Abstract
The iron hormone hepcidin is inhibited by matriptase-2 (MT2), a liver serine protease encoded by the TMPRSS6 gene. Cleaving the bone morphogenetic protein (BMP) coreceptor hemojuvelin (HJV), MT2 impairs the BMP/son of mothers against decapentaplegic homologs (SMAD) signaling pathway, down-regulates hepcidin, and facilitates iron absorption. TMPRSS6 inactivation causes iron-deficiency anemia refractory to iron administration both in humans and mice. Genome-wide association studies have shown that the SNP rs855791, which causes the MT2 V736A amino acid substitution, is associated with variations of serum iron, transferrin saturation, hemoglobin, and erythrocyte traits. In the present study, we show that, in vitro, MT2 736A inhibits hepcidin more efficiently than 736V. Moreover, in a genotyped population, after exclusion of samples with iron deficiency and inflammation, hepcidin, hepcidin/transferrin saturation, and hepcidin/ferritin ratios were significantly lower and iron parameters were consistently higher in homozygotes 736A than in 736V. Our results indicate that rs855791 is a TMPRSS6 functional variant and strengthen the idea that even a partial inability to modulate hepcidin influences iron parameters and, indirectly, erythropoiesis.
Introduction
Hepcidin is the key regulator of iron homeostasis, controlling surface expression of the iron exporter ferroportin on enterocytes and macrophages.1 Inactivation of hepcidin causes severe iron overload in mice and humans, whereas hepcidin overexpression causes iron-deficiency anemia.2 Hepcidin expression is up-regulated in response to increased body iron, through the bone morphogenetic protein (BMP)–hemojuvelin (HJV)–son of mothers against decapentaplegic homologs (SMAD) pathway3 and inhibited by matriptase-2 (MT2), a type II transmembrane serine protease encoded by the TMPRSS6 gene4,5 that in vitro cleaves the BMP coreceptor HJV.6 In vivo, “Mask” mice, which have a deleted serine protease domain,4 and Tmprss6-null mice7 show microcytic anemia because of high hepcidin levels. TMPRSS6 deleterious mutations in humans cause iron-refractory iron-deficiency anemia that is unresponsive to oral iron administration.5 The same mutations show partial inhibition of the hepcidin promoter activity when overexpressed with HJV in vitro in hepatoma cells.6,8
Recent genome-wide association studies reported the association of common genetic variants of TMPRSS6 (rs855791 and rs4820268) with serum iron and transferrin saturation,9-11 hemoglobin, mean corpuscular volume, and mean corpuscular hemoglobin,12,13 highlighting a role for MT2 in the control of iron and erythrocyte parameters. The SNP rs855791 (2321G- > A) causes a nonsynonymous alanine to valine change (A736V) in the catalytic domain, whereas the SNP rs4820268 leads to a synonymous change at 521 and is in linkage disequilibrium with rs855791. Because rs855791 affects the MT2 catalytic domain, a common speculation was that its effects were hepcidin mediated.9,14 We tested this hypothesis using an in vitro assay based on the luciferase reporter gene driven by the hepcidin promoter, and showed that the MT2736A inhibits hepcidin more efficiently than MT2736V. We also demonstrated that this variant affects the hepcidin levels of normal individuals.
Methods
In vitro studies
The in vitro analyses (Western blot, hepcidin promoter activity assay, and hepcidin-binding assay) were reported previously6 and are detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The TMPRSS6 variant encoding 736A (MT2736A) was obtained by mutagenesis of the MT2736V plasmid using the QuikChange site-directed mutagenesis kit (Stratagene).
Human studies
Statistical analysis
Association of TMPRSS6 rs855791 was first analyzed in 655 unrelated (pairwise kinship coefficient < 0.0625) individuals selected using the Greffa software program developed by Falchi et al.17 We included in the final analysis only individuals with hepcidin levels above the lower limit of detection (0.55nM) and subjects with ferritin ≥ 30 ng/mL and C-reactive protein ≤ 1mg/dL (subset 1). Mean values were adjusted for sex, age, squared age, and interaction between them (sex*age, sex*squared age) using ANOVA (95% confidence interval) and SPSS 17.0 software and in house R-2.8.1 scripts (The R Project for Statistical Computing at http://www.r-project.org).
Results and discussion
In vitro function of MT2 A736V variants
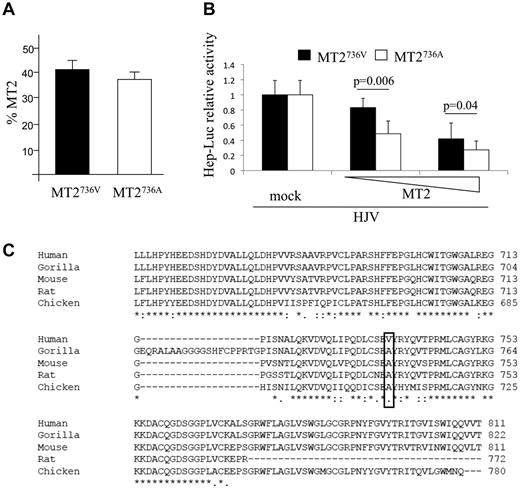
We first demonstrated that the proportion of MT2736A and MT2736V variants expressed on the surface of transfected cells was similar (Figure 1A). We next observed that the MT2736A variant inhibited the luciferase-hepcidin promoter more efficiently than MT2736V, with a dose-dependent effect at low concentrations (Figure 1B). In agreement with the luciferase assay, the release of the serine protease domain, which we have shown to be correlated with protease activity,6,8 was slightly increased in cells transfected with the more active MT2736A variant compared with MT2736V (supplemental Figure 1 top panel). These results suggest that rs855791 is a functional variant. Western blot on cell lysates and phospholipase C were not sensitive enough to detect a significant difference in the cleavage of HJV between the 2 variants (supplemental Figure 1 bottom panel).
In vitro characterization of the function of MT2 variants and evolutionary conservation of the catalytic domain. (A) Quantification of membrane-bound MT2 (MT2) by binding assay. HeLa cells were transiently transfected with the TMPRSS6 cDNA encoding MT2736V, MT2736A, or the empty vector (mock) and analyzed for the percentage of MT2 on the cell surface.6 The amount of surface MT2 was calculated as the ratio between the absorbance of unpermeabilized and permeabilized cells. Error bars indicate SD. (B) Hepcidin promoter activity assay. Hep3B cells were transiently transfected with 0.25 μg of pGL2-basic reporter vector (Promega) containing the 2.9-kb fragment of the human hepcidin promoter23 in combination with pRL-TK Renilla luciferase vector (Promega) and HJV, as described previously.6 Increasing doses (from 0.002 to 0.01 μg/mL) of MT2736V- or MT2736A-expressing vectors were used. Relative luciferase activity was calculated as the ratio of firefly (reporter) to Renilla luciferase activity and is expressed as a multiple of the activity of cells transfected with the reporter alone. Experiments were performed in triplicate. The statistical significance is indicated above the bars. (C) Alignment of part of the serine protease domain of MT2 of different species by multiple sequence alignment ClustalW (EMBL-EBI) program. The sequence is highly conserved. The human 736 and the orthologous position in the other species are boxed.
In vitro characterization of the function of MT2 variants and evolutionary conservation of the catalytic domain. (A) Quantification of membrane-bound MT2 (MT2) by binding assay. HeLa cells were transiently transfected with the TMPRSS6 cDNA encoding MT2736V, MT2736A, or the empty vector (mock) and analyzed for the percentage of MT2 on the cell surface.6 The amount of surface MT2 was calculated as the ratio between the absorbance of unpermeabilized and permeabilized cells. Error bars indicate SD. (B) Hepcidin promoter activity assay. Hep3B cells were transiently transfected with 0.25 μg of pGL2-basic reporter vector (Promega) containing the 2.9-kb fragment of the human hepcidin promoter23 in combination with pRL-TK Renilla luciferase vector (Promega) and HJV, as described previously.6 Increasing doses (from 0.002 to 0.01 μg/mL) of MT2736V- or MT2736A-expressing vectors were used. Relative luciferase activity was calculated as the ratio of firefly (reporter) to Renilla luciferase activity and is expressed as a multiple of the activity of cells transfected with the reporter alone. Experiments were performed in triplicate. The statistical significance is indicated above the bars. (C) Alignment of part of the serine protease domain of MT2 of different species by multiple sequence alignment ClustalW (EMBL-EBI) program. The sequence is highly conserved. The human 736 and the orthologous position in the other species are boxed.
Based on gene-expression analysis, it was proposed that rs4820268, the other TMPRSS6 variant that is significantly associated with iron and erythrocyte traits,10,18 might cause a differential allelic expression (60:40 ratio) of TMPRSS6 mRNA.19 However, it is unlikely that the modest difference observed results in detectable changes of the protease activity. rs4820268 is in linkage disequilibrium (R2 = 0.811 in the VB cohort) with rs855791; therefore, its association with iron and erythrocyte traits might simply be secondary to that of rs855791.
The human database indicates V at the 736 position as the “wild-type” MT2. However, comparative analysis indicates A as the ancestral amino acid, because A is evolutionary conserved in all the species in which an annotated MT2 sequence is available (Figure 1C). This observation suggests that the MT2736V variant, which leads to increased hepcidin production and inhibition of iron absorption, is a recent evolutionary change.
Hepcidin levels of normal carriers of MT2736 variants
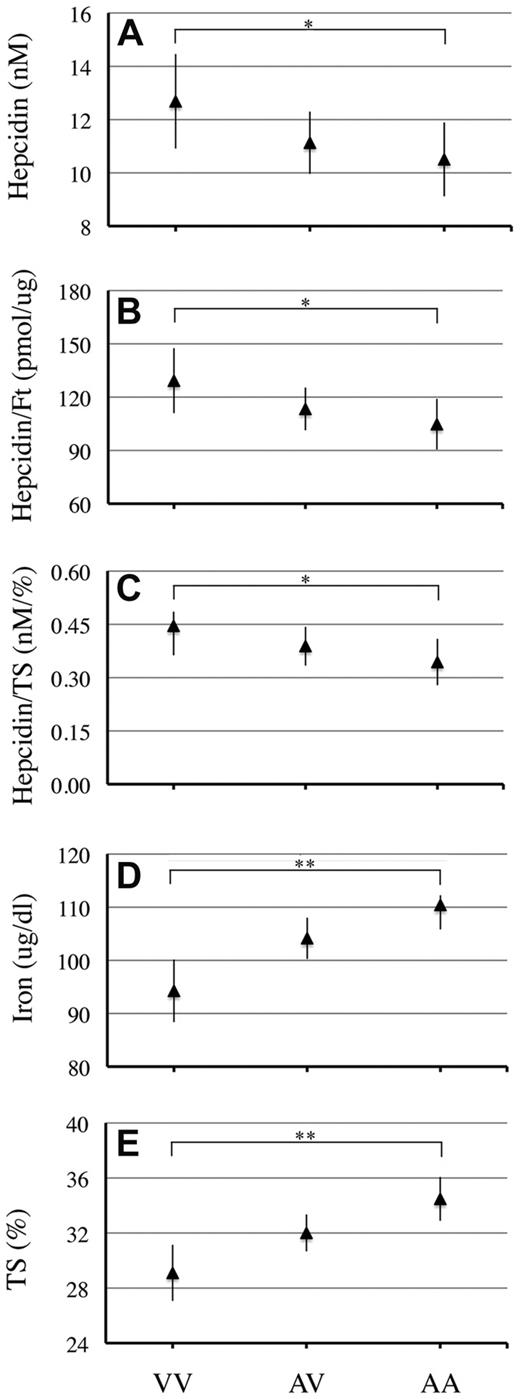
We validated our in vitro results in the VB cohort, which had serum hepcidin levels measured. Because in a genetic isolate, many individuals are related, only a group of 655 unrelated individuals was studied. We also selected 545 normal subjects after excluding iron-deficient individuals (serum ferritin < 30 ng/mL) and individuals with clinically relevant inflammatory conditions (C-reactive protein > 1 mg/dL).11 Serum hepcidin levels were lower in AA compared with VV homozygous individuals. The difference was not significant in the whole series, only in the selected subset (P = .038; Figure 2 and supplemental Table 1). Because hepcidin expression is strongly dependent on both iron stores and plasma iron, we normalized hepcidin on ferritin and on transferrin saturation. In both cases, we confirmed that the normalized values were significantly lower in AA compared with VV homozygotes (P = .038 for hepcidin/ferritin and P = .056 for hepcidin/transferrin saturation, respectively) in subset 1 (Figure 2 and supplemental Table 1). Consistently, iron and transferrin saturation were higher in AA than in VV homozygotes (Figure 2 and supplemental Table 1), as was observed previously.9 Mean corpuscular volume and mean corpuscular hemoglobin showed a similar trend, although the difference did not reach statistical significance (supplemental Figure 2). No difference was found for ferritin, transferrin, or hemoglobin levels (supplemental Figure 1 and not shown).
Hepcidin traits and iron parameter mean levels in individuals from subset 1 classified according to MT2 genotypes (AA, AV, and VV). Hepcidin (A), hepcidin/ferritin ratio (B), hepcidin/transferrin saturation ratio (C), serum iron (D), and transferrin saturation (E) are shown. Data are expressed as mean values and are corrected by sex, age, squared age, and interaction by ANOVA (95% confidence intervals are shown). VV indicates homozygotes for the TMPRSS6 alleles encoding valine; AA, homozygotes for the TMPRSS6 alleles encoding alanine; and AV, compound heterozygotes for the 2 alleles. P values refer to comparison between AA and VV homozygotes; *P < .05; **P < .0005.
Hepcidin traits and iron parameter mean levels in individuals from subset 1 classified according to MT2 genotypes (AA, AV, and VV). Hepcidin (A), hepcidin/ferritin ratio (B), hepcidin/transferrin saturation ratio (C), serum iron (D), and transferrin saturation (E) are shown. Data are expressed as mean values and are corrected by sex, age, squared age, and interaction by ANOVA (95% confidence intervals are shown). VV indicates homozygotes for the TMPRSS6 alleles encoding valine; AA, homozygotes for the TMPRSS6 alleles encoding alanine; and AV, compound heterozygotes for the 2 alleles. P values refer to comparison between AA and VV homozygotes; *P < .05; **P < .0005.
Our results suggest that MT2 influences normal hepcidin response to both plasma and total body iron. Hepcidin regulation is complex.2 In mice, the hepcidin response to isolated increase of transferrin saturation20 or to an acute iron increase21 differs from the response to increased total body iron or to chronic iron treatment. Both responses are based on the same BMP-signaling pathway and on SMAD activation, but only the second entails a BMP6 increase.20,21 The difference in the hepcidin/transferrin saturation and hepcidin/ferritin ratios observed between the two TMPRSS6 genotypes strengthens a role for MT2 in counterbalancing both BMP6-dependent and BMP6-independent hepcidin up-regulation. The reduced activity of MT2736V demonstrated by the in vitro assay is in agreement with the effect observed in vivo.
MT2736V is the less frequent allele, with a frequency of 0.45 in VB, as in other white populations. From the available studies, the distribution among different populations is not homogeneous (supplemental Table 2). Although the samples analyzed are limited, MT2736A seems largely prevalent among blacks (0.80-0.90) compared with whites (0.50)9,11,14 and Japanese (0.40).22 Whether the variant might provide an advantage by enhancing iron absorption in conditions of limited dietary availability or may have conferred protection against certain infections remains to be clarified in future studies.
In conclusion, our data indicate that TMPRSS6 rs855791 has a functional role in determining protease activity and regulating hepcidin expression both in vitro and in normal subjects, suggesting that it influences the hepcidin response to the increase of both circulating and total body iron.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professor Carlos Lopez-Otin (Departamento de Bioquimica y Biologia Molecular-IUOPA, Universidad de Oviedo, Spain) for the kind gift of the full-length human TMPRSS6 cDNA encoding MT2736V.
This work was supported by grants from the Cariplo Foundation (project 2009-2483), e-rare 2009, Regione Lombardia (SAL-11, ID17389) and Telethon GGP08089 (to C.C.); by Fondazione Compagnia di San Paolo; the Italian Health Ministry; Progetti Finalizzati 2008; and Health Ministry Public Health program 2010 to (D.T.).
Authorship
Contribution: A.N. designed the experimental work, performed the research, and wrote the manuscript; A.P., L.S., N.C., and M.C. performed the research and analyzed the data; M.T. and D.T. performed the statistical analysis and wrote the manuscript; D.G. contributed to the experimental design and wrote the manuscript; and C.C. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clara Camaschella, Vita-Salute San Raffaele University, Via Olgettina, 60, 20132 Milan, Italy; e-mail: camaschella.clara@hsr.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal