Abstract
It is unclear how epigenetic changes regulate the induction of erythroid-specific genes during terminal erythropoiesis. Here we use global mRNA sequencing (mRNA-seq) and chromatin immunoprecipitation coupled to high-throughput sequencing (CHIP-seq) to investigate the changes that occur in mRNA levels, RNA polymerase II (Pol II) occupancy, and multiple posttranslational histone modifications when erythroid progenitors differentiate into late erythroblasts. Among genes induced during this developmental transition, there was an increase in the occupancy of Pol II, the activation marks H3K4me2, H3K4me3, H3K9Ac, and H4K16Ac, and the elongation methylation mark H3K79me2. In contrast, genes that were repressed during differentiation showed relative decreases in H3K79me2 levels yet had levels of Pol II binding and active histone marks similar to those in erythroid progenitors. We also found that relative changes in histone modification levels, in particular, H3K79me2 and H4K16ac, were most predictive of gene expression patterns. Our results suggest that in terminal erythropoiesis both promoter and elongation-associated marks contribute to the induction of erythroid genes, whereas gene repression is marked by changes in histone modifications mediating Pol II elongation. Our data map the epigenetic landscape of terminal erythropoiesis and suggest that control of transcription elongation regulates gene expression during terminal erythroid differentiation.
Introduction
The terminal differentiation of erythroid progenitors into mature erythroblasts is a complex and highly regulated process, including the survival of progenitors, 3 to 5 terminal cell divisions, and global chromatin condensation leading ultimately to enucleation. Erythroid gene expression patterns are largely governed by the coordinated interaction of coactivators, chromatin-remodeling factors, Pol II and its transcriptional complex, as well as EpoR-activated transcription factors, such as Foxo3 and Stat5.1 In addition, posttranslational modifications of histones (and their dysregulation) have been implicated in many hematologic disorders, ranging from severe anemias, such as thalassemias and aplastic anemia, to leukemias and other myelodysplastic disorders.2,3 Histone modifications regulating the switch from human fetal to adult globin expression also draw significant clinical interest because reactivation of fetal globin can greatly relieve the symptoms of sickle cell anemia and thalassemias. Thus, systematic in vivo mapping of modified histones during terminal erythropoiesis could broaden our understanding of gene regulation during this terminal developmental state and perhaps uncover other mechanisms for these disorders and possible improved treatment options.
Redistribution of histone variants and posttranslational modifications of histones in both the promoter and enhancer regions are crucial in regulating gene expression in many tissues.4-6 Most global chromatin studies thus far have focused on changes of histone modifications associated with self-renewal of stems cells or their differentiation into mature cell types.7-9 Previous work studying the role of histone modifications on erythropoiesis has centered primarily on the regulation of globin gene expression or the role of specific regulators, such as GATA1 and NF-E2, in erythroid gene expression. Extensive relationships between histone modifications and globin regulation have been established. Extensive histone modification analysis over a single locus, the β-globin locus control region (LCR),10 showed that its role is primarily to enhance the transition from transcriptional initiation to elongation. The role of single histone modifications in erythroid transcriptional regulation was shown through analysis of the effect of H3K9 methylation on elongation at the LCR11 and histone hyperacetylation of the endogenous β-globin locus on high-level β-globin expression.12 Lastly, the relationship between the genome-wide binding patterns of specific regulators, such as GATA113-15 and Ldb1,16,17 and histone modification patterns are beginning to be uncovered. Because erythroid cell differentiation is marked by activation of multiple erythroid-important genes followed by progressive chromatin condensation and associated gene silencing, it remains unknown how histone modifications regulate the selective activation and repression of genes. Our goal was to interrogate and correlate 7 important histone modifications with RNA Pol II binding and RNA-seq during terminal erythropoiesis to draw more comprehensive conclusions about erythroid epigenetics.
Recent genome-wide studies have challenged the once commonly held view that recruitment of the Pol II complex to a promoter is sufficient for gene activation. These studies have discovered that Pol II is frequently stalled at many promoters, supporting the concept that regulation of transcriptional elongation is a critical step in gene expression.9,18,19 Between 12% and 90% of genes have been reported to have paused Pol II at their promoters in different cell types in both mammals and Drosophila. The set of genes with stalled Pol II at their promoters was enriched for developmental regulators and “silent” genes in many eukaryotic genomes.9,18-21 It has been proposed that the presence of paused Pol II allows rapid up-regulation of these genes in response to external signals. However, Pol II pausing is also prevalent in highly expressed genes that are not enriched for particular developmental functions.20,22,23 Whereas Pol II pausing regulates gene expression in erythroid cell fate determination in zebrafish,24 it is unknown whether Pol II pausing is prevalent during terminal erythroid differentiation or whether it plays a significant regulatory role.
Through global profiling of histone modifications, Pol II occupancy, and gene expression of both early committed erythroid progenitors (enriched colony-forming units-erythroid [CFU-Es]) and late erythroblasts, we report here that induced genes are probably regulated at both the level of initiation and elongation, reflected by highly dynamic changes in both active modifications as well as Pol II binding and elongation modifications. Among the global patterns of histone modifications profiled herein, we found that H3K79me2 and H4K16Ac most significantly predicted changes in gene expression for the highly induced and highly repressed genes.
Methods
Primary cell isolation, antibodies, and ChIP
Erythroid TER119-negative cells (enriched for CFU-Es) and TER119-positive cells (enriched late erythroblasts) were sorted using anti-TER119 magnetic beads (BD Biosciences) from the fetal liver of E14.5 C57Bl6 mouse embryos. ChIP was performed as described previously20,25 but in brief: freshly isolated cells were fixed with 1% formaldehyde. The sheared chromatin then was immunoprecipitated using factor-specific antibodies listed in this section, and the resulting DNA was purified, along with an unimmunoprecipitated (input) control. The following antibodies against histone modifications were used: H3K4me3 (Abcam 8580), H3K4me2 (Millipore 07-030), H3K79me2 (Abcam 3594), H4K16ac (Abcam 23352), H3K36me3 (Abcam 9050), H3K27me3 (Millipore 07-449), and RNA polymerase lI (Covance mms-126r).
RNA-seq and computational analysis
Erythroid cells were isolated from E14.5 mouse embryos and sorted into 5 differentiation stages as described previously.26 Total RNA was extracted using an RNeasy kit (QIAGEN) from freshly sorted cells. The library preparation was performed using polyA+ enriched RNA according to the manufacturer's instructions (Illumina) and then sequenced on a Solexa sequencing cell. MAQ was used for mapping reads to the mouse mm9 genome.27 Filtering which reads were to be mapped is described in detail in supplemental Methods (see the Supplemental Materials link at the top of the article). Gene expression values were calculated as “reads per kilobase of exon unit per million mapped reads” (RPKM).28 The RPKM values from R3, R4, and R5 were normalized with respect to R2 and expressed as a fold change relative to R2. DAVID was used to find enriched Gene Ontology (GO) terms in the up-regulated and down-regulated subsets of the top 1000 differentially expressed genes.29
RT-PCR of erythroid-specific genes
Total RNA was isolated with the RNeasy micro kit (QIAGEN) and then reverse-transcribed using the cDNA synthesis kit with random primers (Invitrogen). Quantitative PCR was performed using SYBR Green real-time PCR on the ABI Prism 7900 sequence detection system (Applied Biosciences). Normalization was performed against 18s ribosomal RNA. Primers used were published previously.30
ChIP-seq preparation, density calculation, normalization, computational analysis, and figure generation
Detailed descriptions of ChIP library preparation have been published previously.31 Sequences uniquely mapping to the genome with zero or 1 mismatch were used in further analysis. The analysis methods used were derived from previously published methods,9,32 but in brief, the genome was divided into bins 25 base pairs in width and the ChIP-seq density within each genomic bin was then calculated as the number of ChIP-seq reads mapping within a 500-bp window (± 250 bp) surrounding the middle of that genomic bin. To facilitate comparison of ChIP-seq samples for ratio calculations between different cell stages, quantile normalization was used. Density calculation and this method of normalization are described in detail in supplemental Methods. Enriched CFU-E and late erythroblast samples for each histone mark were subjected to quantile normalization as separate groups, and the ratio of densities of each mark was calculated as the density in erythroblasts over erythroid precursors (more differentiated over less differentiated). A summary file of the density ratios for all histone modifications for each gene is included in supplemental Table 5.
Detailed descriptions of the analyses used to make each type of graph and each type of comparison in the main figures of the paper, including calculation of the traveling ratio, are included in supplemental Methods.
Sequencing data
Both RNA and ChIP sequencing data have been uploaded to the Gene Expression Omnibus database under the accession number GSE27893.
Results
Global expression analysis of erythroid progenitors reveals massive tissue-specific transcription during the TER119 transition
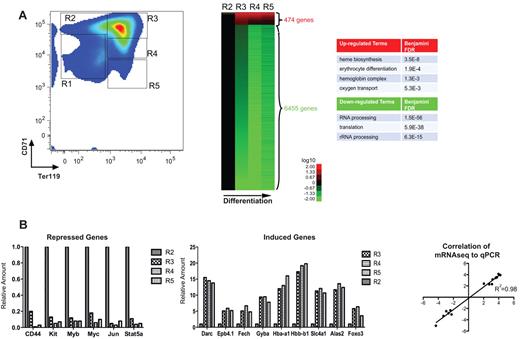
From E12 to E16, mouse fetal liver serves as the primary erythropoietic site for the embryo: cells of the erythroid lineage compose > 90% of total fetal liver cells.30 Using murine fetal liver cells, our laboratory has previously developed a series of methods to monitor erythroid differentiation both in vivo and in vitro, and to purify large quantities of primary erythroid progenitors with a very high purity.26 Mouse fetal liver cells are double-labeled for erythroid-specific TER119 and non–erythroid-specific transferrin receptor (CD71) and then sorted by flow cytometry (Figure 1A). E14.5 fetal livers contain at least 5 distinct populations of cells (R1-R5); as they progressively differentiate, they gain TER119 and then gain and subsequently lose CD71. CFU-E cells and proerythroblasts make up the R1 population; R2 consists of proerythroblasts and early basophilic erythroblasts; R3 includes early and late basophilic erythroblasts; R4 is mostly polychromatophilic and orthochromatophilic erythroblasts; and R5 is composed of late orthochromatophilic erythroblasts and reticulocytes.26
mRNA regulation during erythroid differentiation. (A) FACS plot depicting progressive stages of in vivo fetal liver erythroid differentiation used for RNA-seq; stages R1 through R5 reflect progressively further differentiated erythroid precursors, as they gain TER119 and then gain and lose CD71. The RNA-seq expression patterns of highly induced and repressed genes (rows) at differentiation stages R2 to R5 (columns) are displayed as a heat map. Expression of R3, R4, and R5 is shown as a ratio compared with R2; red represents an increase in expression; and green, a decrease in expression for each gene. The GO for highly induced and repressed genes is shown for terms with false discovery rate < 0.05. The RNA-seq data for each developmental stage were pooled from at least 2 independent experiments. (B) Expression levels of the indicated transcripts were determined by RNA-seq analysis of R2 to R5 cells isolated from D14.5 fetal liver cells. Results are expressed as ratios relative to the normalized read number in the R2 stage. Correlation against quantitative PCR for each transcript was also performed, and the coefficient of determination is shown on the right.
mRNA regulation during erythroid differentiation. (A) FACS plot depicting progressive stages of in vivo fetal liver erythroid differentiation used for RNA-seq; stages R1 through R5 reflect progressively further differentiated erythroid precursors, as they gain TER119 and then gain and lose CD71. The RNA-seq expression patterns of highly induced and repressed genes (rows) at differentiation stages R2 to R5 (columns) are displayed as a heat map. Expression of R3, R4, and R5 is shown as a ratio compared with R2; red represents an increase in expression; and green, a decrease in expression for each gene. The GO for highly induced and repressed genes is shown for terms with false discovery rate < 0.05. The RNA-seq data for each developmental stage were pooled from at least 2 independent experiments. (B) Expression levels of the indicated transcripts were determined by RNA-seq analysis of R2 to R5 cells isolated from D14.5 fetal liver cells. Results are expressed as ratios relative to the normalized read number in the R2 stage. Correlation against quantitative PCR for each transcript was also performed, and the coefficient of determination is shown on the right.
We isolated RNA from fractions R2 to R5 and used second-generation sequencing technology to sequence, on average 10 million reads per sample (supplemental Table 1). Sequenced cDNA reads were mapped to the mouse genome (mm9 version) and the exonic reads were > 100-fold higher than the intronic or intergenic reads (data not shown), suggesting that most reads were obtained from mature mRNA. We compared the expression of each gene at progressive differentiation stages relative to the expression level at the R2 stage. Gene expression was calculated using RPKM, which measures the molar concentration of a transcript by normalizing read counts to the respective mRNA length and the total number of reads in each sample. We based our gene expression pattern analysis on moderately abundant transcripts, defined as RPKM > 1 in at least 1 stage from R2 to R5. This corresponds approximately to 1 copy of RNA per cell.28 Based on this definition, the total number of expressed genes decreased from 9825 in R2 to 7494 in R5.
Figure 1A shows the genes that change most significantly during terminal erythropoiesis; approximately 474 genes increase > 2-fold in expression just as the glycoprotein TER119 is induced, during the R2 to R3 transition. This is the same stage when the most important erythroid genes are also highly induced, including α- and β-globins, heme biosynthetic enzymes, erythroid membrane proteins, and the erythroid-important transcription factors; both RNA-seq levels and their correlation to quantitative PCR (R2 = 98%) are shown for these genes we analyzed by RT-PCR in Figure 1B. GO analysis for highly induced genes yielded the following categories with a false discovery rate < 0.005: erythrocyte differentiation, oxygen transport, and hemoglobin complex; the complete GO classification is included in supplemental Table 2.
Approximately 6000 genes expressed in R2 erythroid progenitors were significantly down-regulated during erythroid differentiation, defined as a > 2-fold decrease in expression (Figure 1A). Representative examples showing gene expression changes of highly repressed erythroid-specific genes are shown in Figure 1B; again, RNA-seq levels and quantitative PCR are strongly correlated. The most significant GO term for the down-regulated genes was rRNA processing.
Going forward, we used a set of the 126 most highly induced (> 2.5-fold) and 536 most highly repressed (> 3-fold) genes to elucidate the interactions between patterns of histone modifications and gene expression; gene IDs and expression values for these very highly regulated genes are listed in supplemental Table 3.
Global chromatin modification patterns reveal that histone marks and Pol II are enriched in the nucleosomes bound to their expected genetic regions
Because of the dramatic change in gene expression we observed from R2 to R3 cells (when erythroblasts gain TER119 and ∼ 500 erythroid-important genes are induced), we decided to interrogate the relationship between global posttranslational histone modifications and gene expression across the TER119 transition. Using enriched CFU-Es (TER119-negative fetal liver cells composed of R1 and R2) and late erythroblasts (TER119-positive fetal liver cells composed of R3-R5 cells, with the majority of cells in the R3 window; Figure 1A), we carried out global location analysis of RNA Pol II binding and a number of posttranslational histone modifications: H3K4me2, H3K4me3, H3K27me3, H3K36me3, H3K79me2, H3K9Ac, and H4K16Ac using ChIP-seq.
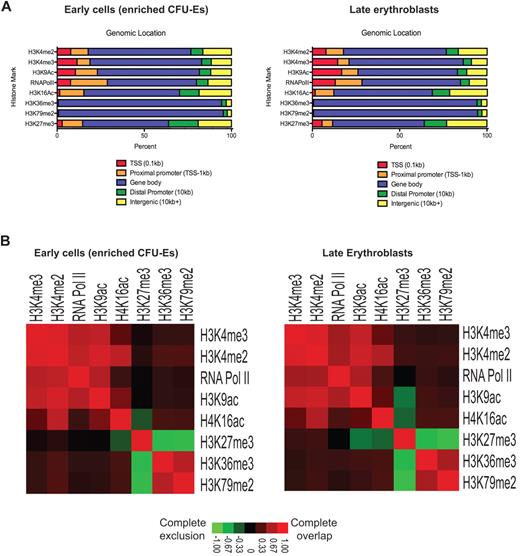
Figure 2 shows a summary of the global location analyses mapped across specific genetic compartments (irrespective of assignment to individual genes). Note first that the various histone modifications do not differ in their distribution within genetic compartments across the enriched CFU-E cell to late erythroblast transition, even though there are major changes in the levels of histone modifications near specific genes. As expected by their known functions, the activation modifications H3K4me2, H3K4me3, H3K9ac, and H4K16ac, repressive mark H3K27me3, as well as RNA Pol II, are found near the transcriptional start site (TSS) and proximal promoter regions of genes and occupy these areas with similar frequencies before and after TER119 induction. Elongation marks, such as H3K36me3 and H3K79me2, are found predominately along the bodies of genes and could serve to stop unintended transcriptional initiation within the gene bodies.33,34
The genetic distribution of various histone modifications. (A) Coverage of genetic compartments bound by various histone marks and RNA Pol II in enriched CFU-Es and late erythroblasts. Compartments are defined as: TSS (−100 bp from the TSS), proximal promoter (from −100 bp to −1 kb from the TSS), distal promoter (from −1 kb up to −10 kb from the TSS), gene body (all coding regions, including introns and exons), or intergenic (past the coding region, > −10 kb from the TSS, or not defined by one of the other categories). (B) Heat maps showing colocalization frequencies of regions in enriched CFU-Es and late erythroblasts bound by one of the various histone marks or RNA Pol II. Colors in the heat map reflect the colocalization frequency of each pair of regulators are delineated in the legend below the heat maps; red represents which regions have the most overlap; and green, exclusion of the regions.
The genetic distribution of various histone modifications. (A) Coverage of genetic compartments bound by various histone marks and RNA Pol II in enriched CFU-Es and late erythroblasts. Compartments are defined as: TSS (−100 bp from the TSS), proximal promoter (from −100 bp to −1 kb from the TSS), distal promoter (from −1 kb up to −10 kb from the TSS), gene body (all coding regions, including introns and exons), or intergenic (past the coding region, > −10 kb from the TSS, or not defined by one of the other categories). (B) Heat maps showing colocalization frequencies of regions in enriched CFU-Es and late erythroblasts bound by one of the various histone marks or RNA Pol II. Colors in the heat map reflect the colocalization frequency of each pair of regulators are delineated in the legend below the heat maps; red represents which regions have the most overlap; and green, exclusion of the regions.
Correspondingly, when we looked at the overlap between these bound regions before and after TER119 induction (Figure 2B), we noted, as expected, that the active methylation marks generally colocalize with each other (red) and that the elongation marks colocalize with each other but are excluded from regions (green) where H3K27me3 is bound, consistent with the exclusive binding specificities of polycomb complexes and methyltransferases.35 However, in late erythroblasts, once the massive induction of such genes as globins and the erythroid membrane proteins has already taken place, regions with H3K9Ac become more excluded from H3K27me3-bound regions and regions with H4K16Ac colocalize more with another active mark H3K4me2. Bound regions are listed in supplemental Table 3.
Both activating and repressive histone modifications are present on highly induced genes before and after gene induction
Prior studies have suggested that certain histone modifications mark specific genes for future expression.36-38 We examined the relationship between global posttranslational histone modifications and gene expression across the TER119 transition by focusing initially on the set of 126 genes that were induced by the greatest amount during erythropoiesis (supplemental Table 3).
Location analysis of histone modifications revealed that H3K4me2, H3K4me3, and H3K9Ac were present at many promoters of highly induced genes, even when they were expressed at relatively low levels in enriched CFU-Es (Figure 3) but, depending on the gene, increased either slightly or significantly on induction. Although individual induced genes vary as to the extent of increase for each histone modification, the average binding densities of the selected highly induced genes reveal that each of these 3 marks increases significantly in this group of genes across the TER119 transition. Pol II binding and the level of the elongation mark H3K79me2 also increased significantly on induction (Figure 4; Table 1).
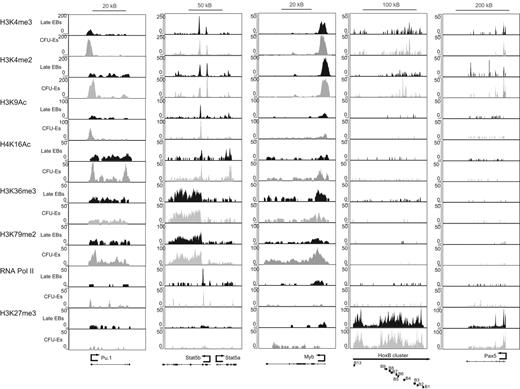
Binding of histone modifications and RNA pol II in highly induced genes. ChIP-seq binding data for each of the histone marks is shown for select erythroid-specific genes known to be induced during differentiation (Foxo3, Epb4.1 [band 4.1], Hbb-b1 [β-globin], Alas2, Slc4a1 [band 3]). Light-colored peaks are in enriched CFU-Es, and dark-colored peaks in late erythroblasts. Numbers shown outside each graph are the scale of each graph (in normalized reads), and genomic scales (in kilobases) are shown for each gene above the set of graphs.
Binding of histone modifications and RNA pol II in highly induced genes. ChIP-seq binding data for each of the histone marks is shown for select erythroid-specific genes known to be induced during differentiation (Foxo3, Epb4.1 [band 4.1], Hbb-b1 [β-globin], Alas2, Slc4a1 [band 3]). Light-colored peaks are in enriched CFU-Es, and dark-colored peaks in late erythroblasts. Numbers shown outside each graph are the scale of each graph (in normalized reads), and genomic scales (in kilobases) are shown for each gene above the set of graphs.
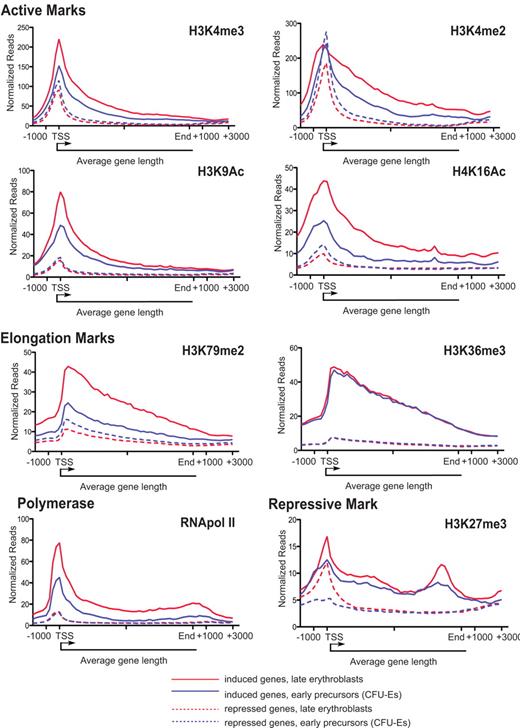
Regulation of histone modifications and RNA pol II binding in highly regulated genes. The mean binding density of each of the indicated modifications is shown averaged across the length of an average gene from 2 kb upstream of the TSS to 3 kb downstream of the gene terminus for the set of 126 induced genes (solid line) and 536 repressed genes (dashed line). Blue lines represent the mean binding density for each mark in enriched CFU-Es; and red lines, densities in late erythroblasts.
Regulation of histone modifications and RNA pol II binding in highly regulated genes. The mean binding density of each of the indicated modifications is shown averaged across the length of an average gene from 2 kb upstream of the TSS to 3 kb downstream of the gene terminus for the set of 126 induced genes (solid line) and 536 repressed genes (dashed line). Blue lines represent the mean binding density for each mark in enriched CFU-Es; and red lines, densities in late erythroblasts.
Quantification and distribution of reads near highly induced and highly repressed genes across the TER119 transition
| Histone mark . | Cell type (across TER119 transition) . | Genes . | |||
|---|---|---|---|---|---|
| Up-regulated . | Down-regulated . | ||||
| Peak ± 95% CI . | Total ± 95% CI . | Peak ± 95% CI . | Total ± 95% CI . | ||
| H3K4me3 | CFU-Es | 152.6 ± 21.3 | 2083.1 ± 367.1 | 115.1 ± 5.8 | 1001.4 ± 89.7 |
| Erythroblasts | 220.2 ± 27.2 | 3225 ± 544.7 | 91.1 ± 4.9 | 723 ± 64.1 | |
| H3K4me2 | CFU-Es | 241.1 ± 26.5 | 4077.5 ± 635.7 | 276.4 ± 10.7 | 2437.4 ± 195 |
| Erythroblasts | 228.6 ± 23.2 | 6383.4 ± 896.1 | 186.4 ± 9.9 | 1656.4 ± 149.1 | |
| H3K9Ac | CFU-Es | 48.5 ± 9.4 | 869.9 ± 135 | 18.4 ± 1.3 | 268 ± 16.3 |
| Erythroblasts | 79.9 ± 16.1 | 1238.6 ± 214.3 | 15.7 ± 0.9 | 209.5 ± 13.1 | |
| H4K16Ac | CFU-Es | 25.3 ± 5.4 | 605.2 ± 98.2 | 14.1 ± 1.3 | 282.4 ± 37.3 |
| Erythroblasts | 43.9 ± 7.9 | 1141.7 ± 184.1 | 9.9 ± 0.6 | 254.2 ± 9.5 | |
| RNA Pol II | CFU-Es | 45.2 ± 10.7 | 611 ± 126.8 | 11.8 ± 1.3 | 198 ± 20.6 |
| Erythroblasts | 77.5 ± 15.8 | 1279.4 ± 296.9 | 12.6 ± 0.9 | 214.2 ± 13.8 | |
| H3K79me2 | CFU-Es | 24.5 ± 6 | 690.1 ± 95.7 | 16.1 ± 1.1 | 443.3 ± 25.7 |
| Erythroblasts | 42.8 ± 7.5 | 1300.8 ± 165.1 | 11.2 ± 0.7 | 336.5 ± 18.7 | |
| H3K36me3 | CFU-Es | 47 ± 8.3 | 1511.7 ± 194.7 | 7.5 ± 0.4 | 259.7 ± 12 |
| Erythroblasts | 48.8 ± 8 | 1558.7 ± 206 | 7.4 ± 0.4 | 240.7 ± 12.7 | |
| H3K27me3 | CFU-Es | 12.5 ± 3.6 | 463.9 ± 113.3 | 5.3 ± 0.7 | 223 ± 21.3 |
| Erythroblasts | 16.9 ± 3.8 | 536.7 ± 125.3 | 12 ± 1.1 | 277.5 ± 26.2 | |
| Histone mark . | Cell type (across TER119 transition) . | Genes . | |||
|---|---|---|---|---|---|
| Up-regulated . | Down-regulated . | ||||
| Peak ± 95% CI . | Total ± 95% CI . | Peak ± 95% CI . | Total ± 95% CI . | ||
| H3K4me3 | CFU-Es | 152.6 ± 21.3 | 2083.1 ± 367.1 | 115.1 ± 5.8 | 1001.4 ± 89.7 |
| Erythroblasts | 220.2 ± 27.2 | 3225 ± 544.7 | 91.1 ± 4.9 | 723 ± 64.1 | |
| H3K4me2 | CFU-Es | 241.1 ± 26.5 | 4077.5 ± 635.7 | 276.4 ± 10.7 | 2437.4 ± 195 |
| Erythroblasts | 228.6 ± 23.2 | 6383.4 ± 896.1 | 186.4 ± 9.9 | 1656.4 ± 149.1 | |
| H3K9Ac | CFU-Es | 48.5 ± 9.4 | 869.9 ± 135 | 18.4 ± 1.3 | 268 ± 16.3 |
| Erythroblasts | 79.9 ± 16.1 | 1238.6 ± 214.3 | 15.7 ± 0.9 | 209.5 ± 13.1 | |
| H4K16Ac | CFU-Es | 25.3 ± 5.4 | 605.2 ± 98.2 | 14.1 ± 1.3 | 282.4 ± 37.3 |
| Erythroblasts | 43.9 ± 7.9 | 1141.7 ± 184.1 | 9.9 ± 0.6 | 254.2 ± 9.5 | |
| RNA Pol II | CFU-Es | 45.2 ± 10.7 | 611 ± 126.8 | 11.8 ± 1.3 | 198 ± 20.6 |
| Erythroblasts | 77.5 ± 15.8 | 1279.4 ± 296.9 | 12.6 ± 0.9 | 214.2 ± 13.8 | |
| H3K79me2 | CFU-Es | 24.5 ± 6 | 690.1 ± 95.7 | 16.1 ± 1.1 | 443.3 ± 25.7 |
| Erythroblasts | 42.8 ± 7.5 | 1300.8 ± 165.1 | 11.2 ± 0.7 | 336.5 ± 18.7 | |
| H3K36me3 | CFU-Es | 47 ± 8.3 | 1511.7 ± 194.7 | 7.5 ± 0.4 | 259.7 ± 12 |
| Erythroblasts | 48.8 ± 8 | 1558.7 ± 206 | 7.4 ± 0.4 | 240.7 ± 12.7 | |
| H3K27me3 | CFU-Es | 12.5 ± 3.6 | 463.9 ± 113.3 | 5.3 ± 0.7 | 223 ± 21.3 |
| Erythroblasts | 16.9 ± 3.8 | 536.7 ± 125.3 | 12 ± 1.1 | 277.5 ± 26.2 | |
Although repressive marks are thought to be mostly associated with silent or repressed genes,7,9,32,38 our analysis indicates that H3K27me3 marks are present near highly induced genes in both enriched CFU-Es and late erythroblasts, albeit at a relatively low level. The average peak height for H3K27me3 was much higher for the constitutively repressed Hox cluster and Pax5 (Figure 4) than for the induced genes shown in Figure 3 and averaged in Figure 5.
Binding of histone modifications and RNA pol II in highly repressed genes. ChIP-seq binding data for each of the histone marks are shown for select erythroid-specific genes known to be repressed during differentiation (Myb, Pu.1, Stat5a and -b, and the constitutively repressed HoxB gene cluster and Pax5). Light-colored peaks are in enriched CFU-Es, and dark-colored peaks in late erythroblasts. Numbers shown outside each graph are the scale of each graph (in normalized reads), and genomic scales (in kilobases) are shown for each gene above the set of graphs.
Binding of histone modifications and RNA pol II in highly repressed genes. ChIP-seq binding data for each of the histone marks are shown for select erythroid-specific genes known to be repressed during differentiation (Myb, Pu.1, Stat5a and -b, and the constitutively repressed HoxB gene cluster and Pax5). Light-colored peaks are in enriched CFU-Es, and dark-colored peaks in late erythroblasts. Numbers shown outside each graph are the scale of each graph (in normalized reads), and genomic scales (in kilobases) are shown for each gene above the set of graphs.
Levels of active acetylation marks H3K9ac and H4K16ac increase significantly in highly induced genes
Previous studies in other cell types have shown that the levels of H3 and H4 acetylation are proportional to the rate of transcription.25,39 We observed that both H3K9Ac and H4K16Ac modifications showed increased enrichment near the TSS of induced genes during erythroid differentiation when binding densities were averaged across the entire set of 126 highly induced genes (Figure 4; Table 1). Of note, H4K16Ac increased to a greater extent along the gene body as well (Figure 4) and correlated better with gene expression changes (Figures 6 and 7). This observation is consistent with the extended histone hyperacetylation observed in the body of the globin genes detailed previously.12
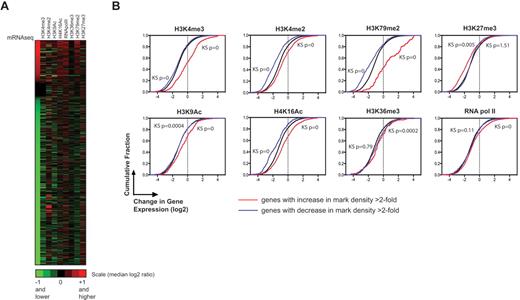
Correlation between gene expression and changes in histone modifications during erythroid differentiation. (A) Clustergram showing the fold change in enrichment of histone marks (columns) for induced and repressed genes (rows) compared with the fold change in expression for the same genes. Both types of ratios are expressed as log2 ratios (late erythroblasts relative to enriched CFU-Es). Genes that are black in the expression column (unchanged ratios) are constitutively expressed genes with RPKM > 2 and < 0.2 fold change between the R2 and R3 stages. (B) Cumulative distribution function plots comparing mRNA expression ratios (late erythroblasts relative to enriched CFU-Es) for the set of all genes with significant enrichment in a specific histone mark (red line) or decrease in that mark (blue line), against the set of all genes (black line). Shifts to the right signify a positive correlation; shifts to the left a negative correlation. K-S test P values are shown on each graph; values < .001 for a line signify that the subset of genes is highly significantly different from the black line. Histone marks are indicated above each graph.
Correlation between gene expression and changes in histone modifications during erythroid differentiation. (A) Clustergram showing the fold change in enrichment of histone marks (columns) for induced and repressed genes (rows) compared with the fold change in expression for the same genes. Both types of ratios are expressed as log2 ratios (late erythroblasts relative to enriched CFU-Es). Genes that are black in the expression column (unchanged ratios) are constitutively expressed genes with RPKM > 2 and < 0.2 fold change between the R2 and R3 stages. (B) Cumulative distribution function plots comparing mRNA expression ratios (late erythroblasts relative to enriched CFU-Es) for the set of all genes with significant enrichment in a specific histone mark (red line) or decrease in that mark (blue line), against the set of all genes (black line). Shifts to the right signify a positive correlation; shifts to the left a negative correlation. K-S test P values are shown on each graph; values < .001 for a line signify that the subset of genes is highly significantly different from the black line. Histone marks are indicated above each graph.
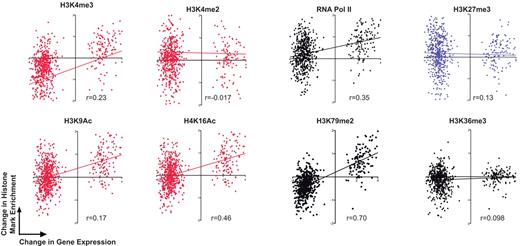
Correlation between gene expression and changes in histone modifications of the most highly changed genes during erythroid differentiation. Correlation curves of the change in enrichment of a histone mark bound to a gene (late erythroblasts relative to enriched CFU-Es) plotted against the change in expression of that gene (late erythroblasts relative to enriched CFU-Es) using the set of 126 highly induced genes and 536 highly repressed genes. Ratios represented for enrichment and expression changes are in a log2 scale. Histone marks are indicated above each graph, and correlation coefficient (Pearson r) is given below each graph. Graphs with red dots represent active marks; black dots, elongation marks and Pol II; and blue, the repressive mark H3K27me3.
Correlation between gene expression and changes in histone modifications of the most highly changed genes during erythroid differentiation. Correlation curves of the change in enrichment of a histone mark bound to a gene (late erythroblasts relative to enriched CFU-Es) plotted against the change in expression of that gene (late erythroblasts relative to enriched CFU-Es) using the set of 126 highly induced genes and 536 highly repressed genes. Ratios represented for enrichment and expression changes are in a log2 scale. Histone marks are indicated above each graph, and correlation coefficient (Pearson r) is given below each graph. Graphs with red dots represent active marks; black dots, elongation marks and Pol II; and blue, the repressive mark H3K27me3.
H3K4me2 levels increase more along the gene body in highly induced genes
Enrichment of H3K4me2 within the gene body is an indicator of actively transcribed genes in yeast,40,41 whereas enrichment around the TSS is more prevalent in mammalian T cells, fibroblasts, and cancer cell lines.32,42 Contrary to the other activation marks profiled, which show increased levels predominantly near the TSS, the level of H3K4me2 near the TSS, averaged across all highly induced genes, is comparable between enriched CFU-Es and late erythroblasts (Figure 4). However, the binding density along the gene body is higher and extends beyond the end of the transcript (Figure 4). This observation probably reflects a tissue-specific regulatory role for H3K4me2 in the specific expression of erythroid genes, similar to that observed in T-cell specific gene induction.43
Levels of the elongation mark H3K79me2 change during gene induction and repression whereas those of H3K36me3 do not
Interestingly, although, in most other tissues, H3K36me3 and H3K79me2 are redundant elongation marks,9,20 our data show that the 2 marks are regulated differently during the induction of erythroid-specific genes during terminal erythropoiesis. Although both bind along the gene body, whereas H3K79me2 is significantly induced, H3K36me3 does not change much near either highly induced or highly repressed genes (Figures 3,Figure 4–5; quantified in Table 1). However, the overall level in these 2 classes of genes differs significantly: highly induced genes show relatively high levels of H3K36me3 even before induction (Figures 3 and 4), whereas H3K79me2 levels seem to correspond to current transcription as well as to levels of bound RNA Pol II.
Highly induced genes appear to be regulated at both transcriptional initiation as well as elongation
Prior studies have provided evidence that both transcriptional initiation and elongation (or promoter-proximal Pol II pause release) are crucial in regulating gene expression. In addition, H3K4me3 and H3K9Ac have been shown to indicate Pol II binding and evidence of transcriptional initiation.9,20 As stated in Figures 3 and 4 and Table 1, we observed that H3K4me3 and H3K9Ac levels increase near the TSS of induced genes during erythropoiesis with a concomitant increase in Pol II, suggesting that these genes are regulated at the level of transcriptional initiation.
The presence of high polymerase density at the TSS relative to the gene body has previously been cited as evidence for Pol II pausing or a related form of postinitiation regulation in many eukaryotic cells.20 We observed that the induced genes were characterized by a relatively high level of Pol II binding near the TSS and a lower level of binding along the coding region (evidence of Pol II pausing) before they were induced (in enriched CFU-Es) with a modest increase in Pol II binding near the TSS and throughout the gene body to the end of the gene (evidence of active transcription) after the gene was induced in later erythroblasts (Figure 4; supplemental Figure 1A). Using the set of all Pol II-bound genes, we calculated the traveling ratio as defined previously20 to compare the relative degree of pausing between the 2 cell types (by comparing the read density of Pol II in the promoter region over that the gene body). Promoters of induced genes showed evidence of more paused Pol II in enriched CFU-Es than late erythroblasts (mean log2 traveling ratio of 4.23 vs 3.36). Using an even more stringent definition of pausing, defining paused Pol II as traveling ratio > 1 SD above the mean,22 we found twice as many Pol II-bound induced genes defined as paused in enriched CFU-Es than late erythroblasts. In addition, the elongation mark H3K79me2 increases dramatically on gene induction. These observations provide evidence that postinitiation steps may be important in regulating gene induction during terminal erythropoiesis.
Repressed genes are also regulated at the level of elongation and gain repressive marks during differentiation
Similar to H3K4me3 and H3K9Ac before and after repression, RNA Pol II binding does not change significantly near the highly repressed genes; indeed, a small peak near the promoter remains in both enriched CFU-Es and late erythroblasts (Figures 4 and 5; quantified in Table 1). The presence of Pol II at the TSS of repressed genes suggests that repression is probably not only regulated at the level of transcriptional initiation, but that inhibiting Pol II pause release could also regulate gene repression. Significant decreases in H3K79me2 during this transition (Figures 4 and 5; quantified in Table 1) also confirm this conclusion. As expected, levels of the repressive mark H3K27me3 increase near the TSS in repressed genes (Figure 4) on repression but also cover large domains of constitutively repressed genes (eg, > 100 kb of the HoxB gene cluster and the entire Pax5 coding region illustrated in Figure 5).
Quantitative changes in the enrichment of certain histone marks, in particular H3K79me2 and H4K16Ac, predict changes in gene expression
In many cell types, particular patterns of histone modifications have been associated with active transcription or repression. To examine the concept that dynamic changes in histone modifications may regulate erythroid transcription, we compared the change in enrichment of all the histone marks that we assayed with the change in expression of the most highly regulated genes (Figure 6A; ratios listed in supplemental Table 5). The absolute presence of these histone modifications in highly induced and repressed genes shows almost indistinguishable patterns; H3K4me2, H3K4me3, and H3K9Ac are present at a measurable level on > 80% of the promoters of both highly induced and highly repressed genes (supplemental Figure 2 showing bound genes). This suggests that the qualitative presence of a specific histone mark is not accurate in predicting changes in gene expression during erythroid differentiation, whereas, in contrast, quantitative changes in specific marks can accurately predict changes in gene expression.
To better quantitate the relative changes in enrichment of each histone modification, we determined the cumulative distribution functions (Figure 6B) of histograms of genes bound by each histone mark. The blue and red lines represent the sets of genes in which each specific histone mark decreased or increased > 2-fold, respectively; the black line represents the set of all genes. Note first that the majority of genes are repressed during this stage of erythropoiesis, so the black line is not symmetric around x = 0 but rather shifted to the left. A shift of the red curve to the right relative to the black curve, as observed for H4K16Ac and H3K79me2, indicates that a higher level of this histone modification correlates with induction of the gene in late erythroblasts. This correlation with increase in the mark is observed for all the marks except H3K36me3 and H3K27me3. Even for genes with a > 2-fold increase in activation-associated histone marks, many of them are repressed during induction of Ter-119 (red lines fall in the negative range for changes in expression). As described earlier (Figure 4; Table 1), whereas the relative level of H3K36me does not change after active transcription of highly induced genes, the absolute level is much higher in induced genes than repressed ones, suggesting that this mark may play a different role than the other elongation mark H3K79me2. Interestingly, H3K27me3 seems to react in the opposite way from all other marks, an increase in the level of this mark during the TER119 transition correlates with a reduction in the level of the corresponding mRNA.
Thus, relative changes in the enrichment of specific histone modifications more accurately predict the direction of gene expression. Moreover, we tested by quantitative correlation how well these histone marks predict the direction of expression of only the most highly induced and highly repressed genes (Figure 7). Consequently, this additional method confirms that elongation mark H3K79me2 and active acetylation H4K16Ac are most highly correlated with the direction of change in gene expression (Pearson coefficients of r = 0.7 and r = 0.46, respectively). However, the levels of histone marks do not correlate with absolute gene expression levels (supplemental Table 6).
Discussion
Chromatin structure affects several processes important to transcription, including transcription factor and polymerase recruitment, and transcriptional initiation, elongation, and cessation.41 Terminal erythropoiesis offers a unique backdrop in which to study the epigenetic regulation of transcription because there is a relatively short period of massive, highly tissue-specific transcription followed by a period of chromatin condensation leading ultimately to enucleation. Ours is the first study to characterize this complex process in intricate detail using deep-sequencing: from mRNA sequencing revealing vast transcriptional changes in this tissue type to extensive histone modification maps overlying these areas of transcription. This work has revealed some important phenomenon concerning the epigenetic regulation of this highly specialized example of tissue-specific transcription.
Previous studies have interpreted the presence of active chromatin modifications on actively transcribed genes in several different ways. Our data support the interpretation that active chromatin modifications could reflect the current status of transcription44 because, even though we detected significant amounts of RNA polymerase, active acetylation marks, elongation marks, and moderate amounts of transcript in enriched CFU-Es when the genes were expressed at a low level, all of these marks increased on induction, reflecting the transcriptional status of the cells. These increased histone modification levels, together with concomitant increases in transcript levels, also support the interpretation that active histone modifications mark genes that are poised for future activation31,45 and are consistent with previous findings showing that significant levels of histone methylation are present on inactive genes, such as β-globin locus, before erythroid development, when the gene is still inactive.45-47
Recent genome-wide studies have uncovered that Pol II is frequently stalled at the promoter, supporting the concept that transcriptional elongation is a critical step in gene regulation.9,18,19 Our data confirm that both transcriptional initiation and elongation probably contribute to the activation of the most highly induced genes. First, RNA polymerase binding at the TSS increases during gene induction, suggesting that recruitment of the initiation apparatus increases to accommodate the elevated rate of transcription. Second, quantification of paused Pol II using the traveling ratios > 1 SD above the mean showed that there was twice as much paused polymerase near highly induced genes in erythroid precursors, before the genes were fully activated, than in late erythroblasts. The presence of stalled Pol II at actively transcribed genes probably facilitates a permissive chromatin environment around the TSS22 and allows for rapid up-regulation of transcription at these sites. Stalled Pol II may also regulate how genes can be rapidly turned off after transcription because repression of the entire genome is required in preparation for extrusion of the nucleus. A similar incidence of paused Pol II in repressed genes in erythroid precursors (data not shown) also supports this hypothesis as these genes were previously turned off earlier in erythropoiesis.
In yeast, H3K79 methylation is associated with gene activation.25 H3K79me2 is also correlated with transcriptional activation in genome-wide location analyses of Drosophila cells48 as well as in higher eukaryotes, such as human ES cells.9 However, location analysis performed on human T cells revealed that the H3K79me2 mark showed no preferential association with either gene activation or repression,32 raising the possibility that there is tissue specificity of the regulation of gene expression by H3K79me2 levels. In our work, dynamic changes in H3K79me2 correlated very well with the direction of changes in expression, in all genes as well as in only the most highly induced and highly repressed transcripts. This observation highlights the important regulatory role that transcriptional elongation may play during terminal erythroid differentiation. Our observations are consistent with recent studies showing that TIF1-γ is critical for erythroid development by recruiting positive elongation factors, such as pTEFb, to erythroid-specific genes that can counteract Pol II pausing.24 Moreover, our results are in agreement with the recent finding that dot1l, encoding an enzyme responsible for H3K79 methylation, is crucial for erythropoiesis.49
It is striking that the other elongation mark, H3K36me3, thought to be redundant to H3K79me2,9,20 did not appear to act in the same manner as H3K79me2. In contrast to H3K79me2, levels of H3K36me3 did not change in either induced or repressed genes between erythroid precursors and late progenitors, nor did this mark correlate with changes in gene expression. Importantly, the magnitude of H3K36me3 enrichment differed dramatically between highly induced and highly repressed genes (Figure 4). This observation suggests that H3K36me3 may play a completely different role in terminal erythroid transcription than in other tissues; indeed, this modification may be marking the coding regions of highly induced genes for future activation much in the same manner that the active modifications mark the promoter region.
In conclusion, quantitative comparisons of histone marks, in particular H3K79me2, are accurate predictors for the direction of gene expression. We found that active promoter marks and elongation marks, such as H3K36me3, are present on highly induced genes before activation, possibly poising these genes for later induction. This finding, along with evidence that transcriptional elongation may contribute to the regulation of erythroid-specific gene repression and induction, suggests that such functions may be acting to facilitate such a highly specialized terminal cell state requiring rapid, highly controlled, massive transcription (eg, of the globin genes) before rapid gene repression and chromatin condensation before enucleation.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Manuel Ortega and Paula Trapman for technical assistance in generating the ChIP-seq data, Prathapan Thiru from WIBR/BARC for initial analysis of the ChIP-seq data and cumulative distribution function plot generation, Vijay Sankaran and Youngtae Jeong for useful discussions, Peter Rahl for critical reading of the manuscript, and Charles Lin for the calculation of Pol II pausing.
This work was supported by the National Institute of Diabetic, Digestive, and Kidney Diseases, National Institutes of Health (S.M.H. and H.F.L.) and the National Heart, Lung, and Blood Institute, National Institutes of Health (H.F.L.) and in part by a Croucher Foundation Postdoctoral Research Grant (P.W.).
National Institutes of Health
Authorship
Contribution: P.W. and S.M.H. designed and performed research, analyzed and interpreted the data, and wrote the manuscript; A.W.C. and G.M.F. analyzed data and reviewed the manuscript; R.A.Y. reviewed and revised the manuscript; and H.F.L. designed research and wrote and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.
References
Author notes
P.W. and S.M.H. contributed equally to this study.

![Figure 3. Binding of histone modifications and RNA pol II in highly induced genes. ChIP-seq binding data for each of the histone marks is shown for select erythroid-specific genes known to be induced during differentiation (Foxo3, Epb4.1 [band 4.1], Hbb-b1 [β-globin], Alas2, Slc4a1 [band 3]). Light-colored peaks are in enriched CFU-Es, and dark-colored peaks in late erythroblasts. Numbers shown outside each graph are the scale of each graph (in normalized reads), and genomic scales (in kilobases) are shown for each gene above the set of graphs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/16/10.1182_blood-2011-03-341404/4/m_zh89991179740003.jpeg?Expires=1769160190&Signature=uopWV-9rFz-2rewfYj5sbxkbr~cLMpT8Q8c6rBrEYQ32P1QbV7f29nwHCjVqwRHXi~lpO9Ls9ZOSvVFTNJHf7EsotySj-~4VoMq7U~lorSlNMU5MDU2MMFliwHrr~VpYce32o~fAahnitfOj1XWtfE8PT5niSUuV93J~QJN~bsZ46YPRF8cecz0Q6PlMgvJA~hhljH6RvyIT~LNR~iirEayhqZJHBLoNn1jpU7PM1Bss39SAPFp9k9HUdo0RI6mYCqV0hbpPJg86BQeewl7py3707fVdSMidDHBgxNBlAqeQfU3YprNtQpoBl9BMYmYmKcY4UtY4Itg8a8jWtvJLMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)