintained within the therapeutic range in a parallel and similar manner.
Secondary outcomes
Secondary efficacy and safety outcomes of anticoagulation are summarized in Table 7. By day 6 of therapy, 40.1% of patients enrolled in the present study had an INR within the therapeutic range, whereas 57.5% had an INR within the therapeutic range by day 9. The proportion of patients reaching the extended INR range of 1.8-3.2 by days 6 and 9 was 60.5% and 78.4%, respectively. Although approximately 20% of patients reached an INR ≥ 4 during the entire study duration, the percentage of time spent with INR ≥ 4 was only 1.2%. Moreover, only 3.6% of patients experienced excessive overanticoagulation with an INR ≥ 5 during the entire study period. The average maintenance dose was 5.54 mg/d and followed the known gene-dose relationship (supplemental Table 2).
Secondary outcomes after dosing with pharmacogenetics-based algorithm
| Initiation phase (day 1-9), n (%) | |
| INR 2-3 within 3 days | 13 (7.8) |
| INR 2-3 within 6 days | 67 (40.1) |
| INR 2-3 within 9 days | 96 (57.5) |
| INR extended 1.8-3.2 within 3 days | 23 (13.8) |
| INR extended 1.8-3.2 within 6 days | 101 (60.5) |
| INR extended 1.8-3.2 within 9 days | 131 (78.4) |
| INR ≥ 5 | 0 (0) |
| 90-day follow-up period* | |
| Time spent in range, % (SD) | 64.8 (19.8) |
| Time spent in extended therapeutic INR range (1.8-3.2), % (SD) | 77.3 (14.4) |
| Time spent in INR ≥ 4, % (SD) | 1.2 (2.9) |
| Number of INR measurements in 90 days, mean ± SD | 12.3 ± 2.7 |
| INR ≥ 5, n (%) | 6 (3.6) |
| Post 30-day follow-up period | |
| Time spent in range, % (SD) | 68.1 (25.0) |
| Initiation phase (day 1-9), n (%) | |
| INR 2-3 within 3 days | 13 (7.8) |
| INR 2-3 within 6 days | 67 (40.1) |
| INR 2-3 within 9 days | 96 (57.5) |
| INR extended 1.8-3.2 within 3 days | 23 (13.8) |
| INR extended 1.8-3.2 within 6 days | 101 (60.5) |
| INR extended 1.8-3.2 within 9 days | 131 (78.4) |
| INR ≥ 5 | 0 (0) |
| 90-day follow-up period* | |
| Time spent in range, % (SD) | 64.8 (19.8) |
| Time spent in extended therapeutic INR range (1.8-3.2), % (SD) | 77.3 (14.4) |
| Time spent in INR ≥ 4, % (SD) | 1.2 (2.9) |
| Number of INR measurements in 90 days, mean ± SD | 12.3 ± 2.7 |
| INR ≥ 5, n (%) | 6 (3.6) |
| Post 30-day follow-up period | |
| Time spent in range, % (SD) | 68.1 (25.0) |
Ninety-day follow-up period excludes initiation phase (days 1-9). The outcomes change slightly when initiation is included.
Dosing algorithm assessment
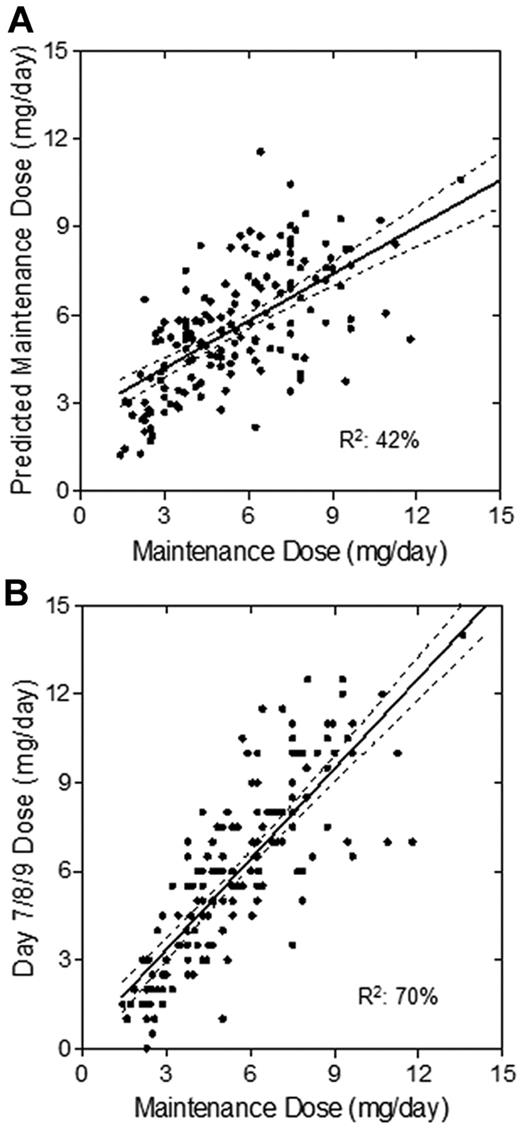
The association between observed maintenance dose, algorithm-predicted dose, and day 7/8/9 dose was determined. The proportion of variance explained by the final maintenance regression was 42% (Figure 4A), whereas the variance explained after INR-guided dose adjustments was 70% (Figure 4B). In addition, there was less bias between day 7/8/9 dose and maintenance dose than that of the algorithm-predicted dose. The mean absolute error (SE) of the final model was 10.4 (0.1) mg/wk, whereas for the INR-adjusted dose, it was 8.5 (0.9) mg/wk, comparable to that of other pharmacogenetics-based nomograms.33
Association of predicted maintenance dose to observed maintenance dose. Scatterplots show the association of algorithm-predicted maintenance dose (A) and day 7/8/9 dose after response-based adjustments (B) with the observed maintenance dose. The solid lines represent the linear regression, and the dashed lines represent its 95% CI.
Association of predicted maintenance dose to observed maintenance dose. Scatterplots show the association of algorithm-predicted maintenance dose (A) and day 7/8/9 dose after response-based adjustments (B) with the observed maintenance dose. The solid lines represent the linear regression, and the dashed lines represent its 95% CI.
Discussion
The clinical benefit of warfarin for decreasing stroke risk among AF patients and treating VTE is well established; however, the unpredictable anticoagulation response for a significant proportion of patients poses a substantial clinical challenge to optimal warfarin therapy. Several studies have examined various initiation strategies for treatment of VTE and AF.1,34-36 Although several of these studies have incorporated loading dose nomograms during initiation, most have been in the setting of VTE,19,20 and few studies incorporating loading dose strategies for other indications have been reported.34,37 Pharmacogenomic studies conducted in the last decade have established the contribution of both VKORC1 and CYP2C9 genetic variations to maintenance dose requirements; however, VKORC1 is a more important modulator of early warfarin response than CYP2C9.24 Not surprisingly, both genes have recently been reported to predict therapeutic doses during the initial weeks of therapy.38,39 With these considerations, we developed and evaluated a practical and universal pharmacogenetics-based loading dose algorithm for both AF and VTE patients.
In contrast to the findings previously observed with nonpharmacogenetics-based dosing,24 we show that use of the WRAPID algorithm eliminated VKORC1 and CYP2C9 genotype-related differences in attainment of first therapeutic INR in both AF and VTE patients. This finding did not change after adjustment for confounding variables. Interestingly, subanalysis of patients in the initial cohort, in which CYP4F2 was not included as a predictor of dose, demonstrated that the c.1297G > A SNP did not significantly influence attainment of therapeutic INR. This was in contrast to a recent study by Zhang et al that examined the role of CYP4F2 as a genetic determinant during initiation in patients dosed according to standard methods.40 The present findings suggest that dosing according to VKORC1 and CYP2C9 genotype is sufficient. This is consistent with the fact that CYP4F2 genotype accounts for only a small portion of the observed maintenance dose variability (0%-4%).12,41 However, a caveat here is that the present study was not powered to detect an association between response and CYP4F2 genotype. Thus, the definitive role of CYP4F2 genotype in individualized warfarin therapy requires further assessment in a powered study of sufficient sample size.
With respect to risk for excessive anticoagulation (INR ≥ 4), several groups have reported that variant carriers of VKORC1 and CYP2C9 are subject to significantly increased risk of overanticoagulation.22 After initiation with a pharmacogenetics-based dosing algorithm, neither the VKORC1 nor the CYP2C9 variant groups had an elevated risk of supratherapeutic INR during the first month of therapy or throughout the entire study period. The present findings contrast with those of a study conducted by Voora et al in which patients were dosed prospectively only according to CYP2C9 genotype.42 In that study, carriers of a variant CYP2C9 allele still exhibited an increased risk for excessive anticoagulation. This may be explained by dosing algorithm and adjustment differences during initiation compared with the present study.
An important measure of variability in anticoagulation quality is the time to stability. Higashi et al reported that variant carriers of CYP2C9 required significantly longer to attain stability in the absence of pharmacogenetics-based dosing.8 The present study demonstrates that differences in time to stability between VKORC1 and CYP2C9 genotype groups can be effectively reduced with the use of pharmacogenetics-guided dosing. Moreover, Wadelius et al23 and Limdi et al22 recently reported that the INR response profile differed between genotype groups during initiation using a standard dosing regimen, in which variant carriers had greater warfarin sensitivity. In contrast, the increase in INR to therapeutic range in the present study was similar among genotype groups.
It cannot be entirely ruled out that the lack of association observed in the present study may have been caused by insufficient sample size; however, calculations showed that we had ample power (> 80%) to detect the association of a causal VKORC1 −1639G > A SNP, with an allele frequency of 35%, with anticoagulation response, for an HR of 2. Because we observed no evidence of an association between VKORC1 genotype and anticoagulation responses, it would be reasonable to conclude that the WRAPID algorithm eliminated the VKORC1-driven response variation. Although we did not observe a significant association between CYP2C9*2 or *3 genotype and response, this may be because of lack of power. When one considers the small proportion of heterozygous and homozygous carriers of CY2C9*2 and *3 allele, the sample size required to detect such individual associations would be very large (∼ 1000). Thus, we assessed the association of pooled CYP2C9 variant status (at least 1 of *2 or *3 allele), a frequency of 30% in the present study population, with anticoagulation response. In this case, calculation showed that sufficient power was achieved (> 80%) to detect the association of CYP2C9 variant status with anticoagulation responses for an HR of 2. Such pooling of CYP2C9*2 and *3 variants has been used previously by other studies for similar reasons.8,41,43 In particular, these studies demonstrated that among patients whose therapy was initiated with standard dosing protocols, CYP2C9 variant carriers spent more time above therapeutic INR, had an elevated risk of overanticoagulation, and a lower dose requirement overall.
To the best of our knowledge, the WRAPID nomogram is the first warfarin initiation algorithm that incorporates both VKORC1 and CYP2C9 genotype-determined loading doses, which differs from the typical doubling of the maintenance dose. Thus far, there have been 2 prospective randomized clinical trials (RCTs) in which pharmacogenetics-based warfarin initiation with loading doses was compared with standard warfarin loading dose initiation, whereas other studies have not incorporated loading doses. In the first trial, control patients were loaded with 5 mg, whereas study patients were loaded according to CYP2C9 genotype.44 In the second trial, control patients were initiated with 10 mg, whereas study patients were initiated with double the maintenance dose determined with VKORC1 and CYP2C9 genotype.45 Evidently, there is a lack of consensus with respect to warfarin initiation, especially concerning loading dose selection from genetic information. The aforementioned trials, albeit small, indicate that pharmacogenetics-guided dosing improves warfarin response in terms of more time spent within the therapeutic range, decreased bleeding events, and faster attainment of therapeutic INR, which supports the use of loading doses for initiation. In addition, some studies recommend that loading dose should be age adjusted because of concern about warfarin sensitivity.34 However, we did not observe a disproportional number of elderly patients with excessive anticoagulation with our loading dose regimen, in which some elderly patients were indeed loaded with 10 mg as per genotype. Thus, the present data do not support age-modified loading doses. We did, however, observe an effect of decreased weight on response sensitivity during initiation.
Limitations of the present study include the inability to determine the influence of a pharmacogenetics-guided dosing algorithm on rare bleeding complications because of insufficient sample size; however, we can comment on the general safety of our dosing regimen. The number of INRs > 5 has been used previously as a measure of the safety of a dosing protocol.19 In the present study, only 3.6% of patients experienced such excessive anticoagulation, which is lower than that observed with other initiation protocols (5.6%-8.6%).19,36 Several RCTs involving larger sample sizes are currently under way to evaluate the safety and efficacy of pharmacogenetics-based dosing compared with standard dosing (www.clinicaltrials.gov; NCT01006733, NCT00839657, and NCT01119300). Interestingly, a proposal has been made for a multicenter trial in Europe that will test pharmacogenetics-guided initiation with the use of genotype-based loading doses to examine the clinical utility of such dosing methods. The present study supports the use of a genotype-guided loading dose during warfarin initiation.
Another limitation is that because the present study lacked a control group (nongenetics-based warfarin initiation), the results may be attributed in part to management of warfarin therapy by anticoagulation clinics. However, the present study was not designed as an RCT; rather, it had the goal of demonstrating the minimization of genotype-dependent differences in early anticoagulation response, because this has not been demonstrated conclusively in the warfarin pharmacogenetics field. Furthermore, several studies published to date have described the contribution of genetic variations to initial warfarin response variability in patients whose treatment was initiated with respective anticoagulation clinic regimens, likely with similar INR response–monitoring schedules as WRAPID.22,23,43 Thus, we believe that pharmacogenetics-based initiation, particularly with the use of loading doses, should result in a safe and similar rise to optimal anticoagulation responses among VKORC1 and CYP2C9 genotype groups. Supportive of the role of genotype-guided initiation for warfarin therapy, a recent study suggested that genotyping for patients in whom therapy was being initiated significantly reduced the hospitalization rate for bleeding or thromboembolic events compared with a control group.46 Furthermore, the results of 5 small RCTs (range from 38-200 patients) completed thus far largely suggest that pharmacogenetics-guided dosing improves warfarin response, in terms of more time spent within the therapeutic range and decreased bleeding events compared with standard dosing,44,45,47,48 with the exception of 1 study in orthopedic patients.49 In that RCT, patients were followed up for only 2-4 weeks, and daily INR monitoring in addition to a similar dose adjustment protocol between the 2 arms may have rendered the effect of genotype-guided dosing nonsignificant. Indeed, larger RCTs that incorporate models such as WRAPID are required to compare adverse event rates between standard and pharmacogenetics-guided dosing of warfarin-based anticoagulation.
To the best of our knowledge, this is the first prospective study to demonstrate the utility of a genotype-guided warfarin initiation algorithm for the minimization of widely recognized VKORC1 and CYP2C9 genotype-associated differences in anticoagulation response for both AF and VTE patients. The pharmacogenetics-based algorithm proposed here is feasible and effective for outpatient management of individuals requiring warfarin-based anti-coagulation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Cameron Ross, BSc; Matilde Leon-Ponte, PhD; Jennifer Clermont; and Barron Gin for their excellent technical assistance.
This study was funded by the Drug Innovation Fund established by the Ministry of Health and Long-Term Care. I.Y.G. is a recipient of an Ontario Graduate Scholarship and Canadian Institutes of Health Research Canada Graduate Scholarships.
Authorship
Contribution: I.Y.G., R.G.T., U.I.S., N.C., G.K.D., A.L., P.S.W., and R.B.K. designed the research study; I.Y.G., N.C., and S.L. were responsible for acquisition of data; I.Y.G., R.G.T., U.I.S., N.C., G.K.D., P.S.W., and R.B.K. analyzed and interpreted data; I.Y.G. and R.B.K. drafted the manuscript; I.Y.G., R.G.T., U.I.S., N.C., G.K.D., N.L., A.L., G.Z., D.R., C.M.S., M.R., MC, M.F., P.S.W., and R.B.K. were responsible for critical revision of the manuscript for important intellectual content; and I.Y.G. and G.Z. were responsible for statistical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard B. Kim, MD, University Hospital, ALL-144, 339 Windermere Road, London, ON N6A 5A5; e-mail: Richard.Kim@lhsc.on.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal