Abstract
Splenic marginal zone lymphoma (SMZL) is one of the few B-cell lymphoma types that remain orphan of molecular lesions in cancer-related genes. Detection of active NF-κB signaling in 14 (58%) of 24 SMZLs prompted the investigation of NF-κB molecular alterations in 101 SMZLs. Mutations and copy number abnormalities of NF-κB genes occurred in 36 (36%) of 101 SMZLs and targeted both canonical (TNFAIP3 and IKBKB) and noncanonical (BIRC3, TRAF3, MAP3K14) NF-κB pathways. Most alterations were mutually exclusive, documenting the existence of multiple independent mechanisms affecting NF-κB in SMZL. BIRC3 inactivation in SMZL recurred because of somatic mutations that disrupted the same RING domain that in extranodal marginal zone lymphoma is removed by the t(11;18) translocation, which points to BIRC3 disruption as a common mechanism across marginal zone B-cell lymphomagenesis. Genetic lesions of NF-κB provide a molecular basis for the pathogenesis of more than 30% of SMZLs and offer a suitable target for NF-κB therapeutic approaches in this lymphoma.
Introduction
Knowledge of the pathogenesis of splenic marginal zone lymphoma (SMZL) is scarce.1,2 On the basis of clinical and epidemiologic data, hepatitis C virus infection might play a role in a fraction of SMZLs.1,2 The contribution of antigen stimulation is suggested by the highly restricted immunoglobulin gene repertoire and by the stereotyped B-cell receptor in ∼ 10% of SMZLs.3,4 Cancer genes involved in SMZL pathogenesis are currently unknown, except for TP53 disruption in ∼ 10%-15% of cases.3,5
Indirect evidence points to nuclear factor-κB (NF-κB) pathway genes as attractive candidates in SMZL. Abnormal marginal zone B-cell expansions are observed frequently in animal models with constitutive NF-κB activation.6-8 In addition, up-regulated expression of NF-κB target genes in SMZL suggests the occurrence of NF-κB activation,9 which in other B-cell malignancies is sustained by cancer-specific genetic lesions.10,11
This rationale prompted the assessment of NF-κB genetic alterations in SMZL. We document that the canonical and noncanonical NF-κB pathways are affected by multiple genetic lesions in > 30% of SMZLs.
Methods
Patient samples
The study was based on 101 SMZLs diagnosed according to World Health Organization classification and SMZL Working Party criteria (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).1,2 Matched germline DNA was extracted from saliva. Patients provided informed consent in accordance with local institutional review board requirements and the Declaration of Helsinki. The study was approved by the ethics committee of the Amedeo Avogadro University.
Molecular studies
Mutation analysis of 20 NF-κB pathway genes (supplemental Table 2; supplemental Figure 1) and TP53 was performed by DNA Sanger sequencing. Copy number abnormalities (CNAs) of NF-κB genes and TP53 were analyzed by FISH (probes in supplemental Table 2) and single-nucleotide polymorphism array (GeneChip Human Mapping 250K NspI, Affymetrix; Gene Expression Omnibus accession number GSE24881).5 Immunoglobulin heavy chain gene rearrangements and stereotyped VH CDR3 were analyzed as reported previously.4
Immunohistochemistry
Presence of active NF-κB was analyzed by immunohistochemistry on formalin-fixed, paraffin-embedded primary SMZL biopsy samples (n = 24) with anti-NFKB1 (p50; Cell Signaling, #3035) and anti-NFKB2 (p52; Cell Signaling, #3017) primary antibodies.10 Cases with > 20% cells showing positive nuclear staining (of 200 cells counted in representative fields) were scored as NF-κB active.10
Western blot studies
B cells from SMZL spleens were purified by negative selection with Dynal magnetic beads (Invitrogen). Proteins were resolved by SDS-PAGE and analyzed by Western blot. Antibodies were anti-NFKB2 (Cell Signaling, #4882), anti-MAP3K14 (Cell Signaling, #4994), and anti-actin (Santa Cruz Biotechnology, #sc-1615). Image acquisition and densitometric analyses were performed with ImageQuant LAS4000 and TL software (GE Healthcare).
Results and discussion
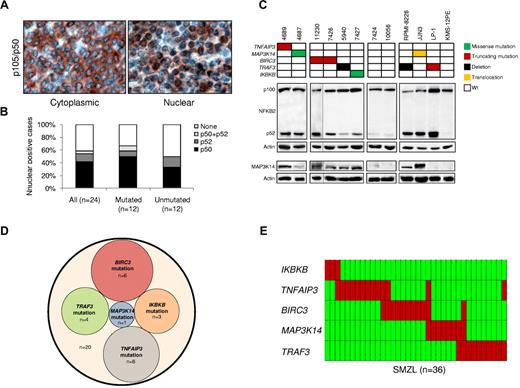
SMZLs were characterized for active NF-κB signaling by immunohistochemical assays that detected nuclear NFKB1 (p50) and NFKB2 (p52).10 Nuclear localization of NF-κB was observed in tumor cells of 14 (58%) of 24 SMZLs (Figure 1A-B). Both canonical (11/24, 46%) and noncanonical (4/24, 17%) NF-κB pathways were activated. One patient showed activation of both pathways. Engagement of the noncanonical NF-κB pathway was confirmed by biochemical detection of NFKB2 processing (Figure 1C).
The NF-κB pathway is activated and frequently targeted by genetic lesions in SMZLs. (A) Immunohistochemical staining of SMZL biopsy samples with anti-NFKB1 (p50) antibody. Nuclear localization of NF-κB denotes active signaling as opposed to inactive cytoplasmic localization. Original magnification ×400. (B) Prevalence of constitutive NF-κB activation scored by immunohistochemistry in SMZLs overall and according to mutation status of NF-κB genes. (C) Western blot analysis showing NFKB2 processing and MAP3K14 expression in purified primary tumor cells from 8 SMZL cases carrying wild-type (Wt) or aberrant NF-κB genes. Case 11230 was run in a different gel using the same conditions, as indicated by the black dividing line. The RPMI-8226 and LP-1 (both multiple myeloma) and JJN3 (plasma cell leukemia) cell lines were used as positive controls for NFKB2 processing. The KMS-12PE cell line (multiple myeloma) was used as negative control for NFKB2 processing. The RPMI-8226 and JJN3 cell lines were used as positive controls for MAP3K14 expression. The LP-1 and KMS-12PE cell lines were used as negative controls for MAP3K14 expression. Actin was used as loading control. (D) Venn diagram illustrating the absence of overlap between NF-κB gene somatic mutations. (E) Genetic lesions (including mutations and CNAs) of NF-κB genes are largely mutually exclusive. In the heat map, rows correspond to identical genes, and columns represent individual patients color-coded on the basis of gene status (green, wild-type; red, mutations of IKBKB, mutations or gain of MAP3K14, mutations and/or deletion of TRAF3, TNFAIP3, and BIRC3).
The NF-κB pathway is activated and frequently targeted by genetic lesions in SMZLs. (A) Immunohistochemical staining of SMZL biopsy samples with anti-NFKB1 (p50) antibody. Nuclear localization of NF-κB denotes active signaling as opposed to inactive cytoplasmic localization. Original magnification ×400. (B) Prevalence of constitutive NF-κB activation scored by immunohistochemistry in SMZLs overall and according to mutation status of NF-κB genes. (C) Western blot analysis showing NFKB2 processing and MAP3K14 expression in purified primary tumor cells from 8 SMZL cases carrying wild-type (Wt) or aberrant NF-κB genes. Case 11230 was run in a different gel using the same conditions, as indicated by the black dividing line. The RPMI-8226 and LP-1 (both multiple myeloma) and JJN3 (plasma cell leukemia) cell lines were used as positive controls for NFKB2 processing. The KMS-12PE cell line (multiple myeloma) was used as negative control for NFKB2 processing. The RPMI-8226 and JJN3 cell lines were used as positive controls for MAP3K14 expression. The LP-1 and KMS-12PE cell lines were used as negative controls for MAP3K14 expression. Actin was used as loading control. (D) Venn diagram illustrating the absence of overlap between NF-κB gene somatic mutations. (E) Genetic lesions (including mutations and CNAs) of NF-κB genes are largely mutually exclusive. In the heat map, rows correspond to identical genes, and columns represent individual patients color-coded on the basis of gene status (green, wild-type; red, mutations of IKBKB, mutations or gain of MAP3K14, mutations and/or deletion of TRAF3, TNFAIP3, and BIRC3).
To investigate the molecular basis of NF-κB activation, NF-κB pathway genes were screened for mutations and CNAs in 101 SMZLs. Mutations that targeted multiple NF-κB genes were detected in 20 (20%) of 101 SMZLs (supplemental Table 3). Mutations affected key regulators of both canonical (IKBKB, TNFAIP3) and noncanonical (BIRC3, TRAF3, MAP3K14) NF-κB pathways12 and were mutually exclusive, which points to multiple molecular mechanisms targeting NF-κB in SMZLs (Figure 1D). By combining mutations and CNAs identified by single-nucleotide polymorphism array and validated by FISH (supplemental Table 4; Figure 1E), NF-κB was targeted in 36 (36%) of 101 SMZLs. The mutually exclusive distribution of NF-κB lesions was confirmed in most cases (29/36, 80%) and when mutations and CNAs were considered together (Figure 1E). These data document that NF-κB gene lesions are recurrent in SMZL, a disease that until now was free from the molecular lesions that affect cancer genes.1
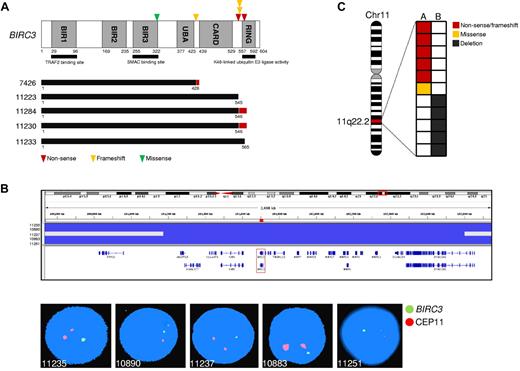
The TRAF3/MAP3K14-TRAF2/BIRC3 regulatory complex of the noncanonical NF-κB pathway12-14 was targeted in 25 (25%) of 101 SMZLs. BIRC3 was affected in 11 (11%) of 101 SMZLs by inactivating mutations (3 frameshift and 2 nonsense), missense mutations (n = 1), and gene deletions (n = 5; supplemental Tables 3-4; Figure 2). All inactivating mutations were somatically acquired, were predicted to generate aberrant transcripts carrying premature stop codons, and caused elimination or truncation of the C-terminal RING domain, the E3 ubiquitin ligase activity of which is essential for MAP3K14 proteasomal degradation by BIRC3 (Figure 2A).8,12,14 The sole BIRC3 missense mutation targeted a conserved Zn binding site within the BIR3 domain that is also occasionally mutated in solid tumors (COSMIC [Catalogue Of Somatic Mutations In Cancer] v52; http://www.sanger.ac.uk/genetics/CGP) and was categorized as probably damaging by PolyPhen-2 (Polymorphism Phenotyping v2; supplemental Table 3; Figure 2A). The identification of BIRC3-inactivating mutations in SMZL points to BIRC3 disruption as a common mechanism across marginal zone B-cell lymphomagenesis. In fact, disruption of the BIRC3 RING domain, which in SMZL is produced by inactivating mutations, in extranodal marginal zone lymphoma is caused by a t(11;18)(q21;q21) translocation that leads to the BIRC3/MALT1 fusion protein, which lacks the RING domain of BIRC3.15
BIRC3 mutations in SMZL. (A) Schematic representation of the BIRC3 protein with its key functional domains. Color-coded symbols indicate the type and position of the mutations on BIRC3. The predicted BIRC3 polypeptides in SMZL cases harboring mutations that truncate the BIRC3 RING domain are aligned below (black bars represent normal coding regions; red bars represent the position of frameshifts and the length of translation before a stop codon is encountered). (B) Graphic representation of segmentation data from SMZL carrying BIRC3 deletions, visualized with Integrative Genomics Viewer (IGV) software (http//www.broadinstitute.org/igv). Each track represents 1 sample, where gray denotes a normal (diploid) copy number and blue indicates region of a copy number loss. Individual genes in the region are aligned in the bottom panel. Dual-color FISH validation of BIRC3 deletions (RP11-177O8-BIRC3–specific probe in green and chromosome 11 centromeric probe in orange). (C) Allelic (A or B) distribution of BIRC3 genetic lesions in individual SMZL samples. Chr11 indicates chromosome 11.
BIRC3 mutations in SMZL. (A) Schematic representation of the BIRC3 protein with its key functional domains. Color-coded symbols indicate the type and position of the mutations on BIRC3. The predicted BIRC3 polypeptides in SMZL cases harboring mutations that truncate the BIRC3 RING domain are aligned below (black bars represent normal coding regions; red bars represent the position of frameshifts and the length of translation before a stop codon is encountered). (B) Graphic representation of segmentation data from SMZL carrying BIRC3 deletions, visualized with Integrative Genomics Viewer (IGV) software (http//www.broadinstitute.org/igv). Each track represents 1 sample, where gray denotes a normal (diploid) copy number and blue indicates region of a copy number loss. Individual genes in the region are aligned in the bottom panel. Dual-color FISH validation of BIRC3 deletions (RP11-177O8-BIRC3–specific probe in green and chromosome 11 centromeric probe in orange). (C) Allelic (A or B) distribution of BIRC3 genetic lesions in individual SMZL samples. Chr11 indicates chromosome 11.
BIRC3 mutations in SMZLs were all monoallelic, in agreement with independent evidence suggesting that BIRC3 genetic lesions may exert a dominant negative effect. First, BIRC3 experimental mutants with a disrupted RING domain abrogate the ability of wild-type BIRC3 to efficiently target proteins for proteasomal degradation.16,17 Second, monoallelic disruption of the RING domain of BIRC3 contributes to BIRC3/MALT1 fusion protein oncogenicity in extranodal marginal zone lymphoma.17 Consistently, all tested SMZL primary cases that harbored monoallelic BIRC3 disruption displayed constitutive NF-κB activation, including MAP3K14 accumulation, and active NFKB2 processing (Figure 1C) and/or NF-κB localization by immunohistochemistry.
TRAF3 was affected in 10 (10%) of 101 SMZLs by inactivating mutations (2 frameshift and 1 nonsense), missense mutations (n = 1), and deletions (n = 7). Inactivating mutations were predicted to generate aberrant transcripts carrying premature stop codons and causing elimination or truncation of the C-terminal MATH domain, which provides the docking site for MAP3K14 and is required for MAP3K14 recruitment to BIRC3 degradation (supplemental Tables 3-4; supplemental Figure 2).6,12-14,18,19 The sole missense mutation of TRAF3 targeted a TRAF domain that interacts with TRAF2,19 was categorized as probably damaging by PolyPhen-2, and was coupled with deletion of the second TRAF3 allele (supplemental Table 3; supplemental Figure 2). Genetic lesions of TRAF3 are also recurrent in multiple myeloma, where they contribute to NF-κB signaling activation.20,21 SMZL primary cells harboring monoallelic TRAF3 inactivation showed NF-κB localization, MAP3K14 accumulation, and active NFKB2 processing (Figure 1C).
MAP3K14 (also known as NIK) was affected by gain (n = 7) or missense (n = 1) mutations in 8 (8%) of 101 SMZLs (supplemental Tables 3-4; supplemental Figure 3). The missense mutation was somatically acquired and mapped near the kinase domain and far from the TRAF3-binding domain, which suggests a potential role in MAP3K14 downstream signaling activation rather than a role in MAP3K14 protein stabilization (supplemental Table 3; Figure 1C; supplemental Figure 3).
BIRC3 and TRAF3, along with TRAF2, cooperate in the same protein complex that negatively regulates MAP3K14, the central activator of noncanonical NF-κB signaling.12-14 In SMZLs, genetic lesions activate noncanonical NF-κB either by disrupting BIRC3 and TRAF3 domains required for functional integrity of the MAP3K14 regulatory complex or directly through MAP3K14 gain or mutation (Figure 1C).12-14,18-20 Notably, animal models that harbor BIRC3 inactivation, conditional deletion of TRAF3, or MAP3K14 overexpression are all characterized by B-cell enhanced proliferation and survival, which leads to abnormal marginal zone B-cell expansions.6-8
In addition to noncanonical signaling, the canonical NF-κB pathway was affected in SMZLs (15/101, 15%) by genetic lesions of TNFAIP3 and IKBKB, which encode for negative and positive regulators of NF-κB signaling, respectively.12 TNFAIP3 was affected in 13 (13%) of 101 SMZLs by inactivating mutations (6 frameshift mutations) and deletions (n = 9), including a focal loss that points to TNFAIP3 as the specific target of deletions (supplemental Tables 3-4; supplemental Figure 4). TNFAIP3 lesions in SMZLs displayed a preferentially monoallelic distribution, as also observed in other indolent B-cell malignancies in which monoallelic TNFAIP3 inactivation is associated with low expression of the residual wild-type allele, which suggests a haploinsufficient role in tumor suppression.22,23
IKBKB was mutated in 3 (3%) of 101 SMZLs carrying a recurrent, somatically acquired missense mutation (K171E) that targeted the IKBKB kinase domain near the activation loop (supplemental Table 3; supplemental Figure 5). This mutation leads to an amino acid substitution within a site that is conserved both intraspecies and interspecies and is predicted as possibly being damaging according to PolyPhen-2 (supplemental Table 3; supplemental Figure 5). The potential pathogenetic relevance of this mutation is supported by the notion that mutations within the activation loop constitutively activate IKBKB tyrosine kinase activity and NF-κB signaling in vitro24 and contribute to B-cell lymphomagenesis in mouse models.25
The preferential monoallelic distribution of NF-κB gene abnormalities in SMZL is consistent with the preferential monoallelic distribution of these lesions in other indolent B-cell malignancies.22,23 The occurrence of NF-κB molecular lesions in SMZLs was observed independent of hepatitis C virus infection (P = .402), IGHV gene mutation status (P = .065), IGHV1-2 gene usage (P = .761), stereotyped VH CDR3 (P = .969), 7q deletion (P = .621), or TP53 disruption (P = .638), which suggests that genetic deregulation of NF-κB is not restricted to specific SMZL clinico-molecular subgroups.1-4
The identification of NF-κB mutations in SMZLs elucidates the molecular basis of a fraction of these lymphomas and expands the number of genetic mechanisms that activate NF-κB in B-cell neoplasia. In addition to its pathogenetic relevance, the identification of NF-κB as a pathway repeatedly involved in SMZL provides the rationale for targeted anti–NF-κB therapeutic approaches in this lymphoma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Professor Mariano Rocchi and Dr Giulia Daniele, University of Bari, Bari, Italy, for the generous gift of several FISH probes used in this study.
This study was supported by AIRC, Special Program in Clinical Molecular Oncology 5 × 1000, No. 10007, Milan, Italy; Progetto FIRB-Programma “Futuro in Ricerca” 2008, PRIN 2008 and PRIN 2009, MIUR, Rome, Italy; Progetto Giovani Ricercatori 2008, Ministero della Salute, Rome, Italy; Novara-Associazione Italiana contro le Leucemie, Linfomi e Mielomi (AIL) Onlus, Novara, Italy; Oncosuisse grant OCS-02034-02-2007; Swiss National Science Foundation grant 205321-112430; Fondazione per la Ricerca e la Cura sui Linfomi, Lugano, Switzerland; and Computational Life Science/Ticino in rete. V.S. has been supported by the Associazione Franca Capurro per Novara Onlus, Novara, Italy.
Authorship
Contribution: D.R. and G.G. designed the study, analyzed and interpreted data, performed statistical analysis, and drafted the manuscript; S.D. and T.V. performed and interpreted biochemical assays; D.D.-S. and C.A. performed and interpreted immunohistochemistry assays; S.R., V.S., A.B., M.C., S.C., and D.C. performed mutation analysis and interpreted data; S.M. performed FISH analysis and interpreted data; M.F., L.A., M.L., R.M., C.T., and F.F. provided well-characterized biologic samples; I.K. and F.B. provided single-nucleotide polymorphism array data; and D.D.-S., S.A.P., R.F., R.D.-F., and L.P. contributed to data interpretation and drafting of the manuscript.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: Davide Rossi, MD, PhD, Division of Hematology, Department of Clinical and Experimental Medicine, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail: rossidav@med.unipmn.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal