Abstract
Protein palmitoylation is a dynamic process that regulates membrane targeting of proteins and protein-protein interactions. We have previously demonstrated a critical role for protein palmitoylation in platelet activation and have identified palmitoylation machinery in platelets. Using a novel proteomic approach, Palmitoyl Protein Identification and Site Characterization, we have begun to characterize the human platelet palmitoylome. Palmitoylated proteins were enriched from membranes isolated from resting platelets using acyl-biotinyl exchange chemistry, followed by identification using liquid chromatography-tandem mass spectrometry. This global analysis identified > 1300 proteins, of which 215 met criteria for significance and represent the platelet palmitoylome. This collection includes 51 known palmitoylated proteins, 61 putative palmitoylated proteins identified in other palmitoylation-specific proteomic studies, and 103 new putative palmitoylated proteins. Of these candidates, we chose to validate the palmitoylation of triggering receptors expressed on myeloid cell (TREM)–like transcript-1 (TLT-1) as its expression is restricted to platelets and megakaryocytes. We determined that TLT-1 is a palmitoylated protein using metabolic labeling with [3H]palmitate and identified the site of TLT-1 palmitoylation as cysteine 196. The discovery of new platelet palmitoyl protein candidates will provide a resource for subsequent investigations to validate the palmitoylation of these proteins and to determine the role palmitoylation plays in their function.
Introduction
Platelets are key mediators of hemostasis and thrombosis, and their acute response at sites of vascular injury requires a rapid and complex series of signaling events. Posttranslational modifications, such as phosphorylation,1 have been shown to influence platelet-signaling pathways, thus regulating platelet function, and palmitoylation, which is also a reversible modification, has been proposed to play an analogous regulatory role.2,3 Palmitoylation is the most common form of S-acylation, which is the covalent attachment of long chain fatty acids via thioester bonds to cysteine residues.4 Palmitoylation involves the attachment of the 16-carbon saturated fatty acid palmitate. Although attachment of palmitate enhances the hydrophobicity of a protein, its function extends beyond that of a simple membrane anchor as palmitoylation has been shown to regulate protein trafficking, sorting, stability, and activity.5 Thus, the addition of this lipid group to a protein has functional consequences. Palmitoylation is unique among the different types of lipid modifications in that it is reversible and does not require a specific sequence motif. The reversible nature of palmitoylation functions as a regulatory mechanism or molecular switch, directing protein-lipid and protein-protein interactions6,7 and plays a role in the regulation of cellular responses to external stimuli.
We have previously shown that platelets possess palmitoylation machinery and require palmitoylation for activation because chemical inhibition of protein palmitoylation results in abrogation of platelet aggregation and decreased incorporation of platelets into thrombi in a murine laser-induced model of vascular injury.8 In resting platelets, we8 and others9-13 observe incorporation of [3H]-palmitate into platelet proteins, which indicates that this fatty acid is being cycled even in the resting state. Although palmitate accounts for 74% of the fatty acids linked to proteins by a thioester bond in the platelet,10 protein palmitoylation remains a poorly understood posttranslational modification in platelets. These observations prompted us to assess the global scope of palmitoylation in platelets.
Proteomics offers a powerful means to study the biology and biochemistry of the anucleate platelet.14,15 Recently, palmitoylomes in yeasts,16 rat neurons,17 and human prostate cancer cells18 have been described. In our efforts to define the platelet palmitoylome, we have purified and identified palmitoylated proteins from membranes of resting platelets using the Palmitoyl Protein Identification and Site Characterization (PalmPISC) method that we recently developed.18 In this study, we present a comprehensive identification of palmitoylated platelet proteins. This strategy identified 215 significantly enriched palmitoylated platelet protein candidates. Of these, 51 are known palmitoylated proteins and 61 were identified in proteomic studies of other cells with medium to high confidence, indicating that they are most likely palmitoylated. The remaining 103 palmitoyl protein candidates have not previously been shown to be palmitoylated. As proof of this concept, we validated the palmitoylation of triggering receptors expressed on myeloid cell (TREM)–like transcript-1 (TLT-1), a platelet- and megakaryocyte-specific protein, and identified its palmitoylation site. These experiments further our understanding of the scope of palmitoylation in platelets and will provide a basis for studying the dynamics and function of palmitoylation in these putative palmitoyl proteins.
Methods
Chemicals and reagents
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich. Complete, EDTA-free protease inhibitor cocktail (11873580001) was obtained from Roche Diagnostics. [3H]Yohimbine (NET 659250UC), guanosine triphosphate (GTP)[γ-35S] (NEG 030H250UC), and [3H]palmitic acid (NET043) were obtained from PerkinElmer Life and Analytical Sciences. Tris(2-carboxyethyl)phosphine (TCEP; 77720) and N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP; 21341) were obtained from Thermo Scientific. Coomassie Brilliant Blue and polyvinylidene fluoride (PVDF) membranes were obtained from Bio-Rad. Iodoacetamide (RPN6302) was purchased from GE Healthcare. MS-grade trypsin (V5280) was obtained from Promega. Anti–human TREML1/TLT-1 primary antibody (AF2394) was obtained from R&D Systems, and anti–human Cdc42 (610928) antibody was obtained from BD Transduction Laboratories. FITC-conjugated secondary antibodies were obtained from Pierce Biotechnology. Anti-Gβ1 primary antibody (sc-379) and Protein A/G PLUS-agarose (sc-2003) were obtained from Santa Cruz Biotechnology.
Platelet and platelet membrane preparation
Platelet-rich plasma that had outdated within 24 hours before use was obtained from the Beth Israel Deaconess Medical Center Blood Bank. Platelets were washed 3 times and assessed by flow cytometry using a P-selectin expression assay.8 Only platelet preparations demonstrating resting P-selectin levels comparable with resting fresh platelets were further processed. An aliquot of the selected platelets was tested to confirm normal P-selectin expression in response to 100μM SFLLRN. Resting, washed platelets were centrifuged, and platelet pellets were resuspended in ice-cold lysis buffer (5mM Tris-HCl, pH 7.5, and 5mM ethylene glyco-bis(b-aminoethyl ester)-N,N,N′,N′-tetraacetic acid) plus protease inhibitor cocktail to 4 mL/g wet weight. Platelets were lysed by successive rounds of sonication; and after removal of intact cells by low speed centrifugation, the lysates were pooled. Membranes were pelleted at 50 000g and flash frozen at −80°C. To assess the quality of the platelet membrane preparation, immunoblotting for the peripheral membrane protein Gβ1 was performed. To further evaluate the integrity of the membrane preparation, [3H]yohimbine binding and GTP[γ-35S] binding19 were evaluated. Only preparations in which the receptors within the membrane retained the ability to bind agonists and stimulate GTP turnover on stimulation were used for liquid chromatography-mass spectrometry (LC-MS) studies.
Platelet palmitoyl protein purification, separation, and trypsin digestion
Proteins were prepared from 100 mg of platelet membranes using our adaptation of acyl-biotinyl exchange (ABE) chemistry as previously described.18 Briefly, membrane proteins were denatured with SDS, reduced with 10mM TCEP, and alkylated with 50mM N-ethylmaleimide (NEM) to block nonpalmitoylated cysteines. After methanol/chloroform precipitations to remove excess NEM, proteins were treated with 0.75M hydroxylamine (HA) and 1mM biotin-HPDP to replace palmitoyl groups with biotinyl groups. The in vitro biotinylated (previously palmitoylated) proteins were enriched by streptavidin affinity purification, specifically eluted by TCEP, and concentrated by methanol/chloroform precipitation. Enriched proteins were separated on a 12% SDS-PAGE gel and stained with Coomassie Brilliant Blue. Each gel lane was cut into 5 slices before reduction with 10mM dithiothreitol, alkylation with 55mM iodoacetamide, and in-gel digestion with trypsin.18
Mass spectrometry
Tryptic peptides were analyzed by on-line nanoflow reversed-phase high performance liquid chromatography (Eksigent nanoLC-2D) connected to an LTQ Orbitrap mass spectrometer (Thermo Scientific) essentially as described.18 Samples were loaded onto an in-house packed C18 column (Magic C18, 5 μm, 200 Å, Michrom Bioresources) with 15-cm length and 100-μm inner diameter, and separated at ∼ 200 nL/min with 60-minute linear gradients from 5% to 35% acetonitrile in 0.2% formic acid. Survey spectra were acquired in the Orbitrap analyzer with the resolution set to a value of 30 000. Lock mass option was enabled in all measurements, and decamethylcyclopentasiloxane background ions (at m/z 371.10123) were used for real-time internal calibration as described previously.20 Up to 5 of the most intense multiply charged ions per cycle were fragmented and analyzed in the linear ion trap.
Database searching, spectral counting, and statistical analysis
The Thermo raw files were deposited at Tranche (https://proteomecommons.org/dataset.jsp?i = 76 216) and will be made publically accessible on publication. Raw data were analyzed using MaxQuant Version 1.0.13.13.21 The parameters were set as follows. In the Quant module, Singlets was selected; oxidation (M), acetyl (protein N-term), carbamidomethyl (C), and N-ethylmaleimide (C) were set as variable modifications; no fixed modifications were allowed; concatenated IPI human database Version 3.52 (74 190 forward sequences and 74 190 reverse sequences) downloaded from www.maxquant.org was used for database searching; all other parameters used were default values. In the Identify module, all parameters used were default values, except that maximal peptide posterior error probability was set as .05. False discovery rates for protein identification and peptide identification were both set at 1%. The relative protein abundance changes between the paired HA+ and HA− samples were determined using a label-free spectral counting approach.22 Statistical analysis was performed as previously described18 with minor modifications. Briefly, the spectral counts were merged over both biologic replicates, and all zeros were replaced with 1 in the merged dataset to avoid division by zero. Given that spectral counting is not accurate for proteins with very low spectral counts, statistical analysis was only performed on the proteins with a spectral count of at least 3. The protein-wise log2-transformed HA+/HA− ratios were calculated and then clustered using a Bayesian information criterion-based Gaussian mixture model. The resulting 2 Gaussian components represent the log-ratio distributions of protein sets largely dominated by contaminating proteins or by palmitoyl proteins. P values were computed based on the distribution of the contaminating protein-dominant dataset.
Western blot analysis
For immunoblotting, separated proteins were transferred to PVDF membranes and blocked in TBS/0.1% Tween-20/5% milk for 1 hour at room temperature. Blots were probed with anti–human TREML1/TLT-1 primary antibody (1:1000) or anti–human Cdc42 (1:500) overnight at 4°C followed by 1-hour incubation at room temperature with FITC-conjugated secondary antibodies (1:1000) to detect TLT-1 or Cdc42. Blots were analyzed with an Amersham Typhoon 9400 molecular imager.
Labeling platelet proteins with [3H]palmitate and immunoprecipitation of TLT-1
Washed platelets (2 mL at 1 × 109 platelets/mL) were radiolabeled with 100 μCi/mL [3H]palmitic acid for 1 hour at 37°C, in PIPES/NaCl buffer with 3.6 mg/mL BSA. Platelets were activated as described in the text, diluted 5 times with PIPES/NaCl buffer, and pelleted to remove unincorporated [3H]palmitic acid. Platelets were then resuspended in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 158mM NaCl, 10mM Tris-HCl, pH 7.6, 1mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) and allowed to lyse on ice for 5 minutes. The lysate was centrifuged for 5 minutes at 16 000g, and the supernatant was used as the RIPA soluble fraction. The platelet lysate was precleared by adding 20 μL of Protein A/G beads and incubating for 30 minutes at 4°C with end-over-end rotation. Beads were pelleted, and TLT-1 was immunoprecipitated from the supernatant using 2.5 mg/mL human TREML1/TLT-1 primary antibody overnight at 4°C with end-over-end rotation. Protein/immune complexes were pulled out by addition of 20 μL of Protein A/G beads and incubation for 60 minutes at 4°C with end-over-end rotation. Beads were then washed 3 times with 1 mL of RIPA buffer, and immunoprecipitates were eluted from the beads by the addition of 20 μL SDS sample buffer. [3H]Palmitate-labeled platelet proteins were then separated by SDS-PAGE and transferred onto a PVDF membrane, which was exposed to a tritium detection screen for 2 weeks and then analyzed using an Amersham Typhoon 9400 molecular imager. Tritium blot band intensities were normalized for the amount of protein loaded.
Identification of TLT-1 palmitoylation site
Enriched palmitoyl proteins were separated by SDS-PAGE. Because the apparent molecular weight of TLT-1 is ∼ 37 kDa, a gel slice containing 30- to 50-kDa proteins was excised. Proteins were tryptically digested in gel without reduction and alkylation. Tryptic peptides were extracted and analyzed by LC tandem MS (LC-MS/MS) as described in “Mass spectrometry.” Free cysteines in the identified peptides are candidate palmitoylation sites because nonpalmitoylated cysteines were blocked by NEM before ABE reaction.
Results
Purification and identification of palmitoylated platelet proteins
Palmitoylated proteins were enriched from platelet membranes that were prepared from resting platelets pooled from 6 aphaeresis packs, and purification of palmitoylated proteins was carried out using our adaption of ABE chemistry.18 After extraction of platelet membrane proteins, all disulfide bonds were reduced with TCEP, a reducing agent that does not cleave thioester bonds, and free thiols were blocked with the alkylating agent, NEM. The sample was split into 2 groups, which were treated with parallel protocols: an experimental group in which the thioester bonds were specifically cleaved with neutral HA+ and a control group (HA−), which was not treated with hydroxylamine. The HA− group represents background binding to the steptavidin beads, which may be the result of endogenous biotinylation, nonspecific biotinylation, or nonspecific binding. After the newly formed thiols were labeled with biotin-HPDP, biotinylated proteins were enriched by streptavidin affinity chromatography and eluted with TCEP. This approach enriched for a large number of proteins (Figure 1A) that were separated and visualized by SDS-PAGE followed by in gel digestion and LC-MS/MS.
Analysis of ABE-purified palmitoylated proteins from platelet membranes. (A) Electrophoretic analysis. Lane 1 indicates molecular weight marker; lane 2, experimental samples (HA+) with hydroxylamine treatment; and lane 3, control samples (HA−) without hydroxylamine treatment. (B) The correlation between proteins identified in 2 separate rounds of spectral counting analysis. Data were fit with a linear function (R2 = 0.8645). (C) Distribution of log(HA+/HA−) ratios. Bayesian Information Criterion-based Gaussian Mixture modeling suggested that the distribution of log-ratios (solid line) is composed of 2 Guassian components (dashed line). The left Gaussian represents the log-ratio distribution of most contaminating proteins and some palmitoyl proteins, whereas the right Guassian represents the log-ratio distribution of most palmitoyl proteins and a small number of contaminants. To distinguish palmitoyl proteins from contaminating proteins, P values were calculated based on the distribution of the left Gaussian. Proteins with P < .05 were treated as significant and accepted as palmitoyl protein candidates.
Analysis of ABE-purified palmitoylated proteins from platelet membranes. (A) Electrophoretic analysis. Lane 1 indicates molecular weight marker; lane 2, experimental samples (HA+) with hydroxylamine treatment; and lane 3, control samples (HA−) without hydroxylamine treatment. (B) The correlation between proteins identified in 2 separate rounds of spectral counting analysis. Data were fit with a linear function (R2 = 0.8645). (C) Distribution of log(HA+/HA−) ratios. Bayesian Information Criterion-based Gaussian Mixture modeling suggested that the distribution of log-ratios (solid line) is composed of 2 Guassian components (dashed line). The left Gaussian represents the log-ratio distribution of most contaminating proteins and some palmitoyl proteins, whereas the right Guassian represents the log-ratio distribution of most palmitoyl proteins and a small number of contaminants. To distinguish palmitoyl proteins from contaminating proteins, P values were calculated based on the distribution of the left Gaussian. Proteins with P < .05 were treated as significant and accepted as palmitoyl protein candidates.
LC-MS/MS and MaxQuant analyses of 2 biologic replicates of enriched palmitoyl proteins resulted in the identification of > 1300 proteins (supplemental Table 3, see the Supplemental Materials link at the top of the article), with a false discovery rate of 1% for both proteins and peptides. Plotting both rounds of spectral counting against each other revealed good agreement between experimental datasets (R2 = 0.8645; Figure 1B). Label-free spectral counting quantitation and statistical analysis showed that 215 nonredundant proteins were significantly enriched (P < .05) in the HA+ sample over that of the HA− control (Figure 1C). To ensure the accuracy of protein identification based on a single unique peptide, the MS/MS spectra of all platelet palmitoyl protein candidates were manually verified (supplemental Figure 1). Proteins that were considered significantly enriched had an HA+/HA− ratio > 3 (P < .05); and of the 215 enriched proteins, 103 proteins are not known to be palmitoylated or have not been described in other palmitoyl proteomic studies (Table 1). The remaining 112 identified proteins are known to be palmitoylated or are palmitoyl protein candidates identified in other proteomic studies (Table 2).
Novel candidates of the platelet palmitoylome
| Protein name . | Gene name . | Ratio . | P . |
|---|---|---|---|
| Apoptosis | |||
| Myc target protein 1 | MYCT1 | 3.10 | .039094 |
| Placenta-derived apoptotic factor | PDAF | 4.00 | .015866 |
| Cell growth and/or maintanence | |||
| Tetraspanin-33 | TSPAN33 | 6.25 | .002368 |
| Claudin-5 | CLDN5 | 3.33 | .030650 |
| Dynein heavy chain 1, axonemal | DNAH1 | 6.00 | .002867 |
| Metalloproteinase inhibitor 1 | TIMP1 | 3.29 | .032192 |
| Lysophosphatidylcholine acyltransferase 1 | LPCAT1 | 4.00 | .015866 |
| Transmembrane protein 97 | TMEM97 | 3.00 | .043487 |
| DNA repair | |||
| Histone H2A | H2AFX | 3.00 | .043487 |
| Immune response | |||
| CKLF-like MARVEL transmembrane domain-containing protein 3 | CMTM3 | 11.00 | .000116 |
| CKLF-like MARVEL transmembrane domain-containing protein 5 | CMTM5 | 7.43 | .001013 |
| Early activation antigen CD69 | CD69 | 5.00 | .006457 |
| Minor histocompatibility protein HA-1 | HMHA1 | 3.00 | .043487 |
| Ion transport | |||
| Calcium release-activated calcium channel protein 1 | ORAI1 | 5.00 | .006457 |
| Metabolism | |||
| [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase | BCKDK | 4.00 | .015866 |
| α-amylase 1 | AMY1A | 3.00 | .043487 |
| ATPase family AAA domain-containing protein 3A | ATAD3A | 3.00 | .043487 |
| Catechol-O-methyltransferase | COMT | 4.50 | .010000 |
| Dihydrolipoamide branched chain transacylase E2 | DBT2 | 8.00 | .000690 |
| Dolichyldiphosphatase 1 | DOLPP1 | 3.00 | .043487 |
| GPI ethanolamine phosphate transferase 1 | PIGN | 3.00 | .043487 |
| Probable cysteinyl-tRNA synthetase | CARS2 | 4.00 | .015866 |
| Protein disulfide-isomerase TMX3 | TMX3 | 15.00 | .000016 |
| Suppressor of Lec15 | MPDU1 | 3.00 | .043487 |
| Thioredoxin-related transmembrane protein 1 | TMX1 | 3.45 | .027084 |
| Thioredoxin-related transmembrane protein 4 | TMX4 | 6.00 | .002867 |
| V-type proton ATPase 116-kDa subunit α isoform 2 | ATP6V0A2 | 3.00 | .043487 |
| Protein metabolism | |||
| 26S proteasome non-ATPase regulatory subunit 11 | PSMD11 | 6.00 | .002867 |
| CAAX prenyl protease 1 homolog | ZMPSTE24 | 4.00 | .015866 |
| Copper chaperone for superoxide dismutase | CCS | 4.00 | .015866 |
| Cystatin-S | CST4 | 3.00 | .043487 |
| E3 ubiquitin-protein ligase MARCH2 | MARCH | 6.00 | .002867 |
| Eukaryotic translation elongation factor 1 ϵ-1 | EEF1E1 | 3.00 | .043487 |
| Eukaryotic translation initiation factor 5A-1 | EIF5A | 6.00 | .002867 |
| GrpE protein homolog 1 | GRPEL1 | 3.00 | .043487 |
| Leucyl/cystinyl aminopeptidase | LNPEP | 4.00 | .015866 |
| RING finger protein 11 | RNF11 | 3.50 | .025869 |
| Signal transduction | |||
| Atlastin-1 | ATL1 | 5.00 | .006457 |
| CKLF-like MARVEL transmembrane domain-containing protein 7 | CMTM7 | 4.00 | .015866 |
| Endothelial cells scavenger receptor | SCARF1 | 10.00 | .000202 |
| Filaggrin | FLG | 3.00 | .043487 |
| GTP-binding protein Rheb | RHEB | 4.00 | .015866 |
| Leucine-rich repeat-containing protein 32 | LRRC32 | 21.00 | .000002 |
| Metalloreductase STEAP3 | STEAP3 | 6.00 | .002867 |
| Nuclear receptor subfamily 4 group A member 2 | NR4A2 | 3.00 | .043487 |
| Platelet-derived endothelial cell growth factor | ECGF1 | 3.00 | .043487 |
| Rho-related GTP-binding protein RhoQ | RHOQ | 3.00 | .043487 |
| Tescalcin | TESC | 18.00 | .000005 |
| Tetraspanin-15 | TSPAN15 | 17.00 | .000007 |
| Transmembrane protein 11 | TMEM11 | 7.00 | .001366 |
| Transmembrane protein 50A | TMEM50A | 3.25 | .033406 |
| Transmembrane protein 55A | TMEM55A | 3.83 | .018614 |
| Transcription regulator activity | |||
| 39S ribosomal protein L12 | MRPL12 | 3.00 | .043487 |
| Oxidoreductase HTATIP2 | HTATIP2 | 3.00 | .043487 |
| Transport | |||
| BET1 homolog | BET1 | 10.00 | .000202 |
| BET1-like protein | BET1L | 11.00 | .000116 |
| Cholesteryl ester transfer protein | CETP | 4.00 | .015866 |
| MLN64 N-terminal domain homolog | STARD3NL | 4.00 | .015866 |
| Nonspecific lipid-transfer protein | SCP2 | 9.00 | .000366 |
| Proteolipid protein 2 | PLP2 | 3.00 | .043487 |
| SID1 transmembrane family member 1 | SIDT1 | 4.00 | .015866 |
| Sodium/hydrogen exchanger 9 | SLC9A9 | 3.67 | .021908 |
| Solute carrier family 43 member 3 | SLC43A3 | 5.00 | .006457 |
| Syntaxin-2 | STX2 | 3.00 | .043487 |
| Syntaxin-10 | STX10 | 4.00 | .015866 |
| Syntaxin-11 | STX11 | 6.18 | .002507 |
| Tandem C2 domains nuclear protein | TC2N | 42.00 | .000000 |
| Vesicle transport protein SFT2C | SFT2D3 | 6.00 | .002867 |
| Vesicle-associated membrane protein 5 | VAMP5 | 15.00 | .000016 |
| Unknown | |||
| Catechol-O-methyltransferase domain-containing protein 1 | COMTD1 | 3.50 | .025869 |
| CD99 antigen-like protein 2 | CD99L2 | 3.00 | .043487 |
| Chronic lymphocytic leukemia deletion region gene 6 protein | CLLD6 | 4.50 | .010000 |
| COMM domain-containing protein 9 | COMMD9 | 3.00 | .043487 |
| Cyclin-Y | CCNY | 9.00 | .000366 |
| Exocyst complex component 3-like protein 2 | EXOC3L2 | 4.00 | .015866 |
| Leptin receptor gene-related protein | LEPROT | 6.00 | .002867 |
| Lipoma HMGIC fusion partner | LHFP | 8.00 | .000690 |
| Lipoma HMGIC fusion partner-like 2 protein | LHFPL2 | 6.40 | .002115 |
| LITAF-like protein | CDIP | 6.00 | .002867 |
| Major facilitator superfamily domain-containing protein 6 | MFSD6 | 4.00 | .015866 |
| Malectin | MLEC | 4.90 | .007035 |
| Metallo-β-lactamase domain-containing protein 2 | MBLAC2 | 12.00 | .000068 |
| Mps one binder kinase activator-like 3 | MOBKL3 | 5.00 | .006457 |
| Oligosaccharyltransferase complex subunit OSTC | OSTC | 3.00 | .043487 |
| PDZK1-interacting protein 1 | PDZK1IP1 | 8.00 | .000690 |
| Prolactin-inducible protein | PIP | 4.00 | .015866 |
| Protein EFR3 homolog A | EFR3A | 9.67 | .000245 |
| Protein FAM78A | FAM78A | 3.00 | .043487 |
| Protein S100-A14 | S100A14 | 3.00 | .043487 |
| Putative uncharacterized protein C1orf150 | C1orf150 | 10.00 | .000202 |
| Putative uncharacterized protein C1orf95 | C1orf95 | 3.00 | .043487 |
| SEC6-like protein C14orf73 | C14orf73 | 24.00 | .000001 |
| Small VCP/p97-interacting protein | SVIP | 3.00 | .043487 |
| Tetraspanin-14 | TSPAN14 | 9.80 | .000227 |
| Transmembrane BAX inhibitor motif-containing protein 1 | TMBIM1 | 6.00 | .002867 |
| Transmembrane protein 222 | TMEM222 | 10.00 | .000202 |
| Transmembrane protein 50B | TMEM50B | 8.00 | .000690 |
| Transmembrane protein with metallophosphoesterase domain | TMPPE | 5.00 | .006457 |
| Uncharacterized protein C22orf25 | C22orf25 | 7.00 | .001366 |
| Uncharacterized protein KIAA2013 | KIAA2013 | 7.00 | .001366 |
| UPF0389 protein FAM162A | FAM162A | 8.50 | .000500 |
| UPF0598 protein C8orf82 | C8orf82 | 11.00 | .003588 |
| UPF0733 protein C2orf88 | C2orf88 | 8.00 | .000116 |
| Protein name . | Gene name . | Ratio . | P . |
|---|---|---|---|
| Apoptosis | |||
| Myc target protein 1 | MYCT1 | 3.10 | .039094 |
| Placenta-derived apoptotic factor | PDAF | 4.00 | .015866 |
| Cell growth and/or maintanence | |||
| Tetraspanin-33 | TSPAN33 | 6.25 | .002368 |
| Claudin-5 | CLDN5 | 3.33 | .030650 |
| Dynein heavy chain 1, axonemal | DNAH1 | 6.00 | .002867 |
| Metalloproteinase inhibitor 1 | TIMP1 | 3.29 | .032192 |
| Lysophosphatidylcholine acyltransferase 1 | LPCAT1 | 4.00 | .015866 |
| Transmembrane protein 97 | TMEM97 | 3.00 | .043487 |
| DNA repair | |||
| Histone H2A | H2AFX | 3.00 | .043487 |
| Immune response | |||
| CKLF-like MARVEL transmembrane domain-containing protein 3 | CMTM3 | 11.00 | .000116 |
| CKLF-like MARVEL transmembrane domain-containing protein 5 | CMTM5 | 7.43 | .001013 |
| Early activation antigen CD69 | CD69 | 5.00 | .006457 |
| Minor histocompatibility protein HA-1 | HMHA1 | 3.00 | .043487 |
| Ion transport | |||
| Calcium release-activated calcium channel protein 1 | ORAI1 | 5.00 | .006457 |
| Metabolism | |||
| [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase | BCKDK | 4.00 | .015866 |
| α-amylase 1 | AMY1A | 3.00 | .043487 |
| ATPase family AAA domain-containing protein 3A | ATAD3A | 3.00 | .043487 |
| Catechol-O-methyltransferase | COMT | 4.50 | .010000 |
| Dihydrolipoamide branched chain transacylase E2 | DBT2 | 8.00 | .000690 |
| Dolichyldiphosphatase 1 | DOLPP1 | 3.00 | .043487 |
| GPI ethanolamine phosphate transferase 1 | PIGN | 3.00 | .043487 |
| Probable cysteinyl-tRNA synthetase | CARS2 | 4.00 | .015866 |
| Protein disulfide-isomerase TMX3 | TMX3 | 15.00 | .000016 |
| Suppressor of Lec15 | MPDU1 | 3.00 | .043487 |
| Thioredoxin-related transmembrane protein 1 | TMX1 | 3.45 | .027084 |
| Thioredoxin-related transmembrane protein 4 | TMX4 | 6.00 | .002867 |
| V-type proton ATPase 116-kDa subunit α isoform 2 | ATP6V0A2 | 3.00 | .043487 |
| Protein metabolism | |||
| 26S proteasome non-ATPase regulatory subunit 11 | PSMD11 | 6.00 | .002867 |
| CAAX prenyl protease 1 homolog | ZMPSTE24 | 4.00 | .015866 |
| Copper chaperone for superoxide dismutase | CCS | 4.00 | .015866 |
| Cystatin-S | CST4 | 3.00 | .043487 |
| E3 ubiquitin-protein ligase MARCH2 | MARCH | 6.00 | .002867 |
| Eukaryotic translation elongation factor 1 ϵ-1 | EEF1E1 | 3.00 | .043487 |
| Eukaryotic translation initiation factor 5A-1 | EIF5A | 6.00 | .002867 |
| GrpE protein homolog 1 | GRPEL1 | 3.00 | .043487 |
| Leucyl/cystinyl aminopeptidase | LNPEP | 4.00 | .015866 |
| RING finger protein 11 | RNF11 | 3.50 | .025869 |
| Signal transduction | |||
| Atlastin-1 | ATL1 | 5.00 | .006457 |
| CKLF-like MARVEL transmembrane domain-containing protein 7 | CMTM7 | 4.00 | .015866 |
| Endothelial cells scavenger receptor | SCARF1 | 10.00 | .000202 |
| Filaggrin | FLG | 3.00 | .043487 |
| GTP-binding protein Rheb | RHEB | 4.00 | .015866 |
| Leucine-rich repeat-containing protein 32 | LRRC32 | 21.00 | .000002 |
| Metalloreductase STEAP3 | STEAP3 | 6.00 | .002867 |
| Nuclear receptor subfamily 4 group A member 2 | NR4A2 | 3.00 | .043487 |
| Platelet-derived endothelial cell growth factor | ECGF1 | 3.00 | .043487 |
| Rho-related GTP-binding protein RhoQ | RHOQ | 3.00 | .043487 |
| Tescalcin | TESC | 18.00 | .000005 |
| Tetraspanin-15 | TSPAN15 | 17.00 | .000007 |
| Transmembrane protein 11 | TMEM11 | 7.00 | .001366 |
| Transmembrane protein 50A | TMEM50A | 3.25 | .033406 |
| Transmembrane protein 55A | TMEM55A | 3.83 | .018614 |
| Transcription regulator activity | |||
| 39S ribosomal protein L12 | MRPL12 | 3.00 | .043487 |
| Oxidoreductase HTATIP2 | HTATIP2 | 3.00 | .043487 |
| Transport | |||
| BET1 homolog | BET1 | 10.00 | .000202 |
| BET1-like protein | BET1L | 11.00 | .000116 |
| Cholesteryl ester transfer protein | CETP | 4.00 | .015866 |
| MLN64 N-terminal domain homolog | STARD3NL | 4.00 | .015866 |
| Nonspecific lipid-transfer protein | SCP2 | 9.00 | .000366 |
| Proteolipid protein 2 | PLP2 | 3.00 | .043487 |
| SID1 transmembrane family member 1 | SIDT1 | 4.00 | .015866 |
| Sodium/hydrogen exchanger 9 | SLC9A9 | 3.67 | .021908 |
| Solute carrier family 43 member 3 | SLC43A3 | 5.00 | .006457 |
| Syntaxin-2 | STX2 | 3.00 | .043487 |
| Syntaxin-10 | STX10 | 4.00 | .015866 |
| Syntaxin-11 | STX11 | 6.18 | .002507 |
| Tandem C2 domains nuclear protein | TC2N | 42.00 | .000000 |
| Vesicle transport protein SFT2C | SFT2D3 | 6.00 | .002867 |
| Vesicle-associated membrane protein 5 | VAMP5 | 15.00 | .000016 |
| Unknown | |||
| Catechol-O-methyltransferase domain-containing protein 1 | COMTD1 | 3.50 | .025869 |
| CD99 antigen-like protein 2 | CD99L2 | 3.00 | .043487 |
| Chronic lymphocytic leukemia deletion region gene 6 protein | CLLD6 | 4.50 | .010000 |
| COMM domain-containing protein 9 | COMMD9 | 3.00 | .043487 |
| Cyclin-Y | CCNY | 9.00 | .000366 |
| Exocyst complex component 3-like protein 2 | EXOC3L2 | 4.00 | .015866 |
| Leptin receptor gene-related protein | LEPROT | 6.00 | .002867 |
| Lipoma HMGIC fusion partner | LHFP | 8.00 | .000690 |
| Lipoma HMGIC fusion partner-like 2 protein | LHFPL2 | 6.40 | .002115 |
| LITAF-like protein | CDIP | 6.00 | .002867 |
| Major facilitator superfamily domain-containing protein 6 | MFSD6 | 4.00 | .015866 |
| Malectin | MLEC | 4.90 | .007035 |
| Metallo-β-lactamase domain-containing protein 2 | MBLAC2 | 12.00 | .000068 |
| Mps one binder kinase activator-like 3 | MOBKL3 | 5.00 | .006457 |
| Oligosaccharyltransferase complex subunit OSTC | OSTC | 3.00 | .043487 |
| PDZK1-interacting protein 1 | PDZK1IP1 | 8.00 | .000690 |
| Prolactin-inducible protein | PIP | 4.00 | .015866 |
| Protein EFR3 homolog A | EFR3A | 9.67 | .000245 |
| Protein FAM78A | FAM78A | 3.00 | .043487 |
| Protein S100-A14 | S100A14 | 3.00 | .043487 |
| Putative uncharacterized protein C1orf150 | C1orf150 | 10.00 | .000202 |
| Putative uncharacterized protein C1orf95 | C1orf95 | 3.00 | .043487 |
| SEC6-like protein C14orf73 | C14orf73 | 24.00 | .000001 |
| Small VCP/p97-interacting protein | SVIP | 3.00 | .043487 |
| Tetraspanin-14 | TSPAN14 | 9.80 | .000227 |
| Transmembrane BAX inhibitor motif-containing protein 1 | TMBIM1 | 6.00 | .002867 |
| Transmembrane protein 222 | TMEM222 | 10.00 | .000202 |
| Transmembrane protein 50B | TMEM50B | 8.00 | .000690 |
| Transmembrane protein with metallophosphoesterase domain | TMPPE | 5.00 | .006457 |
| Uncharacterized protein C22orf25 | C22orf25 | 7.00 | .001366 |
| Uncharacterized protein KIAA2013 | KIAA2013 | 7.00 | .001366 |
| UPF0389 protein FAM162A | FAM162A | 8.50 | .000500 |
| UPF0598 protein C8orf82 | C8orf82 | 11.00 | .003588 |
| UPF0733 protein C2orf88 | C2orf88 | 8.00 | .000116 |
To date, these 103 proteins have not been shown to be palmitoylated and represent putative palmitoylated platelet proteins. Data are the result of 2 independent experiments, and proteins are listed according to the biologic process they mediate as defined by the HPRD. Also shown is the HA+/HA− ratio and corresponding P value.
Known palmitoylated proteins of the platelet palmitoylome
| Protein name . | Gene name . | Ratio . | P . | Source . |
|---|---|---|---|---|
| Cell growth and/or maintenance | ||||
| Claudin-3 | CLDN3 | 16.50 | .000009 | Proteomic studies18 |
| Desmoplakin | DSP | 2.90 | .048440 | Proteomic studies17 |
| Flotillin-1 | FLOT1 | 14.80 | .000018 | Manual |
| 55-kDa erythrocyte membrane protein | MPP1 | 3.00 | .043487 | Uniprot |
| Immune response | ||||
| Complement component 1 Q subcomponent-binding protein | C1QBP | 3.00 | .043487 | Proteomic studies17 |
| HLA class I histocompatibility antigen, B-27 α-chain | HLA-B | 9.00 | .000366 | Manual |
| Interferon-induced transmembrane protein 3 | IFITM3 | 3.00 | .043487 | Uniprot |
| Linker for activation of T-cell family member 1 | LAT | 7.33 | .001082 | Uniprot |
| Protein folding | ||||
| Calnexin | CANX | 2.88 | .049774 | Proteomic studies18 |
| Metabolism | ||||
| 3-mercaptopyruvate sulfur transferase | MPST | 3.33 | .030650 | Proteomic studies17 |
| 4-aminobutyrate aminotransferase | ABAT | 4.00 | .015866 | Proteomic studies17 |
| ADP-ribosyl cyclase 1 | CD38 | 5.00 | .006457 | Proteomic studies17 |
| Acid ceramidase | ASAH1 | 5.25 | .005232 | Proteomic studies18 |
| ATP synthase subunit γ | ATP5L | 4.00 | .015866 | Proteomic studies17 |
| CTD small phosphatase-like protein | CTDSPL | 8.25 | .000586 | Proteomic studies18 |
| Cytochrome b-c1 complex subunit Rieske | UQCRFS1 | 3.00 | .043487 | Proteomic studies17 |
| Glutaminase kidney isoform | GLS | 3.00 | .043487 | Proteomic studies17 |
| NAD(P) transhydrogenase | NNT | 7.92 | .000726 | Proteomic studies17 |
| Probable phospholipid-transporting ATPase IF | ATP11B | 11.00 | .000116 | Proteomic studies18 |
| Sn1-specific diacylglycerol lipase-β | DAGLB | 29.00 | 1.29E-07 | Proteomic studies18 |
| Protein metabolism | ||||
| Cation-dependent mannose-6-phosphate receptor | M6PR | 7.00 | .001366 | Manual |
| DnaJ homolog subfamily C member 5 | DNAJC5 | 27.00 | 2.29E-07 | Uniprot |
| Endothelin-converting enzyme 1 | ECE1 | 35.00 | 2.72E-08 | Manual |
| F-box/LRR-repeat protein 20 | FBXL20 | 6.00 | .002867 | Proteomic studies18 |
| Signal transduction | ||||
| Adenylate cyclase type 6 | ADCY6 | 3.40 | .028628 | Proteomic studies17 |
| ADP-ribosylation factor 5 | ARF5 | 3.00 | .043487 | Proteomic studies17 |
| ADP-ribosylation factor-like protein 15 | ARL15 | 5.25 | .005232 | Proteomic studies18 |
| Casein kinase I γ1 isoform | CSNK1G1 | 3.00 | .043487 | Proteomic studies17 |
| Casein kinase I isoform γ3 | CSNK1G3 | 8.00 | .000690 | Proteomic studies17 |
| CD151 antigen | CD151 | 3.00 | .043487 | Uniprot |
| CD63 antigen | CD63 | 3.67 | .021908 | Manual |
| CD82 antigen | CD82 | 4.00 | .015866 | Manual |
| CDC42 small effector protein 1 | CDC42SE1 | 3.00 | .043487 | Uniprot |
| CDC42 small effector protein 2 | CDC42SE2 | 9.00 | .000366 | Uniprot |
| Cell division control protein 42 homolog | CDC42 | 3.00 | .043487 | Manual |
| Choline transporter-like protein 2 | CTL2 | 27.00 | 2.29E-07 | Proteomic studies18 |
| Disks large-associated protein 4 | DLGAP4 | 6.00 | .002867 | Proteomic studies17 |
| Disheveled-associated activator of morphogenesis 1 | DAAM1 | 9.00 | .000366 | Proteomic studies17 |
| Dystroglycan | DAG1 | 4.00 | .015866 | Proteomic studies17 |
| Erbb2-interacting protein | ERBB2IP | 8.88 | .000395 | Manual |
| Flotillin-2 | FLOT2 | 10.55 | .000149 | Manual |
| G protein-coupled receptor kinase 6 | GRK6 | 8.00 | .000690 | Probable |
| G(i) α-1 | GNAI1 | 3.80 | .019226 | HPRD |
| G(i) α-2 | GNAI2 | 3.25 | .033301 | Manual |
| G(i) α-3 | GNAI3 | 4.86 | .007300 | HPRD |
| G(q) α | GNAQ | 5.17 | .005609 | HPRD |
| G(s) α | GNAS | 5.80 | .003352 | Manual |
| Gα-11 | GNA11 | 6.00 | .002867 | HPRD |
| Gα-13 | GNA13 | 7.45 | .000998 | Uniprot |
| Gα-15 | GNA15 | 4.00 | .015866 | Manual |
| GTPase Hras | HRAS | 7.00 | .001366 | Uniprot |
| GTPase Nras | NRAS | 5.25 | .005232 | Uniprot |
| Junctional adhesion molecule C | JAM3 | 5.14 | .005722 | Proteomic studies17 |
| Kalirin | KALRN | 7.50 | .000965 | Proteomic studies17 |
| Linker for activation of T-cell family member 2 | LAT2 | 7.00 | .001366 | Probable |
| Mitochondrial import inner membrane translocase subunit TIM50 | TIMM50 | 4.00 | .015866 | Proteomic studies24 |
| Phosphatidylinositol 4-kinase type 2-α | PI4K2A | 10.00 | .000202 | Manual |
| Phosphatidylinositol 4-kinase type 2-β | PI4K2B | 3.00 | .043487 | Manual |
| Phospholipid scramblase 1 | PLSCR1 | 20.50 | .000002 | Potential |
| Platelet glycoprotein 4 | CD36 | 6.14 | .002569 | Uniprot |
| Prostacyclin receptor | PTGIR | 10.00 | .000202 | Uniprot |
| Proto-oncogene tyrosine-protein kinase Fyn | FYN | 4.91 | .006980 | HPRD |
| Proto-oncogene tyrosine-protein kinase Yes | YES1 | 10.00 | .000202 | Proteomic studies18 |
| Raftlin | RFTN1 | 5.00 | .006457 | Probable |
| Ras-related protein Rap-2a | RAP2A | 28.00 | .000000 | by similarity |
| Ras-related protein Rap-2b | RAP2B | 3.93 | .017042 | by similarity |
| Ras-related protein Rap-2c | RAP2C | 11.50 | .000089 | by similarity |
| Ras-related protein R-Ras | RRAS | 7.44 | .001002 | Manual |
| Regulator of G-protein signaling 19 | RGS19 | 15.00 | .000016 | Uniprot |
| Semaphorin-4D | SEMA4D | 6.00 | .002867 | Proteomic studies17 |
| Sortilin | SORT1 | 3.00 | .043487 | Proteomic studies17 |
| Stomatin | STOM | 3.29 | .031966 | Uniprot |
| Tetraspanin-9 | TSPAN9 | 6.67 | .001737 | Proteomic studies18 |
| Thromboxane A2 receptor | TBXA2R | 4.00 | .015866 | Manual |
| Transmembrane protein 55B | TMEM55B | 3.50 | .025869 | Proteomic studies17 |
| Type I inositol-1,4,5-trisphosphate 5-phosphatase | INPP5A | 10.67 | .000139 | Potential |
| Tyrosine-protein kinase Lyn | LYN | 5.00 | .006457 | by similarity |
| Transcription regulator activity | ||||
| Histone H2B type 1-N | HIST1H2BN | 6.00 | .002867 | Proteomic studies17 |
| Transport | ||||
| AFG3-like protein 2 | AFG3L2 | 4.00 | .015866 | Proteomic studies17 |
| ATP-binding cassette subfamily B member 6 | ABCB6 | 5.00 | .006457 | Proteomic studies18 |
| Choline transporter-like protein 1 | CTL1 | 14.20 | .000024 | Proteomic studies18 |
| Cytochrome b5 type B | CYB5B | 3.67 | .021908 | Proteomic studies17 |
| Golgin subfamily A member 7 | GOLGA7 | 17.00 | .000007 | Uniprot |
| Multidrug resistance-associated protein 4 | ABCC4 | 3.00 | .043487 | Proteomic studies18 |
| Phospholipid scramblase 3 | PLSCR3 | 13.00 | .000042 | Probable |
| Phospholipid scramblase 4 | PLSCR4 | 7.00 | .001366 | Probable |
| Pituitary tumor-transforming gene 1 protein-interacting protein | PTTG1IP | 19.50 | .000003 | Proteomic studies18 |
| PRA1 family protein 2 | PRAF2 | 14.00 | .000026 | Proteomic studies17 |
| Protein tweety homolog 3 | TTYH3 | 11.00 | .000116 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 1 | SCAMP1 | 10.25 | .000175 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 2 | SCAMP2 | 3.95 | .016681 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 3 | SCAMP3 | 16.00 | .000011 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 4 | SCAMP4 | 3.00 | .043487 | Proteomic studies17 |
| Sodium/potassium-transporting ATPase subunit α-1 | ATP1A1 | 3.00 | .043487 | Proteomic studies17 |
| SNAP-23 | SNAP23 | 3.74 | .020385 | HPRD |
| Syntaxin-8 | STX8 | 11.00 | .000116 | Manual |
| Syntaxin-12 | STX12 | 4.00 | .015866 | Proteomic studies17 |
| Trafficking protein particle complex subunit 3 | TRAPPC3 | 8.67 | .000450 | by similarity |
| Transferrin receptor protein 1 | TFRC | 3.00 | .043487 | Uniprot |
| Vesicle-associated membrane protein 3 | VAMP3 | 8.43 | .000523 | Proteomic studies18 |
| Vesicle-associated membrane protein 4 | VAMP4 | 10.00 | .000202 | Proteomic studies17 |
| Vesicle-associated membrane protein 7 | VAMP7 | 3.69 | .021361 | Proteomic studies18 |
| Unknown | ||||
| 3-oxoacyl-[acyl-carrier-protein] synthase | OXSM | 6.00 | .002867 | Proteomic studies17 |
| Abhydrolase domain-containing protein FAM108B1 | FAM108B1 | 8.33 | .000556 | Proteomic studies18 |
| Coiled-coil domain-containing protein 109A | CCDC109A | 4.50 | .010000 | Proteomic studies17 |
| Endoplasmic reticulum-Golgi intermediate compartment protein 3 | ERGIC3 | 3.00 | .043487 | Proteomic studies17 |
| Abhydrolase domain-containing protein FAM108A1 | FAM108A1 | 7.50 | .000965 | Proteomic studies23 |
| Protein FAM49B | FAM49B | 21.00 | .000002 | Proteomic studies18 |
| Protein LYRIC gene-1 protein | LYRIC | 18.00 | .000005 | Proteomic studies17 |
| Transmembrane protein 63A | TMEM63A | 7.29 | .001118 | Proteomic studies18 |
| Transmembrane protein 63B | TMEM63B | 3.00 | .043487 | Proteomic studies18 |
| UPF0404 protein C11orf59 | C11orf59 | 5.71 | .003588 | Proteomic studies18 |
| Protein name . | Gene name . | Ratio . | P . | Source . |
|---|---|---|---|---|
| Cell growth and/or maintenance | ||||
| Claudin-3 | CLDN3 | 16.50 | .000009 | Proteomic studies18 |
| Desmoplakin | DSP | 2.90 | .048440 | Proteomic studies17 |
| Flotillin-1 | FLOT1 | 14.80 | .000018 | Manual |
| 55-kDa erythrocyte membrane protein | MPP1 | 3.00 | .043487 | Uniprot |
| Immune response | ||||
| Complement component 1 Q subcomponent-binding protein | C1QBP | 3.00 | .043487 | Proteomic studies17 |
| HLA class I histocompatibility antigen, B-27 α-chain | HLA-B | 9.00 | .000366 | Manual |
| Interferon-induced transmembrane protein 3 | IFITM3 | 3.00 | .043487 | Uniprot |
| Linker for activation of T-cell family member 1 | LAT | 7.33 | .001082 | Uniprot |
| Protein folding | ||||
| Calnexin | CANX | 2.88 | .049774 | Proteomic studies18 |
| Metabolism | ||||
| 3-mercaptopyruvate sulfur transferase | MPST | 3.33 | .030650 | Proteomic studies17 |
| 4-aminobutyrate aminotransferase | ABAT | 4.00 | .015866 | Proteomic studies17 |
| ADP-ribosyl cyclase 1 | CD38 | 5.00 | .006457 | Proteomic studies17 |
| Acid ceramidase | ASAH1 | 5.25 | .005232 | Proteomic studies18 |
| ATP synthase subunit γ | ATP5L | 4.00 | .015866 | Proteomic studies17 |
| CTD small phosphatase-like protein | CTDSPL | 8.25 | .000586 | Proteomic studies18 |
| Cytochrome b-c1 complex subunit Rieske | UQCRFS1 | 3.00 | .043487 | Proteomic studies17 |
| Glutaminase kidney isoform | GLS | 3.00 | .043487 | Proteomic studies17 |
| NAD(P) transhydrogenase | NNT | 7.92 | .000726 | Proteomic studies17 |
| Probable phospholipid-transporting ATPase IF | ATP11B | 11.00 | .000116 | Proteomic studies18 |
| Sn1-specific diacylglycerol lipase-β | DAGLB | 29.00 | 1.29E-07 | Proteomic studies18 |
| Protein metabolism | ||||
| Cation-dependent mannose-6-phosphate receptor | M6PR | 7.00 | .001366 | Manual |
| DnaJ homolog subfamily C member 5 | DNAJC5 | 27.00 | 2.29E-07 | Uniprot |
| Endothelin-converting enzyme 1 | ECE1 | 35.00 | 2.72E-08 | Manual |
| F-box/LRR-repeat protein 20 | FBXL20 | 6.00 | .002867 | Proteomic studies18 |
| Signal transduction | ||||
| Adenylate cyclase type 6 | ADCY6 | 3.40 | .028628 | Proteomic studies17 |
| ADP-ribosylation factor 5 | ARF5 | 3.00 | .043487 | Proteomic studies17 |
| ADP-ribosylation factor-like protein 15 | ARL15 | 5.25 | .005232 | Proteomic studies18 |
| Casein kinase I γ1 isoform | CSNK1G1 | 3.00 | .043487 | Proteomic studies17 |
| Casein kinase I isoform γ3 | CSNK1G3 | 8.00 | .000690 | Proteomic studies17 |
| CD151 antigen | CD151 | 3.00 | .043487 | Uniprot |
| CD63 antigen | CD63 | 3.67 | .021908 | Manual |
| CD82 antigen | CD82 | 4.00 | .015866 | Manual |
| CDC42 small effector protein 1 | CDC42SE1 | 3.00 | .043487 | Uniprot |
| CDC42 small effector protein 2 | CDC42SE2 | 9.00 | .000366 | Uniprot |
| Cell division control protein 42 homolog | CDC42 | 3.00 | .043487 | Manual |
| Choline transporter-like protein 2 | CTL2 | 27.00 | 2.29E-07 | Proteomic studies18 |
| Disks large-associated protein 4 | DLGAP4 | 6.00 | .002867 | Proteomic studies17 |
| Disheveled-associated activator of morphogenesis 1 | DAAM1 | 9.00 | .000366 | Proteomic studies17 |
| Dystroglycan | DAG1 | 4.00 | .015866 | Proteomic studies17 |
| Erbb2-interacting protein | ERBB2IP | 8.88 | .000395 | Manual |
| Flotillin-2 | FLOT2 | 10.55 | .000149 | Manual |
| G protein-coupled receptor kinase 6 | GRK6 | 8.00 | .000690 | Probable |
| G(i) α-1 | GNAI1 | 3.80 | .019226 | HPRD |
| G(i) α-2 | GNAI2 | 3.25 | .033301 | Manual |
| G(i) α-3 | GNAI3 | 4.86 | .007300 | HPRD |
| G(q) α | GNAQ | 5.17 | .005609 | HPRD |
| G(s) α | GNAS | 5.80 | .003352 | Manual |
| Gα-11 | GNA11 | 6.00 | .002867 | HPRD |
| Gα-13 | GNA13 | 7.45 | .000998 | Uniprot |
| Gα-15 | GNA15 | 4.00 | .015866 | Manual |
| GTPase Hras | HRAS | 7.00 | .001366 | Uniprot |
| GTPase Nras | NRAS | 5.25 | .005232 | Uniprot |
| Junctional adhesion molecule C | JAM3 | 5.14 | .005722 | Proteomic studies17 |
| Kalirin | KALRN | 7.50 | .000965 | Proteomic studies17 |
| Linker for activation of T-cell family member 2 | LAT2 | 7.00 | .001366 | Probable |
| Mitochondrial import inner membrane translocase subunit TIM50 | TIMM50 | 4.00 | .015866 | Proteomic studies24 |
| Phosphatidylinositol 4-kinase type 2-α | PI4K2A | 10.00 | .000202 | Manual |
| Phosphatidylinositol 4-kinase type 2-β | PI4K2B | 3.00 | .043487 | Manual |
| Phospholipid scramblase 1 | PLSCR1 | 20.50 | .000002 | Potential |
| Platelet glycoprotein 4 | CD36 | 6.14 | .002569 | Uniprot |
| Prostacyclin receptor | PTGIR | 10.00 | .000202 | Uniprot |
| Proto-oncogene tyrosine-protein kinase Fyn | FYN | 4.91 | .006980 | HPRD |
| Proto-oncogene tyrosine-protein kinase Yes | YES1 | 10.00 | .000202 | Proteomic studies18 |
| Raftlin | RFTN1 | 5.00 | .006457 | Probable |
| Ras-related protein Rap-2a | RAP2A | 28.00 | .000000 | by similarity |
| Ras-related protein Rap-2b | RAP2B | 3.93 | .017042 | by similarity |
| Ras-related protein Rap-2c | RAP2C | 11.50 | .000089 | by similarity |
| Ras-related protein R-Ras | RRAS | 7.44 | .001002 | Manual |
| Regulator of G-protein signaling 19 | RGS19 | 15.00 | .000016 | Uniprot |
| Semaphorin-4D | SEMA4D | 6.00 | .002867 | Proteomic studies17 |
| Sortilin | SORT1 | 3.00 | .043487 | Proteomic studies17 |
| Stomatin | STOM | 3.29 | .031966 | Uniprot |
| Tetraspanin-9 | TSPAN9 | 6.67 | .001737 | Proteomic studies18 |
| Thromboxane A2 receptor | TBXA2R | 4.00 | .015866 | Manual |
| Transmembrane protein 55B | TMEM55B | 3.50 | .025869 | Proteomic studies17 |
| Type I inositol-1,4,5-trisphosphate 5-phosphatase | INPP5A | 10.67 | .000139 | Potential |
| Tyrosine-protein kinase Lyn | LYN | 5.00 | .006457 | by similarity |
| Transcription regulator activity | ||||
| Histone H2B type 1-N | HIST1H2BN | 6.00 | .002867 | Proteomic studies17 |
| Transport | ||||
| AFG3-like protein 2 | AFG3L2 | 4.00 | .015866 | Proteomic studies17 |
| ATP-binding cassette subfamily B member 6 | ABCB6 | 5.00 | .006457 | Proteomic studies18 |
| Choline transporter-like protein 1 | CTL1 | 14.20 | .000024 | Proteomic studies18 |
| Cytochrome b5 type B | CYB5B | 3.67 | .021908 | Proteomic studies17 |
| Golgin subfamily A member 7 | GOLGA7 | 17.00 | .000007 | Uniprot |
| Multidrug resistance-associated protein 4 | ABCC4 | 3.00 | .043487 | Proteomic studies18 |
| Phospholipid scramblase 3 | PLSCR3 | 13.00 | .000042 | Probable |
| Phospholipid scramblase 4 | PLSCR4 | 7.00 | .001366 | Probable |
| Pituitary tumor-transforming gene 1 protein-interacting protein | PTTG1IP | 19.50 | .000003 | Proteomic studies18 |
| PRA1 family protein 2 | PRAF2 | 14.00 | .000026 | Proteomic studies17 |
| Protein tweety homolog 3 | TTYH3 | 11.00 | .000116 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 1 | SCAMP1 | 10.25 | .000175 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 2 | SCAMP2 | 3.95 | .016681 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 3 | SCAMP3 | 16.00 | .000011 | Proteomic studies17 |
| Secretory carrier-associated membrane protein 4 | SCAMP4 | 3.00 | .043487 | Proteomic studies17 |
| Sodium/potassium-transporting ATPase subunit α-1 | ATP1A1 | 3.00 | .043487 | Proteomic studies17 |
| SNAP-23 | SNAP23 | 3.74 | .020385 | HPRD |
| Syntaxin-8 | STX8 | 11.00 | .000116 | Manual |
| Syntaxin-12 | STX12 | 4.00 | .015866 | Proteomic studies17 |
| Trafficking protein particle complex subunit 3 | TRAPPC3 | 8.67 | .000450 | by similarity |
| Transferrin receptor protein 1 | TFRC | 3.00 | .043487 | Uniprot |
| Vesicle-associated membrane protein 3 | VAMP3 | 8.43 | .000523 | Proteomic studies18 |
| Vesicle-associated membrane protein 4 | VAMP4 | 10.00 | .000202 | Proteomic studies17 |
| Vesicle-associated membrane protein 7 | VAMP7 | 3.69 | .021361 | Proteomic studies18 |
| Unknown | ||||
| 3-oxoacyl-[acyl-carrier-protein] synthase | OXSM | 6.00 | .002867 | Proteomic studies17 |
| Abhydrolase domain-containing protein FAM108B1 | FAM108B1 | 8.33 | .000556 | Proteomic studies18 |
| Coiled-coil domain-containing protein 109A | CCDC109A | 4.50 | .010000 | Proteomic studies17 |
| Endoplasmic reticulum-Golgi intermediate compartment protein 3 | ERGIC3 | 3.00 | .043487 | Proteomic studies17 |
| Abhydrolase domain-containing protein FAM108A1 | FAM108A1 | 7.50 | .000965 | Proteomic studies23 |
| Protein FAM49B | FAM49B | 21.00 | .000002 | Proteomic studies18 |
| Protein LYRIC gene-1 protein | LYRIC | 18.00 | .000005 | Proteomic studies17 |
| Transmembrane protein 63A | TMEM63A | 7.29 | .001118 | Proteomic studies18 |
| Transmembrane protein 63B | TMEM63B | 3.00 | .043487 | Proteomic studies18 |
| UPF0404 protein C11orf59 | C11orf59 | 5.71 | .003588 | Proteomic studies18 |
Known palmitoylated proteins as catalogued by the Uniprot or HPRD databases or by review of published palmitoylation-related research articles (manual). Also included in this list are proteins characterized as being palmitoylated by similarity, probably palmitoylated, or potentially palmitoylated as defined by the Uniprot database. A total of 61 of the identified proteins were discovered as palmitoyl protein candidates in other proteomic studies.17,18,24 Data are the result of 2 independent experiments, and proteins are grouped according to the biologic process they mediate as defined by the HPRD. Also shown is the HA+/HA− ratio and corresponding P value.
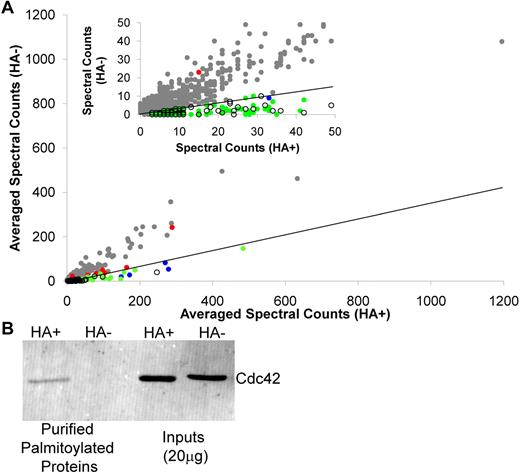
For graphic representation of the proteins identified by LC-MS/MS, the summed spectral counts for each protein in the HA+ sample were plotted against the summed spectral counts for each protein in the HA− sample (Figure 2A). For the majority of proteins identified, there was substantial representation in both the HA+ and HA− samples, which resulted in a HA+/HA−ratio < 3 (gray dots, Figure 2A). Clustered around the x-axis, with the known palmitoylated proteins (green circles, Figure 2A), are the newly identified palmitoyl protein candidates (open black circles, Figure 2A). Well-established, known palmitoylated platelet proteins identified in our study are also indicated (blue dots, Figure 2A) and include the Gα subunits Gq,11,12 Gi,11,12 and G13,12 platelet glycoprotein 4,25 and CD6326 (Table 2). Several highly abundant platelet proteins known to be palmitoylated were identified in the list of 1300 purified proteins but failed to demonstrate a HA+/HA− ratio ≥ 3 (red dots, Figure 2A). This group includes P-selectin,27 tubulin,28 platelet glycoprotein Ib,11 PECAM,29 Gαz,11 platelet glycoprotein IX,11 and CD926 (supplemental Table 1). Included among the known palmitoylated proteins is the palmitoylated form of Cdc42, which previously had been described in neurons17 and identified as a putative palmitoyl protein in human prostate cancer cells.18 To confirm that we were observing the palmitoylated form of Cdc42 in our preparation, samples were stained using antibodies directed against Cdc42. We found Cdc42 only in the HA palmitoyl-protein and not the HA− sample, indicating that platelets contain the palmitoylated form of Cdc42 (Figure 2B).
Global analysis of the platelet palmitoylome. (A) Graphical depiction of the 1300 proteins identified in resting platelet membranes. Gray dots represent identified proteins not meeting the criteria for significance. Candidate palmitoyl proteins (○) cluster around the x-axis with known palmitoylated proteins and putative palmitoyl proteins identified in other palmitoylation-specific proteomic studies (green dots). Also shown are well-established palmitoylated platelet proteins identified (blue dots) and not identified (red dots) as being palmitoylated in this study. Inset: Expanded view of the graph for proteins with < 50 spectral counts. Diagonal line in each graph indicates the HA+/HA− cut-off. (B) Western blotting analysis of ABE-purified proteins from platelet membranes, as prepared for proteomic analysis, in the presence and absence of HA using antibodies directed against Cdc42. Also shown is the total protein input for the HA+ and HA− samples.
Global analysis of the platelet palmitoylome. (A) Graphical depiction of the 1300 proteins identified in resting platelet membranes. Gray dots represent identified proteins not meeting the criteria for significance. Candidate palmitoyl proteins (○) cluster around the x-axis with known palmitoylated proteins and putative palmitoyl proteins identified in other palmitoylation-specific proteomic studies (green dots). Also shown are well-established palmitoylated platelet proteins identified (blue dots) and not identified (red dots) as being palmitoylated in this study. Inset: Expanded view of the graph for proteins with < 50 spectral counts. Diagonal line in each graph indicates the HA+/HA− cut-off. (B) Western blotting analysis of ABE-purified proteins from platelet membranes, as prepared for proteomic analysis, in the presence and absence of HA using antibodies directed against Cdc42. Also shown is the total protein input for the HA+ and HA− samples.
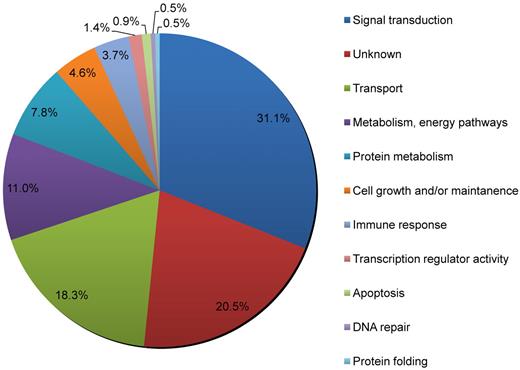
Using the Human Protein Reference Database (HPRD),30 we classified all the known and putative palmitoylated proteins by biologic process (Figure 3). We find that the largest percentage of palmitoylated platelet protein candidates (31.3%) is involved in signal transduction processes. A substantial number of proteins (18.3%) involved in transport processes, which includes the movement of vesicles and small molecules and ions within or out of the cell, were found to be palmitoylated as well. Although close to half of these transport proteins are known to be palmitoylated or have been described in other proteomic studies, the rest are newly identified palmitoyl candidates. This group includes syntaxins-2, -10, and -11 and VAMP-5. These results indicate that signaling and transport proteins represent approximately half of all palmitoylated platelet proteins identified in this study.
The biologic processes mediated by proteins of the platelet palmitoylome. Analysis of the platelet palmitoylome indicates that many of the identified platelet proteins are involved in signal transduction pathways and transport processes. Each protein was assigned a biologic process as defined by the HPRD, which is Gene Ontology compliant.
The biologic processes mediated by proteins of the platelet palmitoylome. Analysis of the platelet palmitoylome indicates that many of the identified platelet proteins are involved in signal transduction pathways and transport processes. Each protein was assigned a biologic process as defined by the HPRD, which is Gene Ontology compliant.
TLT-1 is a palmitoyl protein candidate
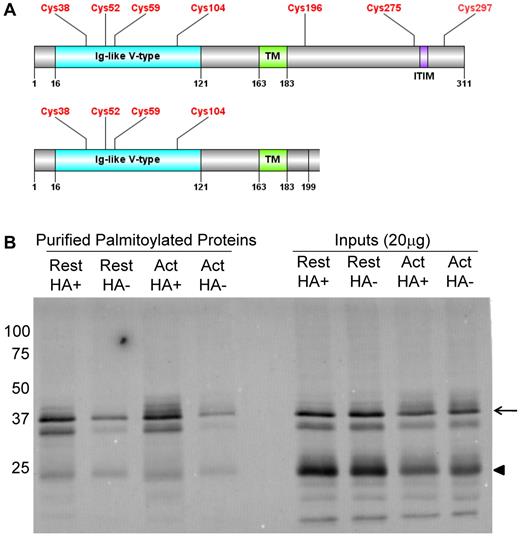
Because the expression of TLT-1 is restricted to megakaryocytes and platelets, we chose it for further analysis. TLT-1 demonstrated an HA+/HA− ratio of 2.5 (P = .076). The full-length form of TLT-1 has a 126-amino acid cytosolic C-terminal tail, which includes an immunoreceptor tyrosine-based inhibitory motif (ITIM) region and a proline-rich region that may mediate protein-protein interactions.31 The cytosolic tail also contains 3 cysteine residues (Figure 4A), and analysis of the sequence using the palmitoylation site predicting software, CSS-Palm 2.0, indicated that TLT-1 was probably palmitoylated only on cysteine 196 (Cys196).32 TLT-1 exists as 2 species: a full-length form with a molecular mass of 35 kDa and a splice variant with a molecular mass of 25 kDa. The splice variant differs from full-length TLT-1 in that it has a truncated cytosolic region, which encodes a short, 14-amino acid tail (Figure 4A).31 In addition, amino acids 190 to 199 of the splice variant differ from the full-length sequence. Instead of GNRLGVCGRF, the splice variant encodes for ESLLSGPPRQ, which does not contain the predicted palmitoylation site at Cys196 (Uniprot database).
TLT-1 is enriched in HA+ samples in resting and activated platelets. (A) Domain organization of full-length TLT-1 and the TLT-1 splice variant depicting an Ig-like V-type domain (blue) with 4 cysteine residues followed by a single-pass transmembrane (TM) domain (green). The TLT-1 splice variant has a truncated cytoplasmic tail and does not contain the 3 intracellular cysteines or ITIM region.32 (B) Western blot analysis of TLT-1 using ABE-purified palmitoylated proteins from resting and thrombin-activated platelets. Arrow indicates full-length TLT-1; and arrowhead, a 25 kDa TLT-1 splice variant. Bands seen directly below full-length TLT-1 and bands seen below the TLT-1 splice variant are degradation products.33 Also shown are 20 μg of the sample inputs that represent the protein sample added to the streptavidin agarose beads.
TLT-1 is enriched in HA+ samples in resting and activated platelets. (A) Domain organization of full-length TLT-1 and the TLT-1 splice variant depicting an Ig-like V-type domain (blue) with 4 cysteine residues followed by a single-pass transmembrane (TM) domain (green). The TLT-1 splice variant has a truncated cytoplasmic tail and does not contain the 3 intracellular cysteines or ITIM region.32 (B) Western blot analysis of TLT-1 using ABE-purified palmitoylated proteins from resting and thrombin-activated platelets. Arrow indicates full-length TLT-1; and arrowhead, a 25 kDa TLT-1 splice variant. Bands seen directly below full-length TLT-1 and bands seen below the TLT-1 splice variant are degradation products.33 Also shown are 20 μg of the sample inputs that represent the protein sample added to the streptavidin agarose beads.
To confirm that TLT-1 is palmitoylated and to determine which TLT-1 species is palmitoylated, we directly assessed samples purified by ABE chemistry for TLT-1. Platelets from outdated aphaeresis packs were washed and divided into 2 equal aliquots. One was left resting and the other was stimulated with 1 U/mL α-thrombin for 5 minutes at room temperature. Prostaglandin E1 was added to both samples, and the platelets were immediately pelleted and dissolved in lysis buffer. Palmitoylated proteins were subsequently purified using the ABE method from membranes prepared from resting and thrombin-activated platelets, and were separated by gel electrophoresis and transferred to a PVDF membrane (Figure 4B). Immunoblotting of ABE-purified TLT-1 demonstrated 3 major bands corresponding to full-length TLT-1, a previously described degradation product,33 and the splice variant. Full-length TLT-1 and its degradation product were palmitoylated because the HA+ samples were enriched for full-length TLT-1 in resting and activated platelets compared with the HA− samples (Figure 4B). In contrast, the TLT-1 splice variant was not enriched. Bands seen for TLT-1 in the HA− sample represent background binding to the streptavidin beads, which may be the result of improper biotinylation or nonspecific binding. Western blot analysis of the ABE-purified samples in the resting and activated HA+ lanes did not demonstrate a change in the palmitoylation state of TLT-1 on platelet activation under the conditions in which this assay was performed (Figure 4B). These results suggest that full-length TLT-1, but not the TLT-1 splice variant, is palmitoylated in platelets.
TLT-1 is a novel palmitoylated platelet protein
To determine whether TLT-1 is a bona fide palmitoylated protein, we metabolically labeled platelets with [3H]palmitic acid and assayed for incorporation of [3H]palmitate. Resting platelets were labeled with [3H]palmitic acid for 1 hour at 37°C and then divided into 2 equal portions. One portion remained resting, and the other was activated with 1 U/mL α-thrombin. Samples were lysed and TLT-1 was immunoprecipitated (Figure 5A). The minor band seen at 33 kDa represents a TLT-1 degradation product.33 Subsequent audioradiography revealed that full-length TLT-1 incorporated [3H]palmitate in both resting and activated platelets (Figure 5B). The 25-kDa splice variant, which was immunoprecipitated with full-length TLT-1 (Figure 5A lanes 3 and 4), did not incorporate [3H]palmitate (Figure 5B), as predicted by the absence of the cytosolic cysteine residues and studies of TLT-1 using ABE chemistry (Figure 4). These studies confirm that TLT-1 is a platelet palmitoyl protein.
[3H]Palmitate incorporation confirms that TLT-1 is a palmitoylated protein. Platelets were metabolically labeled with [3H]palmitic acid and separated into resting (lane 1) and thrombin-activated (lane 2) samples and lysed. TLT-1 was then immunoprecipitated from resting (lane 3) and thrombin-activated platelet lysates (lane 4). Samples were subjected to Western blot analysis (A) or audioradiography to assess [3H] palmitate incorporation (B). Full-length TLT-1 is the band corresponding to 37 kDa. This blot is representative of 3 independent experiments.
[3H]Palmitate incorporation confirms that TLT-1 is a palmitoylated protein. Platelets were metabolically labeled with [3H]palmitic acid and separated into resting (lane 1) and thrombin-activated (lane 2) samples and lysed. TLT-1 was then immunoprecipitated from resting (lane 3) and thrombin-activated platelet lysates (lane 4). Samples were subjected to Western blot analysis (A) or audioradiography to assess [3H] palmitate incorporation (B). Full-length TLT-1 is the band corresponding to 37 kDa. This blot is representative of 3 independent experiments.
To further characterize the palmitoylation of TLT-1, we used PalmPISC to identify the candidate palmitoylation site. In this approach, palmitoylated cysteines can be readily distinguished from nonpalmitoylated cysteines because nonpalmitoylated cysteines are irreversibly blocked by NEM, whereas palmitoylated cysteines become free cysteines after the ABE reaction and TCEP elution. The resulting spectrum of a peptide derived from TLT-1, LGVCGR, shows a free cysteine residue based on the m/z difference of 103 between the y2 and y3 fragment ion (Figure 6). The fact that this cysteine (Cys196) is free and not NEM-modified indicates that Cys196 is a putative palmitoylation site. This result is in agreement with the CSS-Palm prediction that Cys196 is a candidate palmitoylation site of TLT-1.
Representative tandem mass spectrum of a candidate palmitoyl peptide derived from TLT-1 protein. Enriched palmitoyl proteins were separated by SDS-PAGE and a gel slice containing 30- to 50-kDa proteins excised. Proteins were digested in gel, followed by the extraction of tryptic peptides, which were analyzed by LC-MS/MS. Free cysteines in the purified peptides are candidate palmitoylation sites.
Representative tandem mass spectrum of a candidate palmitoyl peptide derived from TLT-1 protein. Enriched palmitoyl proteins were separated by SDS-PAGE and a gel slice containing 30- to 50-kDa proteins excised. Proteins were digested in gel, followed by the extraction of tryptic peptides, which were analyzed by LC-MS/MS. Free cysteines in the purified peptides are candidate palmitoylation sites.
Discussion
Our analysis of the platelet palmitoylome resulted in the identification of > 1300 proteins. We found that 215 of these proteins were significantly enriched in the HA+ sample, suggesting that they are palmitoyl proteins. However, there are several reasons to think that even more palmitoylated proteins exist in platelets. First, we did not observe some of the known palmitoylated proteins as expected in the platelet palmitoylome. These proteins include members of the acyl-transferase family, known as DHHC proteins, which have been observed in other proteomic studies.18 The reason for this could lie in the low abundance of these proteins. Second, to avoid falsely identifying palmitoyl protein candidates, we used strict cut-off values to define the HA+ enriched dataset and excluded actual palmitoyl proteins. For example, TLT-1, which was not significantly enriched in the HA+ sample, was confirmed to be a palmitoylated platelet protein (Figure 5). Similarly, we did not identify all of the platelet proteins that have previously been found to be subject to palmitoylation. P-selectin, tubulin, platelet glycoproteins IX and Ib, PECAM, Gαz, and CD9 were identified (Figure 1B red dots; supplemental Table 2), but their HA+/HA− ratios did not meet the stringent threshold (ratio ≥ 3, P < .05) that was used to identify candidate palmitoyl proteins. It is possible that, although these proteins are highly abundant in platelets, their palmitate-modified, membrane-associated forms are not. Another potential source of error is the copurification of contaminating proteins, a common issue for affinity purification-based methods. Nonspecific copurification of nonpalmitoylated proteins in the HA− sample will lower the HA+/HA− ratio and is a limitation of the PalmPISC method. In addition, the widely used spectral counting quantitation approach is only semiquantitative. Development of better methods to reduce the background contribution of contaminating proteins is currently ongoing and will increase the sensitivity and specificity for identification of palmitoyl proteins in platelets. Yield of palmitoyl proteins could also be improved with better solubilization of the membranes to release more integral membrane proteins. Studies are currently underway to test an alternative method using pressure cycling technology34 to extract more proteins from platelet membranes.
Approximately 50% of identified palmitoyl proteins mediate signal transduction and transport processes. Because palmitoylation is a dynamic lipid modification, the palmitoylation state of platelet proteins may present another layer of regulation governing the localization and protein-protein interactions necessary to carry out these processes. Included in our list of signaling proteins is the palmitoylated form of Cdc42. This result was intriguing because Cdc42 exists as 2 isoforms: a prenylated form that has ubiquitous expression and a palmitoylated form that has been described previously in the rat neuronal palmitoyl proteome.17 Immunoblotting of the ABE-purified palmitoylated proteins demonstrated the HA dependence of Cdc42 (Figure 2C) in our preparations and indicates that platelets possess this palmitoylated isoform. Unlike metabolic labeling with [3H]palmitate, the ABE method provides a snapshot of all the palmitoylated proteins in a cell at a particular moment in time. Although ABE methodology cannot give information about the rate of palmitate turnover, when combined with a global proteomic approach, it can provide the identity of proteins for further targeted study.
We chose TLT-1 as a palmitoyl protein candidate for further study because its palmitoylation has not previously been described and its expression is restricted to megakaryocytes and platelets.35 The limited expression pattern of TLT-1 suggests that it may play a specific role in platelet function and makes it a potential target for the modulation of hemostasis and thrombosis because antibodies directed against TLT-1 are able to block thrombin-mediated platelet aggregation.33 Furthermore, TLT-1 knockout mice demonstrate extended tail bleeding times and are predisposed to hemorrhage in an inflammatory model. Platelets from these mice are defective in aggregation in response to adenosine diphosphate and U46619 stimulation.36 TLT-1 is part of a family of receptors termed triggering receptors expressed on myeloid cells (TREMs)37 ; and although TLT-1 is homologous to TREMs in its V-type immunoglobulin-like extracellular domain, TLT-1 has a longer cytoplasmic tail with a proline-rich region and contains an ITIM instead of an immunoreceptor tyrosine-based activation motif (Figure 3). It has been shown in vitro that the ITIM region of TLT-1 is capable of being phosphorylated and can recruit SH2-domain–containing protein tyrosine phosphatases 1 and 2.31,38
We present evidence here that TLT-1 is a novel palmitoylated protein and is palmitoylated on Cys196. Full-length TLT-1 has 7 cysteine residues.39 Four of these reside in the extracellular domain and form disulfide bonds.40 Of the 3 intracellular cysteine residues, Cys196 occurs just after the transmembrane domain. Our identification of Cys196 as a putative palmitoylation site is in agreement with the observation that the likelihood of a cysteine being palmitoylated increases after stretches of hydrophobic amino acids, such as a transmembrane domains. As expected, the truncated 25-kDa TLT-1 splice variant did not incorporate [3H]palmitate (Figure 5B), consistent with the lack of intracellular cysteines. The role that palmitoylation serves in TLT-1 function is unknown. Many of the palmitoyl proteins that we found in the platelet had previously been identified in our analysis of palmitoyl proteins in lipid rafts (supplemental Table 2). We have found that a fraction of TLT-1 incorporates into lipid rafts (supplemental Figure 2). TLT-1 palmitoylation could affect its incorporation into rafts, as has been demonstrated for the immunoreceptor tyrosine-based activation motif-containing protein PECAM-1.29 Alternatively, palmitoylation may influence phosphorylation of TLT-1, as observed in tissue factor41 and linker for activation of T cells (LAT).42 Our methodologies and experimental conditions did not demonstrate activation-induced palmitoylation of TLT-1. However, activation could affect palmitate cycling, and presently we cannot separate palmitoylation and depalmitoylation activities. Therefore, we cannot conclude whether or not activation-induced TLT-1 palmitoylation occurs. We are currently developing methods to study activation-dependent platelet palmitoylation and determine the significance of TLT-1 palmitoylation in platelets.
We compared the platelet palmitoylome with other palmitoylation-specific proteome studies16-18,23,24,43 and identified 103 new palmitoyl protein candidates. In our analysis of the platelet palmitoylome, we determined the overlap between it and the rat neuronal palmitoylome.17 We chose the neuronal palmitoylome for comparison because megakaryocytes/platelets and neurons share structural and functional characteristics, and there are numerous studies in which platelets have been proposed as a model for neuronal function.44 Of the 215 proteins that compose the platelet palmitoylome, 75 proteins overlap between our study and the rat neuronal palmitoylome (Figure 7). We also find that, of the 51 known palmitoylated proteins identified in the platelet palmitoylome, 37 overlap with the 68 known palmitoylated proteins identified in the neuronal palmitoylome (Figure 7).17 Of particular interest was our identification of the palmitoylated form of Cdc42. Although Cdc42 null platelets display a complex phenotype,45 Cdc42 is implicated in platelet cytoskeletal remodeling46,47 and filopodia formation.48 In neurons, the palmitoylated form of Cdc42 induces the formation of dendritic spines.17 It is tempting to speculate that the palmitoylated form of Cdc42 may play a similar role in megakaryocytes or platelets contributing to proplatelet formation or morphologic changes seen on platelet activation. Studies are currently underway to assess the implications of this finding.
Venn diagrams representing the overlap between the platelet and rat neuronal palmitoylomes. Number of overlapping proteins identified in the platelet (solid line) and rat neuronal (dashed line) palmitoylomes (A). Number of overlapping known palmitoylated proteins identified in the platelet and rat neuronal palmitoylomes (B).
Venn diagrams representing the overlap between the platelet and rat neuronal palmitoylomes. Number of overlapping proteins identified in the platelet (solid line) and rat neuronal (dashed line) palmitoylomes (A). Number of overlapping known palmitoylated proteins identified in the platelet and rat neuronal palmitoylomes (B).
This study is the first comprehensive description of the platelet palmitoylome and expands our understanding of the scope of palmitoylation in platelets. However, given the importance of palmitoylation in regulating protein function, our findings extend beyond platelet biology as our global characterization of the platelet palmitoylome resulted in the identification of 103 novel palmitoyl protein candidates. The validation of the palmitoylation of these candidate proteins will provide opportunities for future studies aimed at increasing our understanding of the role palmitoylation plays in platelet function. The platelet palmitoylome will establish a platform, providing an essential first step to the determination of how palmitoylation alters the function of the novel palmitoyl protein candidates.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Valance Washington (University of Puerto Rico–Mayaguez) for helpful discussions and the Beth Israel Deaconess Medical Center Blood Bank for providing a source of platelets.
This work was supported by the National Institutes of Health (grant HL87203, R.F.) and the United States Army (grant PC093459, M.R.F. and H.S.). L.D. was supported by the National Institutes of Health (grant T32 HL07917). R.F. is a recipient of an Established Investigator Award from the American Heart Association.
National Institutes of Health
Authorship
Contribution: L.D. designed and performed research, analyzed data, and wrote the paper; W.Y. designed and performed research, analyzed data, and contributed to writing the paper; M.R.F. provided reagents and contributed to writing the paper; H.S. facilitated the mass spectrometric analysis and contributed to writing the paper; and R.F. conceived of study, analyzed data, and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Flaumenhaft, Division of Hemostasis and Thrombosis, Department of Medicine, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston, MA 02215; e-mail: rflaumen@bidmc.harvard.edu.
References
Author notes
L.D and W.Y. contributed equally to this study.


![Figure 5. [3H]Palmitate incorporation confirms that TLT-1 is a palmitoylated protein. Platelets were metabolically labeled with [3H]palmitic acid and separated into resting (lane 1) and thrombin-activated (lane 2) samples and lysed. TLT-1 was then immunoprecipitated from resting (lane 3) and thrombin-activated platelet lysates (lane 4). Samples were subjected to Western blot analysis (A) or audioradiography to assess [3H] palmitate incorporation (B). Full-length TLT-1 is the band corresponding to 37 kDa. This blot is representative of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/13/10.1182_blood-2011-05-353078/4/m_zh89991178670005.jpeg?Expires=1769104366&Signature=lJkXj5BUmVXypKuezrMYUcTdhI3ADyYXfakl0NzfUeI7xG4jPWoZekRRW~SY1RK~4dG~u-pCBceLQ9y-BTuJJCJwwibhfltdr1eUaldBN2lsJ9Exal6DPThvfsPEIHhHF-jTppHXeUVSt9kK4XQstVtH0uVPpsJG6Wbb4afN48ayNleMK-HjyohBOzaZ9GWS0MDFuSwTKd3SlshyIKut9IOAJWxae2nXmJ1vowbecB5sB17CIgsAEGaakrrz2b0Gj90T~2dKfWPkaPfaQTSJCODYHItCzWmDJEgAvzkXEIplO4xkhnbCylUqGbdZxOELkfdVjEcgPOv2J7UKMeJa1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
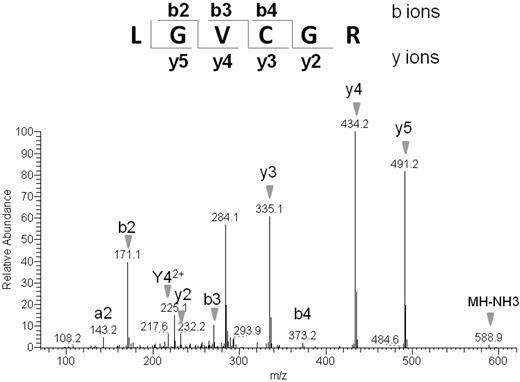
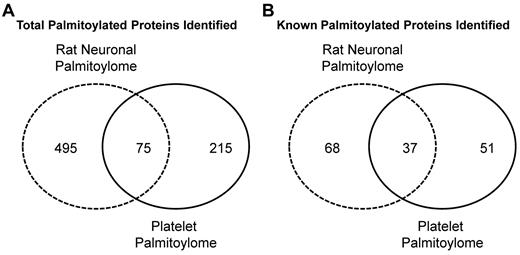
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal