Abstract
The timing and developmental sequence of events for BCR-ABL1+ acute lymphoblastic leukemia (ALL), usually associated with IKAROS (IKZF1) deletions, are unknown. We assessed the status of BCR-ABL1 and IKZF1 genes in 2 pairs of monozygotic twins, one pair concordant, the other discordant for Philadelphia chromosome positive (Ph+) ALL. The twin pair concordant for ALL shared identical BCR-ABL1 genomic sequence indicative of monoclonal, in utero origin. One twin had IKZF1 deletion and died after transplantation. The other twin had hyperdiploidy, no IKZF1 deletion, and is still in remission 8 years after transplantation. In the twin pair discordant for ALL, neonatal blood spots from both twins harbored the same clonotypic BCR-ABL1 sequence. Low level BCR-ABL1+ cells were present in the healthy co-twin but lacked the IKZF1 deletion present in the other twin's leukemic cells. The twin with ALL relapsed and died after transplantation. The co-twin remains healthy and leukemia free. These data show that in childhood Ph+ ALL, BCR-ABL1 gene fusion can be a prenatal and possibly initiating genetic event. In the absence of additional, secondary changes, the leukemic clone remains clinically silent. IKZF1 is a secondary and probable postnatal mutation in these cases, and as a recurrent but alternative copy number change is associated with poor prognosis.
Introduction
Childhood acute lymphoblastic leukemia (ALL) has very diverse genetics,1 but there is substantial evidence that segregates its development into a prenatal initiation phase followed by later acquisition of other mutations, presumed to be more proximal to diagnosis.2 This developmental sequence is most clearly defined for ALL in which ETV6-RUNX1 fusion is the prenatal and probable initiating event, but it is also probable to hold for hyperdiploid ALL3 and other, although not necessarily all,4 subtypes. Much of the evidence for the developmental sequence of acquired genetic events in ALL has been derived from the study of monozygotic twins that are concordant or discordant for ALL.5 In both such instances, the initiating lesion and premalignant clone is shared by the twins as a consequence of intraplacental vascular anastomoses and blood cell chimerism. The twin data are endorsed by backtracking of prenatal-initiating genetic lesions in the archived blood spots, or Guthrie cards, of patients with ALL.6,7
BCR-ABL1 (Philadelphia chromosome positive [Ph+]) ALL is a relatively infrequent (∼ 5%) subtype of pediatric ALL. It traditionally had a poor prognosis with conventional chemotherapy,8 but the introduction of the selective kinase inhibitor imatinib has significantly improved early event-free survival.9 IKZF1 deletions are common (∼ 85%) in Ph+ ALL10,11 and in high-risk (HR) ALL without BCR-ABL1 fusion (∼ 28%)12 and are associated with adverse outcome.12,13 The developmental timing or sequence of these “coupled” genetic events in Ph+ ALL is however unknown.
We report here 2 identical twin pairs, one concordant, the other discordant, for Ph+ ALL. In these patients it was possible to define the sequence of genetic events underlying the development of leukemia and infer the contribution that these mutations make to clonal progression and adverse prognosis.
Methods
Patients
The BM, peripheral blood samples, and neonatal bloodspots were obtained with informed consent in accordance with the Declaration of Helsinki and with local ethical committee approval from the Institute of Cancer Research (CCR 2108 and CCR 2285).
DNA extraction
Mononuclear cells in DMSO were defrosted in a 37°C water bath, spun, and washed with Dulbecco PBS. DNA was extracted with the use of the Puregene DNA isolation kit (QIAGEN).
Cytogenetics and FISH analyses
Mononuclear cells were separated from peripheral blood with Lymphoprep density gradient (Axis-Shield). CD19+ cells were then isolated with the use of a magnetic bead cell separation technique (Miltenyi Biotec), and cytospins were prepared. Interphase FISH was performed on acetone-fixed cells as described previously.14 Cells already labeled with CD19 were further labeled with anti–mouse biotin (Cambridge Bioscience) followed by Avidin D AMCA (Vector Labs). The BCR/ABL1 extra signal (ES) probe (Vysis) was used in conjunction with a bacterial artificial chromosome probe for the region of interest (IKAROS), which was obtained from the BACPAC Resource Center (Children's Hospital, Oakland Research Institute; http://bacpac.chori.org). Clone RP11-663L2 was labeled with biotin-16-dUTP, hybridized, and detected with streptavidin Cy5 (GE Healthcare). Fluorescent signals were viewed using a Zeiss Axioskop fluorescence microscope, and images were captured and analyzed using a Zeiss Plan-Neofluor 100d×/1.30 oil objective, high-resolution ccd digital camera (Hamamatsu Photonics), and SmartCapture Version 2.6.2 software (Digital Scientific) at room temperature. The expected signal pattern from a normal cell nucleus with the BCR/ABL1 ES probe is 2 green and 2 red signals, corresponding to 2 normal copies each of BCR and ABL1, respectively. A cell was considered positive for the BCR-ABL1 fusion gene if the small extra red signal was also present.
RT-PCR and real-time quantitative PCR
cDNA was synthesized from 1 μg of total RNA in 20-μL total volume with the use of random hexamers. RT-PCR for BCR-ABL1 fusion gene was performed as previously described.15 Real-time quantitative PCR (RQ-PCR) for p190 transcript was performed according to the protocol of Europe Against Cancer action.16
Genome mapping analysis
Mapping analysis was performed with 500 ng of tumor and germline DNA. DNA was processed according to manufacturer's instructions with the use of the GeneChip mapping 500K assay protocol for hybridization to GeneChip Mapping 250K Nsp and Sty arrays (Affymetrix). Briefly, genomic DNA was digested in parallel with restriction endonucleases NspI and StyI, ligated to an adaptor, and subjected to PCR amplification with adaptor-specific primers. The PCR products were digested with DNaseI and labeled with a biotinylated nucleotide analog. The labeled DNA fragments were hybridized to the microarray, stained by streptavidin-phycoerythrin conjugates, washed with the Affymetrix Fluidics Station 450, and then scanned with a GeneChip scanner 3000 7G. The raw array data files are available at http://www.icr.ac.uk/array/array.html.
Copy number and LOH analysis
Single nucleotide polymorphism (SNP) genotypes were obtained with the use of Affymetrix GCOS Version 1.4 software to obtain raw feature intensity and Affymetrix GTYPE Version 4.0 software with the BRLMM algorithm to derive SNP genotypes. Samples were analyzed with CNAG 3.0 (http://plaza.umin.ac.jp/genome/) with the use of paired tumor (test) samples with the self-reference control (patient DNA at morphologic remission) samples to determine copy number and loss of heterozygosity (LOH) caused by imbalance.17 The position of regions of LOH and gain were identified with the University of California Santa Cruz Genome Browser, May 2004 Assembly (http://genome.ucsc.edu/cgi-bin/hgGateway).
Detection and amplification of BCR-ABL1 genomic breakpoints
For detection and amplification of DNA breakpoints, ranging from 300 bp to 12 kbp, the Expand Long Template PCR kit (Roche) with System 2 was used, with an annealing temperature of 64°C. To cover the BCR and ABL1 regions, within which breakpoints can occur, 21 BCR forward primers and 20 ABL1 reverse primers were used in multiplex, combining each BCR forward primer with 4 mixes of 5 ABL1 reverse primers, as described elsewhere.18
PCR analysis of Guthrie cards
Guthrie blood spot pieces to be amplified were incubated in 0.5 mL of double-distilled water for 30 minutes each, then dried in a vacuum dessicator before PCR amplification reaction. PCR amplification of the blood spot segments (one-eighth of spot) was conducted in 0.6-mL thin-walled PCR tubes at 50-μL final volume of the following: 1 × Ampdirect Plus (Shimadzu), 50 pmol each of 5′ and 3′ oligonucleotide primers, and 2.5 units of Platinum Taq enzyme mix (Invitrogen) for 10 minutes at 95°C, then 40 cycles of 30 seconds at 94°C, 1 minute at 50.2°C, and 1 minute at 72°C, followed by 7 minutes of extension at 72°C.7 PCR products were routinely size-fractionated on 2% agarose gels with molecular weight DNA markers (Bioline) and visualized by ethidium bromide staining. The PCR products were cleaned with the Illustra GFX purification kit (GE Healthcare). To sequence the PCR products, each 10-μL asymmetric PCR reaction contained 5 μL of purified PCR product, 4 μL of Big Dye Terminator v1.1 reaction mix (Applied Biosystems), 3.2μM of upstream or downstream primer, and was run through 30 cycles of 30 seconds at 95°C, 30 seconds at 55°C, 1 minute at 70°C, and subsequently sequenced on a Applied Biosystems 3130 × l genetic analyzer.
Results
Twin pair 1 (A,B) concordant for Ph+ ALL
The twin boys 1A and 1B shared a single monochorionic placenta in utero and were born at 36 weeks by cesarean section. Twin 1B remained at the Special Care Baby Unit for 3 months after birth because of pyloric stenosis. Twin 1A presented with ALL at age 3.8 years, and twin 1B with ALL at age 4.1 years. Cytogenetic analysis was informative only for twin 1B: 46,XX,t(9;22)(q34;q11)[26] but BCR-ABL1 (e1a2) fusion was identified by RT-PCR in both twins.
Both twins were enrolled in the HR arm of the AIEOP-BFM ALL2000 protocol (Associazione Italiana Ematologia Oncologia Pediatrica-Berlin Frankfurt Munster ALL2000). An allogeneic stem cell (allo-SC) transplant from the same HLA-identical sibling donor was administered to both twins, using the same conditioning regimen (busulphan-thiotepa-melphalan) but different immunosuppressive treatment as a prevention of GVHD (cyclosporine for twin 1A; tacrolimus for twin 1B).
Seven months after allo-SC transplantation, twin 1B had an isolated BM relapse and died during the administration of third-line therapy. Conversely, twin 1A is in good health and in complete remission 8 years after SC transplantation.
Twin pair 2 (A,B) discordant for Ph+ ALL
The twin girls 2A and 2B shared a single monochorionic placenta in utero and were born by emergency cesarean section for growth retardation at 33 weeks. They remained at the Special Care Baby Unit for 4 weeks without requiring further medical intervention.
At 5 years, twin 2A presented with ALL with BCR-ABL1 fusion, using Vysis dual fusion (FISH) probes. BCR-ABL1 (e1a2) fusion was confirmed by RQ-PCR. The patient was initially started on the UKALL2003 protocol (United Kingdom ALL2003), but after confirmation of Ph+ ALL she was switched to the EsPhALL protocol (European Intergroup Study on Post Induction Treatment of Philadelphia Positive Acute Lymphoblastic Leukemia with Imatinib) with concomitant imatinib treatment. She received a matched unrelated BM transplantation in first remission. She had an isolated BM relapse 6 months after transplantation. The patient received palliation in the form of steroids and dasatinib but, unfortunately, died of progressive disease. The co-twin, 2B, remains healthy and leukemia-free 28 months after the diagnosis of ALL in twin 2A.
BCR-ABL1 genomic fusions sequencing
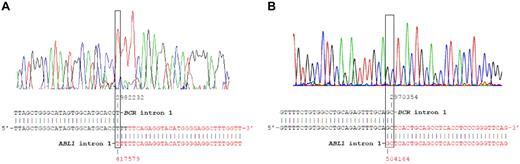
The BCR-ABL1 genomic breakpoints from twin 1A and twin 2A with ALL were initially amplified with BCR forward and the ABL1 reverse primer mix.18 When PCR with the primer mix produced a PCR product, a split out PCR was run with the individual primers contained within the mix to amplify a band with BCR F and ABL1 R. The breakpoints were confirmed by re-amplification and sequencing in the patient samples with BCR F and a twin- and breakpoint-specific ABL1 reverse primers (Figure 1). More specifically, the breakpoints were designated as a fusion between BCR intron 1 (nucleotide 2982232 in genomic sequence NT_011520) and ABL1 intron 1 (nucleotide 417579 in genomic sequence NT_035014) in twin 1A and between BCR intron 1 (nucleotide 2970354 in genomic sequence NT_011520) and ABL1 intron 1 (nucleotide 504164 in genomic sequence NT_035014) in twin 2A.
Sequence of the BCR-ABL1 rearrangements from twins 1A and 2A at diagnosis. The chromatogram and NCBI nucleotide blasts analysis of the BCR-ABL1 breakpoint DNA sequence with BCR intron 1 (NT_011520, black) and ABL1 intron 1 (NT_035014, red) sequences are shown for twins 1A (A) and 2A (B).
Sequence of the BCR-ABL1 rearrangements from twins 1A and 2A at diagnosis. The chromatogram and NCBI nucleotide blasts analysis of the BCR-ABL1 breakpoint DNA sequence with BCR intron 1 (NT_011520, black) and ABL1 intron 1 (NT_035014, red) sequences are shown for twins 1A (A) and 2A (B).
Paired twins shared the same genomic BCR/ABL breakpoint
Twins 1A and 1B.
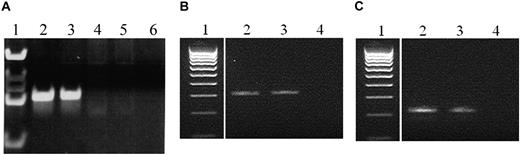
The diagnostic leukemic samples of both twin 1A and twin 1B were screened for the clonotypic BCR-ABL1 genomic sequence detected in twin 1A, using specific primers and TaqMan probe. The primers and probe were designed by Primer Express Version 3.0 software (Applied Biosystems); forward (F1, TTAGCTGGGCATAGTGGCATG) and reverse (R1, GGGAGCAGCACCCAGGT) primers were used in combination with the FAM-labeled TaqMan probe (P, CTTTGGTTTGGTTATGACAGTGCTACAGCCA). The same BCR-ABL1 genomic breakpoint detected in twin 1A was shown in the leukemic diagnostic sample of twin 1B (Figure 2A), indicating that the BCR-ABL1 fusion was shared by the twins and therefore presumably arose in utero in one of them to then be clonally disseminated via intraplacental anastomoses.5
PCR amplification of the genomic BCR-ABL1 rearrangements from twins. (A) Genomic BCR-ABL1 rearrangements from twins 1A and 1B at diagnosis; lanes: 1, marker; 2, diagnosis twin 1A; 3, diagnosis twin 1B; 4 and 5, negative controls; 6, no DNA control. (B) Genomic BCR-ABL1 rearrangements from twin 2A at diagnosis and relapse; lanes: 1, marker; 2, diagnosis twin 2A; 3, relapse twin 2A; 4, no DNA control. (C) Genomic BCR-ABL1 rearrangements from twin 2A and healthy twin 2B (Guthrie specimens); lanes: 1, marker; 2, Guthrie card DNA twin 2A; 3, Guthrie card DNA twin 2B; 4, no DNA control.
PCR amplification of the genomic BCR-ABL1 rearrangements from twins. (A) Genomic BCR-ABL1 rearrangements from twins 1A and 1B at diagnosis; lanes: 1, marker; 2, diagnosis twin 1A; 3, diagnosis twin 1B; 4 and 5, negative controls; 6, no DNA control. (B) Genomic BCR-ABL1 rearrangements from twin 2A at diagnosis and relapse; lanes: 1, marker; 2, diagnosis twin 2A; 3, relapse twin 2A; 4, no DNA control. (C) Genomic BCR-ABL1 rearrangements from twin 2A and healthy twin 2B (Guthrie specimens); lanes: 1, marker; 2, Guthrie card DNA twin 2A; 3, Guthrie card DNA twin 2B; 4, no DNA control.
Twins 2A and 2B.
The diagnostic leukemic sample, relapse leukemic sample, and archived neonatal blood spot (Guthrie card) of both twins were screened for the clonotypic BCR-ABL1 genomic sequence with the use of specific primers. The primers were designed by Primer 3 software.19 Forward (F1, AGTAACAGGTGGGATCTG) and reverse (R1, GGCAAAAATATGAAAATTAG) primers were used for the diagnostic and relapse DNA (Figure 2B). Nested primers (F2, ACATGACAGTTTCGAGTTT; R2, TACCTGTAATCCCAGCTAC) were used for blood spot PCR.
Guthrie card analysis.
Putative preleukemic clone in the peripheral blood of the healthy co-twin (2B).
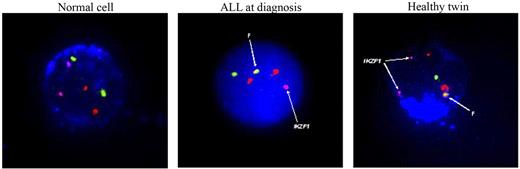
The neonatal blood spot test indicated that the BCR-ABL1 fusion was shared by the twins 2A and 2B, as it was for twin pair 1. We assessed the healthy co-twin (2B) for persistence of these cells shortly after her sister's condition was diagnosed with ALL. A low level of peripheral blood CD19+ cells, 0.12%-0.45% (average, 0.17%) detected (on 4 separate occasions over a 15-month period), were positive for the BCR-ABL1 fusion, but all cells retained 2 copies of IKZF1 (Figure 3). We assume these cells are clonal derivatives of the initial preleukemic clone initiated by BCR-ABL1 fusion in utero (in either twin 2A or twin 2B), but we have no functional assessment of their full leukemogenic potential. The level of risk of ALL developing in the healthy co-twin is uncertain, but, given the calculated rate of discordance of B-cell precursor ALL in monozygotic twins of 10%-15%,5 it may be of the order of 10%.
Immunostained for CD19AMCA, leukemic cell nucleus stained with DAPI. Probes used include Vysis BCR/ABL1 ES probe (BCR/chromosome 22 = green, ABL1/chromosome 9 = red) Ikarosbiotin-Cy5 = pink. F = BCR-ABL1 fusion.
Immunostained for CD19AMCA, leukemic cell nucleus stained with DAPI. Probes used include Vysis BCR/ABL1 ES probe (BCR/chromosome 22 = green, ABL1/chromosome 9 = red) Ikarosbiotin-Cy5 = pink. F = BCR-ABL1 fusion.
SNP array–defined copy number changes
The diagnostic leukemic cell DNA of twins 1A, 1B, and 2A was interrogated by high-resolution (500K) SNP arrays for copy number alterations (CNAs). The results, summarized in Table 1, indicated a constellation of genetic changes, which are common in Ph+ ALL.10,11
Copy number alterations in leukemic cells of twins
| Chromosome . | CNA . | Genes . | Twin . | ||
|---|---|---|---|---|---|
| 1A . | 1B . | 2A . | |||
| Associated with the translocation | |||||
| 9 | amp 9q(34.12–34.3) | ABL1 and several | x | ||
| 9 | del 9q(34.12) | ABL1 | x | x | |
| 22 | amp22q(11.1–11.23) | BCR and several | x | ||
| 22 | del 22q(11.23) | BCR | x | x | |
| “Neutral” (IgH/TcR somatic rearrangements) | |||||
| 7 | del 7p(12.2) | TCRG | x | x | |
| 14 | del 14q(32–33) | IgH* | x | x | |
| 14 | del 14q(32–33) | TCRD | x | ||
| 22 | del 22q(11.22) | IgL | x | ||
| “Driver” aberrations | |||||
| 5 | del 5q(33.3) | EBF1 | x | ||
| 9 | del 9p(12–23) | Several, including CDKN2A, PAX5 | x | ||
| 9 | gp 21.3 | CDKN2A biallelic deletion | x | ||
| 10 | del 10q(24.1) | PIK3AP1 | x | ||
| 14 | del 14q(11.2) | IKZF1 | x | x | |
| 21 | del 21q(22.13) | DSCR5 | x | ||
| Trisomy 4, 6, 9, 14, 17, X | x | ||||
| Tetrasomy 21 | x | ||||
| Chromosome . | CNA . | Genes . | Twin . | ||
|---|---|---|---|---|---|
| 1A . | 1B . | 2A . | |||
| Associated with the translocation | |||||
| 9 | amp 9q(34.12–34.3) | ABL1 and several | x | ||
| 9 | del 9q(34.12) | ABL1 | x | x | |
| 22 | amp22q(11.1–11.23) | BCR and several | x | ||
| 22 | del 22q(11.23) | BCR | x | x | |
| “Neutral” (IgH/TcR somatic rearrangements) | |||||
| 7 | del 7p(12.2) | TCRG | x | x | |
| 14 | del 14q(32–33) | IgH* | x | x | |
| 14 | del 14q(32–33) | TCRD | x | ||
| 22 | del 22q(11.22) | IgL | x | ||
| “Driver” aberrations | |||||
| 5 | del 5q(33.3) | EBF1 | x | ||
| 9 | del 9p(12–23) | Several, including CDKN2A, PAX5 | x | ||
| 9 | gp 21.3 | CDKN2A biallelic deletion | x | ||
| 10 | del 10q(24.1) | PIK3AP1 | x | ||
| 14 | del 14q(11.2) | IKZF1 | x | x | |
| 21 | del 21q(22.13) | DSCR5 | x | ||
| Trisomy 4, 6, 9, 14, 17, X | x | ||||
| Tetrasomy 21 | x | ||||
Different sequences in twin pairs 1A,B.
Twins 1A and 1B.
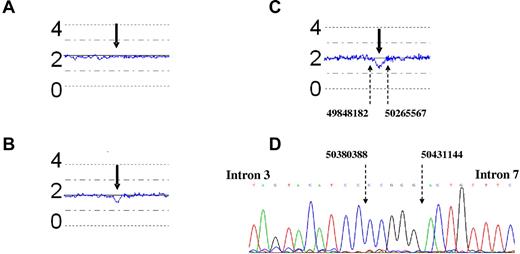
Some CNAs were common in both twins, such as the deletion of ABL exon 1 and BCR exons 2-6, which are frequently associated with the t(9;22) translocation.20 Although not sequenced, the homology in both twins indicate that their origin is associated with the mechanism of the chromosomal translocation. Other lesions were specific for the single twin; more precisely, twin 1A showed a subclone with trisomy of chromosomes 4, 6, 9, 14, 17, and X; tetrasomy of chromosome 21; and gain of chromosome 22q11.1-11.23 region. Although twin 1A showed the deletion of PIK3AP1 and DSCR5 genes, twin 1B carried the deletion of EBF1 gene locus. Twin 1B, and not twin 1A, showed monoallelic IKZF1 deletion (Figure 4A-B).
Copy number analysis of the IKZF1 locus in DNA at diagnosis with the use of CNAG 3.0. The blue lines indicate the mean CNA of 5 contiguous SNPs. The numbers 0, 2, and 4 refer to the genomic copy number. (A-C) The bold arrows point to the IKZF1 locus. (C) The fine dotted arrows indicate the genomic breakpoints (base pair location, NCBI35/Hg17) around the IKZF1 gene. (A) Twin 1A, (B) twin 1B, (C) twin 2A, and (D) twin 1A, IKZF1 sequence.
Copy number analysis of the IKZF1 locus in DNA at diagnosis with the use of CNAG 3.0. The blue lines indicate the mean CNA of 5 contiguous SNPs. The numbers 0, 2, and 4 refer to the genomic copy number. (A-C) The bold arrows point to the IKZF1 locus. (C) The fine dotted arrows indicate the genomic breakpoints (base pair location, NCBI35/Hg17) around the IKZF1 gene. (A) Twin 1A, (B) twin 1B, (C) twin 2A, and (D) twin 1A, IKZF1 sequence.
Twin 2A.
IKZF1 deletion is secondary to BCR-ABL1 fusion
The IKZF1 deletion boundaries in diagnostic DNA of twin 1B were amplified by PCR and sequenced (Figure 4D). The deletion maps to 2 heptamer recombination signals in intron 2 (CACAGTG) and intron 7 (TGCTGTG) are indicative of a probable involvement of RAG-mediated recombination, as previously reported for IKZF1 deletions.10 A highly sensitive and patient-specific genomic RQ-PCR assay did not show the presence of this IKZF1 deletion in twin 1A. Similarly, IKZF1 deletion was detected by FISH in leukemic cells of twin 2A at diagnosis but not in the BCR-ABL1+ cells of the healthy twin 2B (Figure 3).
Discussion
The DNA breakpoints of fusion genes in leukemia are clustered but highly variable with the result that in any individual patient the breakpoints and resultant fusion sequence is unique or clone specific.2 Our finding that BCR-ABL1 fusion sequences are shared in pairs of identical twins parallels what was observed earlier with MLL-AF421 and ETV6-RUNX1 in ALL22,23 and is compatible with an origin in one cell in one fetus followed by twin-twin transfusion of resultant progeny cells via intraplacental metastases in utero.5 Twin pairs with concordant or discordant leukemia are rare; therefore, data derived from them are somewhat anecdotal. Nevertheless, they are highly informative of the natural history of leukemia, and the inferences derived from such studies are largely confirmed by data from non-twin cases2 and modeling with murine24 and human25 cells.
The current 2 pairs of twins indicate that the BCR-ABL1 fusion can originate prenatally in childhood Ph+ ALL. Whether this is the case in most cases of childhood Ph+ ALL will require further studies, including scrutiny of a series of archived neonatal blood spots.
The twin pairs recorded here shed light on the role of BCR-ABL1 and other genetic abnormalities on the development of overt leukemia, its clinical course, and response to therapy. In the pair (no. 2) discordant for ALL, the fact that the healthy co-twin has BCR-ABL1+ cells in her blood at a low level (∼ 10−4) but remains leukemia free suggests that p190 BCR-ABL1 by itself may have a benign effect on clonal advantage and pathology, further genetic alterations being essential for clonal expression and overt development of ALL. This would be in accord with sequential evolutionary models for ALL2 and cancer in general.26 We cannot exclude that the BCR-ABL1+, putative premalignant cells in the healthy co-twin harbored other genetic changes, but they lacked the IKZF1 deletions present in the other twin with ALL. This suggests that IKZF1 deletion was secondary to BCR-ABL1 in this twin pair and probably arose postnatally in twin 2A with ALL along with the other CNAs detected by SNP arrays.
Twin pair 1A and 1B also shared the same prenatally generated BCR-ABL1 genomic breakpoint but had distinctive or divergent, additional genetic changes in their respective leukemic subclones, again indicative of a secondary, and probably postnatal, origin of IKZF1 deletion. That IKZF1 deletion should be subclonal to BCR-ABL1 (p190) in ALL is perhaps anticipated because it is secondary to BCR-ABL1 (p210) in chronic myelogenous leukemia.10 This parallels the divergent “driver” CNAs observed in monozygotic twins with ETV6-RUNX1+ ALL.27 Ph+ ALL in both children and adults has a variable but generally poor prognosis.28,29 The presence of an IKZF1 deletion appears to be associated with poor outcome12 in B-precursor ALL without BCR-ABL1 fusion and may similarly contribute to drug resistance in Ph+ ALL.10,11 Our twin pair 1A and 1B is informative in this respect. They share the same BCR-ABL1–initiated clone, were treated similarly, but had very different outcomes. The twin (1B) with IKZF1 deletion relapsed and died, whereas the co-twin (1A) with no IKZF1 deletion but hyperdiploidy has survived > 8 years in remission, after transplantation. In the twin pair 2A and 2B, discordant for ALL, the twin (2A) with ALL with BCR-ABL1 plus IKZF1 deletion also relapsed quickly and died after transplantation.
Although IKZF1 is not the only CNA or mutation in these cases, these twin-based data support the conclusion that secondary genetic changes are critical to the evolution of Ph+ ALL and that IKZF1 deletion, as an alternative CNA, may be associated with an adverse prognosis, in the context of BCR-ABL1 fusion,13 because it appears to be with other HR ALL subtypes.12 This conclusion accords with mouse modeling which indicates that homozygous loss of IKZF1 greatly accelerates BCR-ABL1–driven leukemogenesis30 and functional studies showing that IKZF1 protein normally directs BCR-ABL1 signaling toward cell cycle exit.31
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Fondazione Tettamanti, Fondazione Cariplo, AIRC, and MURST (Italy); by a specialist program grant from Leukaemia & Lymphoma Research (United Kingdom); and The Kay Kendall Leukaemia Fund (United Kingdom). L.L.N. is supported by a My First AIRC Grant (MFAG).
Authorship
Contribution: G.C., F.W.v.D., A.B., and M.G. designed the research, analyzed the data, and wrote the paper; G.C., F.W.v.D., L.L.N., A.M.F., J.S., I.I., E.M., and S.C. performed the research; and M.T. provided biologic material and clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giovanni Cazzaniga, Centro Ricerca Tettamanti, Clinica Pediatrica University of Milano Bicocca, Fondazione Monza e Brianza per il Bambino e la sua Mamma, c/o Ospedale San Gerardo, via Pergolesi, 33, 20052 Monza (MB), Italy; e-mail: gianni.cazzaniga@hsgerardo.org; or Mel Greaves, Section of Haemato-Oncology, The Institute of Cancer Research, Brookes Lawley Building, 15 Cotswold Room, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: mel.greaves@icr.ac.uk.
References
Author notes
G.C. and F.W.v.D. are joint first authors of this study.
A.B. and M.G. are joint senior authors of this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal