Abstract
Anti-HLA donor-specific Abs (DSAs) have been reported to be associated with graft failure in mismatched hematopoietic stem cell transplantation; however, their role in the development of graft failure in matched unrelated donor (MUD) transplantation remains unclear. We hypothesize that DSAs against a mismatched HLA-DPB1 locus is associated with graft failure in this setting. The presence of anti-HLA Abs before transplantation was determined prospectively in 592 MUD transplantation recipients using mixed-screen beads in a solid-phase fluorescent assay. DSA identification was performed using single-Ag beads containing the corresponding donor's HLA-mismatched Ags. Anti-HLA Abs were detected in 116 patients (19.6%), including 20 patients (3.4%) with anti-DPB1 Abs. Overall, graft failure occurred in 19 of 592 patients (3.2%), including 16 of 584 (2.7%) patients without anti-HLA Abs compared with 3 of 8 (37.5%) patients with DSA (P = .0014). In multivariate analysis, DSAs were the only factor highly associated with graft failure (P = .0001; odds ratio = 21.3). Anti-HLA allosensitization was higher overall in women than in men (30.8% vs 12.1%; P < .0001) and higher in women with 1 (P = .008) and 2 or more pregnancies (P = .0003) than in men. We conclude that the presence of anti-DPB1 DSAs is associated with graft failure in MUD hematopoietic stem cell transplantation.
Introduction
Hematopoietic stem cell transplantation is an effective treatment for a broad range of hematologic malignancies.1 Approximately 70% of patients do not have a matched related donor available for transplantation.2 For these patients, a matched unrelated donor (MUD) transplantation is preferred because of similar outcomes.3,4 HLA typing is generally performed for 4 HLA loci (HLA-A, HLA-B, HLA-C, and HLA-DRB1), because mismatches at HLA-DQB1 and HLA-DPB1 did not have a major effect on outcomes.5 Approximately 86% of MUD transplantations are mismatched in at least 1 HLA-DPB1 Ag in the host-versus-graft (HVG) direction.6
Graft failure occurs more frequently in alternative donor transplantations, with an incidence that varies between 4% in MUD transplantations up to 20% in cord blood and T cell–depleted haploidentical stem cell transplantations.7-9 Despite advances in HLA matching and supportive care, graft failure remains an important problem because of the high treatment-related mortality associated with this event.
The etiology of graft failure remains elusive in most patients. We have recently identified a high risk of graft rejection in patients with anti-HLA donor-specific Abs (DSAs) undergoing T cell–depleted haploidentical stem cell transplantation.10 Patients had DSAs directed against high-expression loci (HEL), including HLA class I (HLA-A and HLA-B) and class II (HLA-DRB1) Ags.10 It is unclear whether anti-HLA Abs directed against low-expression loci (LEL; HLA-DPB1 and HLA-DQB1) are associated with graft failure in this setting. We hypothesized that anti-HLA Abs directed against LEL are deleterious to engraftment. Therefore, in the present study we investigated the role of anti-DPB1 DSAs in a cohort of patients who underwent a MUD stem cell transplantation at a single institution.
Methods
Patients
Consecutive patients (N = 604) who received a MUD stem cell transplantation at The University of Texas M. D. Anderson Cancer Center between January 2005 and October 2009 were enrolled in the study. Of these, 592 (98%) had prospective anti-HLA Ab testing performed. Characteristics of these patients are presented in Table 1. Patients received either a myeloablative (41%) or a reduced-intensity/nonmyeloablative conditioning regimen (defined as busulfan < 520 mg/m2, melphalan < 140 mg/m2, and total-body irradiation < 6 Gy). All patients received anti-thymocyte globulin on days −3, −2, and −1. A total of 516 (87.1%) patients were matched in 8 of 8 alleles at HLA-A, HLA-B, HLA-C, and HLA-DRB1 in both directions; 4 additional patients presented with a single mismatch at these loci in the GVH direction only, whereas the other 5 patients presented with the mismatch only in the HVG vector. Of the 520 patients matched in 8 of 8 alleles of HLA-A/HLA-B/HLA-C/HLA-DRB1 loci in the HVG vector, 73.8% (n = 384) were mismatched in 1 (n = 280) or 2 (n = 104) alleles of DPB1 in the same vector. Twenty-nine (5.6%) patients matching in 8 of 8 alleles in the HVG vector presented 1 mismatch DQB1. In the 8 of 8 group, we observed 24 patients mismatched in DRB3 (1 double DRB3 mismatch) and 5 patients with a single mismatch in DRB4 (1 allele level mismatch; 4 HVG vector only). There were 76 patients that matched in 7 of 8 alleles of HLA-A/HLA-B/HLA-C/HLA-DRB1 loci (67 with the mismatch in both directions, 5 with the mismatch in the HVG direction only, and 4 with the mismatch in the GVH direction only). Among the 72 patients matched in 7 of 8 alleles of HLA-A/HLA-B/HLA-C/HLA-DRB1 loci in the HVG vector, 75.0% (n = 54) were mismatched in 1 (n = 38) or 2 (n = 16) alleles of DPB1 in the same vector. In the 7 of 8 HVG-matched group, we observed 1 patient with only a mismatch in DQB1 and 4 patients with mismatches in DRB3; in this category, there were no patients with a mismatch in DRB4. Among the DRB1-mismatched patients, there were no mismatches in DQB1 alleles; 8 of 9 DRB1 patients presented allele-level mismatches in the subtypes of HLA-DRB1*11 (n = 6), DRB1*01, and DRB1*14. One patient presented with an Ag-level mismatch in DRB1 in the HVG vector only; this patient carried DRB1*04:01 and DQB1*03:01 and the donor carried DRB1*04:01 and DRB1*12:01. That patient also presented with an additional mismatch in DRB3 and was likely to present an additional mismatch in DQA1 alleles because DRB1*04 associates with DQA1*03 and DRB1*12:01-DQB1*03:01 associates with DQA1*05; the patient's serum did not present any anti-HLA reactivity.
Characteristics of the study population
| Characteristic . | N = 592 . |
|---|---|
| Median, age, y (range) | 54 (8-74) |
| Sex, n (%) | |
| Male | 356 (60.1) |
| Female | 236 (39.9) |
| Race, n (%) | |
| White | 512 (86.5) |
| Other | 43 (13.5) |
| Diagnosis, n (%) | |
| AML/MDS | 255 (43.1) |
| Lymphoma/CLL | 212 (35.8) |
| CML/MPD | 54 (9.1) |
| ALL | 42 (7.1) |
| Other | 29 (4.9) |
| Preparative regimen, n (%) | |
| Myeloablative | 351 (59.3) |
| Reduced-intensity/nonmyeloablative | 241 (40.7) |
| Source of stem cells, n (%) | |
| Peripheral blood | 358 (60.5) |
| BM | 234 (39.5) |
| CD34+ cells infused, median, n (range)* | 4.6 (0.8-36.4) |
| HLA-DPB1 mismatch, n (%) | 430 (72.6%) |
| HLA match (HVG direction), n (%) | |
| 10/10 | 495 (83.6%) |
| 0-DP mismatch | 140 |
| 1-DP mismatch | 257 |
| 2-DP mismatch | 98 |
| 9/10 | 97 (16.4%) |
| 0-DP mismatch | 21 |
| 1-DP mismatch | 52 |
| 2-DP mismatch | 24 |
| Distribution of LEL mismatches in 8/8 and 7/8 transplants, n (%) | 8/8 HVG (n = 520) 7/8 HVG (n = 72) |
| 0 DPB1 HVG mismatch | 136 (26.2%) 18 (25%) |
| 1 DPB1 HVG mismatch | 280 (53.8%) 38 (52.8%) |
| 2 DPB1 HVG mismatch | 104 (20.0%) 16 (22.2%) |
| DQB1 HVG mismatch | 29 (5.6%) 1 (1.4%) |
| DRB3 HVG mismatch | 24 (4.6%) 4 (5.6%) |
| DRB4 HVG mismatch | 5 (1.0%) 0 (0.0%) |
| DRB5 HVG mismatch | 0 (0.0%) 0 (0.0%) |
| Sex mismatch between the donor and recipient, n (%)† | |
| No | 416 (70.9) |
| Yes | 170 (29.1) |
| CMV mismatch between the donor and recipient, n (%)† | |
| No | 312 (53.9) |
| Yes | 267 (46.1) |
| ABO mismatch between the donor and recipient, n (%)† | |
| No | 243 (41.1) |
| Yes | 348 (58.9) |
| Pregnancies, median, n (range) | 2 (0-7) |
| Engrafted, n (%) | 573 (96.8) |
| Time to ANC 500, median, d | 12 |
| Time to PLT 20 000, median, d | 14 |
| Relapsed, n (%) | |
| No | 421 (71.1) |
| Yes | 171 (28.9) |
| Alive at the last follow-up, n (%) | |
| Yes | 274 (46.3) |
| No | 318 (53.7) |
| Characteristic . | N = 592 . |
|---|---|
| Median, age, y (range) | 54 (8-74) |
| Sex, n (%) | |
| Male | 356 (60.1) |
| Female | 236 (39.9) |
| Race, n (%) | |
| White | 512 (86.5) |
| Other | 43 (13.5) |
| Diagnosis, n (%) | |
| AML/MDS | 255 (43.1) |
| Lymphoma/CLL | 212 (35.8) |
| CML/MPD | 54 (9.1) |
| ALL | 42 (7.1) |
| Other | 29 (4.9) |
| Preparative regimen, n (%) | |
| Myeloablative | 351 (59.3) |
| Reduced-intensity/nonmyeloablative | 241 (40.7) |
| Source of stem cells, n (%) | |
| Peripheral blood | 358 (60.5) |
| BM | 234 (39.5) |
| CD34+ cells infused, median, n (range)* | 4.6 (0.8-36.4) |
| HLA-DPB1 mismatch, n (%) | 430 (72.6%) |
| HLA match (HVG direction), n (%) | |
| 10/10 | 495 (83.6%) |
| 0-DP mismatch | 140 |
| 1-DP mismatch | 257 |
| 2-DP mismatch | 98 |
| 9/10 | 97 (16.4%) |
| 0-DP mismatch | 21 |
| 1-DP mismatch | 52 |
| 2-DP mismatch | 24 |
| Distribution of LEL mismatches in 8/8 and 7/8 transplants, n (%) | 8/8 HVG (n = 520) 7/8 HVG (n = 72) |
| 0 DPB1 HVG mismatch | 136 (26.2%) 18 (25%) |
| 1 DPB1 HVG mismatch | 280 (53.8%) 38 (52.8%) |
| 2 DPB1 HVG mismatch | 104 (20.0%) 16 (22.2%) |
| DQB1 HVG mismatch | 29 (5.6%) 1 (1.4%) |
| DRB3 HVG mismatch | 24 (4.6%) 4 (5.6%) |
| DRB4 HVG mismatch | 5 (1.0%) 0 (0.0%) |
| DRB5 HVG mismatch | 0 (0.0%) 0 (0.0%) |
| Sex mismatch between the donor and recipient, n (%)† | |
| No | 416 (70.9) |
| Yes | 170 (29.1) |
| CMV mismatch between the donor and recipient, n (%)† | |
| No | 312 (53.9) |
| Yes | 267 (46.1) |
| ABO mismatch between the donor and recipient, n (%)† | |
| No | 243 (41.1) |
| Yes | 348 (58.9) |
| Pregnancies, median, n (range) | 2 (0-7) |
| Engrafted, n (%) | 573 (96.8) |
| Time to ANC 500, median, d | 12 |
| Time to PLT 20 000, median, d | 14 |
| Relapsed, n (%) | |
| No | 421 (71.1) |
| Yes | 171 (28.9) |
| Alive at the last follow-up, n (%) | |
| Yes | 274 (46.3) |
| No | 318 (53.7) |
AML indicates acute myeloid leukemia; MDS, myelodysplastic syndromes; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; MPD, myeloproliferative diseases; ALL, acute lymphoblastic leukemia; PRBCs, packed red blood cells; and PLT, platelet count.
Data were available for 590 patients.
Donor sex known for 586 patients, donor CMV known for 579 patients, and donor ABO known for 591 patients.
Types of mismatches in DRB3, DRB4, and DQB1 loci
There were 32 transplantation patients with mismatches in DRB3, of which 31 included a single mismatch (22 bidirectional, 5 in the HVG direction only, and 4 in the GVH direction only), and 1 donor recipient pair presented with mismatches in both alleles of DRB3. The count of single and double mismatches resulted in 32 allele mismatches in DRB3 and the above-mentioned case for which the donor carried DRB3 that was also present in the patient. The mismatches in the pair of alleles DRB3*01:01/DRB3*02:02 was the most common (n = 29); 3 patients included the mismatches DRB3*02:02/DRB3*03:01, DRB3*02:02/DRB3*02:01, and DRB3*02:02/DRB3*02:03. The 32 mismatches in alleles of DRB3 included patients and donors matching in DRB1; these included patients and donors carrying DRB1*13:01 (n = 17), DRB1*03:01 (n = 10), DRB1*11:01 (n = 2), DRB1*14:01/14:54 (n = 2), and DRB1*13:02 (n = 1).
The difference in the expressed and nonexpressed alleles was the most common mismatch in DRB4. In the present study, there were 10 patients with a single mismatch in DRB4; 1 of these cases included the allele level mismatch DRB4*01:03/01:02; the remaining 9 patients included the presence or absence of the null allele DRB4*01:03N; 4 of the mismatches occurred in the HVG vector and 5 of them in the GVH vector.
In total, there were 31 patients that presented with a mismatch in DQB1; of these, 30 had the mismatch in both directions and only 1 presented the mismatch in DQB1 only in the GVH vector. The DQB1 mismatches occurred in transplantations in which the patient and donor matched in DRB1. The mismatches in DQB1 included 18 allele-level mismatches; 8 of these involved DQB1*03 alleles with a different associated serologic split (DQ7 and DQ8) and may be distinguished by allo-Abs. There were 12 patients presenting with a DQ-Ag level mismatch; 9 of these included the mismatch in the specificities DQ2 (DQB1*02:02)/DQ9 (DQB1*03:03). The remaining DQ-Ag level mismatches included single cases of the mismatches DQ5 9DQB1*05:01) /DQ6 (DQB1*06:04), DQ2 (DQB1*02:02)/DQ8 (DQB1*03:02), and DQ4 (DQB1*04:02)/DQ8 (DQB1*03:02).
Among the patients with mismatches in DQB1, the anti-HLA Ab tests showed that 19 of them did not have anti-HLA Abs. Of the 11 patients with DQB1 mismatches and anti-HLA Abs, 9 did not have any anti-HLA class II Abs. There were 2 patients who presented with mismatches in DQB1 whose serum showed anti-DQ reactivity; there was no reactivity with Ag preparations bearing the donor's DQB1-mismatched allele in either of these, and neither patient's sera showed anti-DQA1 specificity.
The objectives of this study were to determine whether there is an association among anti-HLA Abs, DSAs, and primary graft failure (n = 9), or early death without engraftment beyond the median time to engraftment (n = 10) in MUD hematopoietic stem cell transplantation. We also assessed the impact of sex and number of prior pregnancies on the probability of allosensitization. This retrospective study protocol and a waiver of informed consent were approved by The University of Texas M. D. Anderson Cancer Center Institutional Review Board.
Measurement of anti-HLA Ab levels
The presence of DSAs was determined by testing the patients' sera collected before transplantation prospectively with a panel of fluorescent beads coated with single HLA Ag preparations (LABScreen Single-Antigen; One Lambda).11 The anti-HLA Ab reactivity was detected in a Luminex platform.10 The reactivity of HLA Abs was defined by testing 2 panels of HLA molecules, which included a set of single-Ag preparations for assignment of specificity, and was confirmed by comparison with the reactivity against a panel of HLA class I or class II Ag preparations extracted from lymphocytes from single subjects. Over the course of this study, several lots of reagents were used; these were validated before use with specimens tested with lots used previously. The sera from 20 patients showed anti-DP activity; these sera were tested with more than one lot, showing concordant results in terms of specificity from lot to lot. The specificity of anti-DP Abs was examined in the context of DPB1 alleles present in the donor; the patterns of anti-DP reactivity were also interpreted in an attempt to evaluate possible epitopes recognized in the patient's sera. The putative epitopes recognized by DSAs are listed in Table 2. The sera of 20 patients reacted with HLA-DP molecules; 19 of these recognized unambiguously epitopes present in the DPB1 subunit and 1 recognized an epitope present in DPA1 subunits bearing the alleles DPA1*02:01 and DPA1*04:01. The final DSA was assessed by comparing the high-resolution typing of the donor and Ag panels. Ab levels were interpreted as normalized fluorescence intensity (FI) as defined by the kit's manufacturer against DSA mismatch. FI < 500 was considered negative; positive scores of 500-1500 positive, 1500-3000, 3000-7000, and > 7000 were considered weak, intermediate, strong, and very strong, respectively. HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DRB3/4/5, HLA-DQB1, and HLA-DPB1 high-resolution typing was accomplished by PCR amplification combined with nucleotide sequencing. DPA1 and DQA1 were not typed; possible DSA against alleles of these loci were examined by inferring the alleles likely to be present at these loci from the tight associations with alleles at the contiguous DPB1 and DRB1/DQB1 loci, respectively.12,13 No transplantations were performed in patients presenting with DSAs against HLA-A, HLA-B, HLA-C, or HLA-DRB1 Ags. There were no transplantations performed across DSAs directed against Ag preparations bearing molecules including DQB1 or DRB3/4/5 alleles present in the donor.
Type and specificity of donor-specific anti-HLA Abs in matched unrelated donor transplant recipients
| n . | Anti-DP DSA . | MFI value before first SCT . | Subunit . | Epitope* . | Ags within epitope, n† . | MFI for epitope‡ . | Engrafted . | Second SCT . | MFI second SCT . | Engrafted . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0401 | 1558 | DPB1 | A 56 | 4 | 880-1558 | Y | N | N/A | N/A |
| 2 | 0301 | 4341 | DPB1 | EAV 85-87 | 9 | 2723-8531 | N | N | N/A | N/A |
| 3 | 0301 | 2759 | DPB1 | EAV 85-87 | 11 | 971-3072 | N | Y | 0 | Y |
| 4 | 0101 | 3733 | DPB1 | EAV 85-87 | 11 | 1787-7781 | Y | N | N/A | N/A |
| 5 | 0101 | 4924 | DPB1 | EAV 85-87 | 11 | 552-4924 | Y | N | N/A | N/A |
| 1101 | 1019 | |||||||||
| 6 | 1401 | 5597 | DPB1 | E 56 | 8 | 4835-8419 | Y | N | N/A | N/A |
| 7 | 2001 | 3990 | DPB1 | K/R 69-M 76 | 6 | 1583-6411 | N | N | N/A | N/A |
| 8 | 1001 | 9485 | DPB1 | EAV 85-87 | 10 | 2320-9485 | Y | N | N/A | N/A |
| n . | Anti-DP DSA . | MFI value before first SCT . | Subunit . | Epitope* . | Ags within epitope, n† . | MFI for epitope‡ . | Engrafted . | Second SCT . | MFI second SCT . | Engrafted . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0401 | 1558 | DPB1 | A 56 | 4 | 880-1558 | Y | N | N/A | N/A |
| 2 | 0301 | 4341 | DPB1 | EAV 85-87 | 9 | 2723-8531 | N | N | N/A | N/A |
| 3 | 0301 | 2759 | DPB1 | EAV 85-87 | 11 | 971-3072 | N | Y | 0 | Y |
| 4 | 0101 | 3733 | DPB1 | EAV 85-87 | 11 | 1787-7781 | Y | N | N/A | N/A |
| 5 | 0101 | 4924 | DPB1 | EAV 85-87 | 11 | 552-4924 | Y | N | N/A | N/A |
| 1101 | 1019 | |||||||||
| 6 | 1401 | 5597 | DPB1 | E 56 | 8 | 4835-8419 | Y | N | N/A | N/A |
| 7 | 2001 | 3990 | DPB1 | K/R 69-M 76 | 6 | 1583-6411 | N | N | N/A | N/A |
| 8 | 1001 | 9485 | DPB1 | EAV 85-87 | 10 | 2320-9485 | Y | N | N/A | N/A |
SCT indicates stem cell transplantation; and N/A, not applicable.
Amino acid residues recognized by the DSAs in the donor.
Number of single Ag preparations sharing the DSA epitope.
Range of MFIs detected on single Ag preparations bearing the epitope detected by the DSA.
Flow cytometric studies
BM aspirates and peripheral blood stem cell specimens were collected, suspended in DMSO, and kept frozen in a liquid nitrogen tank. The samples were thawed at 37°C, washed in PBS, and incubated with mAbs for 10 minutes at 4°C. CD3-FITC, CD34-PerCP5.5, CD45-500, HLA-ABC-PE, and HLADR-V450 Abs were obtained from BD Biosciences. HLA-DP (H1461; Fitzgerald Industries) were labeled with Alexa Fluor 647 (Molecular Probes) according to the manufacturer's instructions. HLA-DP–Alexa 647 was serially diluted and the condition was tested in normal BM B cells, monocytes, and CD3+ T cells. Samples were acquired on FACSCanto II instruments (BD Biosciences); 100 000 events or all available cells were acquired. Data were analyzed using FCS Express software (De Novo Software). Nonviable cells, debris, and aggregates were excluded based on forward scatter-height/forward scatter-area. Autofluorescent controls and internal positive and negative controls were used for gating and assessment of HLA-A, HLA-B, HLA-C, HLA-DR, and HLA-DP expression.
Statistical analysis
Descriptive statistics, including frequency (percentage) for categorical variables and median (range) for quantitative variables, were used to summarize patient characteristics. Fisher exact test and generalized Fisher exact tests were used to assess possible associations among anti-HLA Abs, DSAs, and graft failure.14 Univariate and multivariate logistic Bayesian regression models were fit to assess the associations between various risk factors and graft failure.15 The association between number of pregnancies and anti-HLA Abs was assessed by fitting a logistic regression model for just the female patients and also for all patients. The cumulative incidence of absolute neutrophil count (ANC) recovery (ie, reach of ANC 500/μL for 3 consecutive days) was calculated, where death without ANC recovery was treated as the competing event.16 Computations for all statistical analyses were conducted in SAS for Windows Version 9.0 and Winbugs Version 1.4.17
Results
Association among anti-HLA Abs, DSAs, and graft failure
Of 592 patients evaluated, 116 (19.6%) were found to have anti-HLA Abs that were not reactive with donor loci, whereas 8 patients (1.4%) had DSAs. All DSAs were directed against the HLA-DPB1 molecule. Specificities of these Abs are presented in Table 2.
Table 2 also includes possible epitopes recognized by allo-Abs and the ranges of mean fluorescent intensity (MFI) observed in individual Ag preparations. The reactivity of the patient's sera was correlated with specific amino acid replacements on DPB1 alleles present in the donor and in other alleles.
In addition to the above-mentioned patients, we identified 12 other patients whose sera displayed anti-DP reactivity; all them achieved allogeneic engraftment. The serum of 11 of these patients showed reactivity that was exquisitely correlated with epitopes present in the DPB1 alleles; the serum of 1 patient showed reactivity that was correlated with the recognition of an epitope present in DPA1 alleles. In the 11 patients with reactivity against DPB1 there was no reactivity against Ag preparations bearing the DPB1 Ags present in the donor, and none of the DPB1 alleles present in the donors bore the putative epitopes. The likely anti-DPA1 reactivity was defined in one serum based on the exact correlation with the presence of an epitope shared by the DPA1*02 and DPA1*04:01 alleles (defined by either residues R 50 and/or A 83) with weak to intermediate strength (1323-2667 MFI). Although DPA1 typing was not performed, it was judged through knowledge of the tight DPA1-DPB1 associations in Europeans that the donor and patient were not likely to be homozygous for DPA1*01:03 and would not present any mismatch in DPA1. The presence of anti-DPA1 reactivity in the patient's serum was not likely to result in DSAs. The putative DPA1 epitope identified by the patient's serum is allelic to DPA1*01:03; therefore, the pattern reactivity defined by the patient's serum was consistent with the patient's homozygosity at this locus.
Overall, graft failure occurred in 19 of 592 transplantations (3.2%). This event occurred in 16/584 (2.7%) patients without anti-HLA Abs, compared with 3 of 8 (37.5%) of allosensitized patients with DSAs (P = .0014). Ignoring DSAs, 7 of 116 (6.0%) allosensitized patients (patients with anti-HLA Abs) had graft failure versus 12 of 476 (2.5%) patients without anti-HLA Abs (P = .0732).
No association was identified between graft failure and the type of conditioning chemotherapy, either myeloablative or nonmyeloablative/ reduced-intensity; 233 (96.7%) versus 340 (96.9%) patients engrafted in the 2 groups, respectively (P = NS). In terms of time to engraftment, a nonsignificant trend toward a higher engraftment rate was found for the 8 of 8 compared with the 7 of 8 HLA allele–matched donor (P = .053, log rank test; Figure 1). In the 7 of 8 group, there were no patients with DSAs; all patients with DSAs were in the 8 of 8 group.
Cumulative incidence of ANC recovery based on HLA mismatches in HEL, LEL, anti-HLA Abs, and donor-specific anti-HLA Abs.
Cumulative incidence of ANC recovery based on HLA mismatches in HEL, LEL, anti-HLA Abs, and donor-specific anti-HLA Abs.
Multivariate Bayesian logistic regression models were fit for the probability of engraftment based on mismatches in HEL, LEL, anti-HLA Abs, and DSAs (Table 3). In these models, an effect may be considered significant if the posterior probability of a beneficial effect is either very small (P < .01) or very large (P > .99), with nonsignificant values between P = .20 and P = .80. These analyses showed that the probability of DSA being harmful to engraftment was P > .99, whereas matching at HEL and LEL had a posterior probability of P = .857. This reiterated the non-model-based conclusion given above, that the presence of both anti-HLA Abs and DSAs was strongly predictive of graft failure. To assess the effect of all risk factors known to be associated with graft failure in hematopoietic stem cell transplantation patients, we fit univariate Bayesian logistic regression models for the probability graft failure, considering recipients' sex, race, number of CD34+ cell numbers infused, CMV, ABO and sex mismatch between the donor and recipient, number of HELs and LELs in the HVG direction, and the presence of anti-HLA Abs and DSAs (Table 4). The univariate analyses revealed that the only factors significantly associated with graft failure were the presence of DSAs (probability of a harmful effect P < .001, odds ratio [OR] = 0.05) and ABO mismatch (probability of a harmful effect P = .015, OR = 0.22). However, as mentioned above, a borderline effect was seen for the presence of anti-HLA Abs in the absence of DSAs (probability of a harmful effect = .048, OR = 0.42)
Impact of HLA matching and anti-HLA Abs on the probability of engraftment using Bayesian logistic regression models
| Variable . | Mean . | SD . | Posterior 95% CI . | Probability of a beneficial effect . | |
|---|---|---|---|---|---|
| 2.50% . | 97.50% . | ||||
| Full model | |||||
| Intercept | 3.296 | 0.658 | 2.140 | 4.674 | |
| HEL match (vs HEL mismatch) | 0.432 | 0.687 | -1.021 | 1.682 | .761 |
| LEL match (vs LEL mismatch) | 0.413 | 0.675 | -0.772 | 1.888 | .729 |
| AHA+, DSA+ (vs AHA−, DSA−) | −3.114 | 0.835 | −4.703 | −1.470 | < .001 |
| AHA+, DSA− (vs AHA−, DSA−) | −0.315 | 0.598 | −1.456 | 0.919 | .285 |
| Combining AHA+DSA− with AHA− | |||||
| Intercept | 3.246 | 0.689 | 2.065 | 4.758 | |
| HEL match (vs HEL mismatch) | 0.376 | 0.731 | −1.156 | 1.663 | .719 |
| LEL match (vs LEL mismatch) | 0.467 | 0.698 | −0.768 | 1.922 | .758 |
| AHA+, DSA+ (vs DSA−) | −3.070 | 0.809 | −4.647 | −1.302 | < .001 |
| Combining HEL and LEL mismatch | |||||
| Intercept | 3.506 | 0.264 | 3.027 | 4.050 | |
| HEL and LEL match (vs any mismatch in HEL or LEL) | 0.876 | 0.860 | −0.539 | 2.769 | .857 |
| AHA+, DSA+ (vs DSA−) | −2.923 | 0.849 | −4.507 | −1.144 | .001 |
| Variable . | Mean . | SD . | Posterior 95% CI . | Probability of a beneficial effect . | |
|---|---|---|---|---|---|
| 2.50% . | 97.50% . | ||||
| Full model | |||||
| Intercept | 3.296 | 0.658 | 2.140 | 4.674 | |
| HEL match (vs HEL mismatch) | 0.432 | 0.687 | -1.021 | 1.682 | .761 |
| LEL match (vs LEL mismatch) | 0.413 | 0.675 | -0.772 | 1.888 | .729 |
| AHA+, DSA+ (vs AHA−, DSA−) | −3.114 | 0.835 | −4.703 | −1.470 | < .001 |
| AHA+, DSA− (vs AHA−, DSA−) | −0.315 | 0.598 | −1.456 | 0.919 | .285 |
| Combining AHA+DSA− with AHA− | |||||
| Intercept | 3.246 | 0.689 | 2.065 | 4.758 | |
| HEL match (vs HEL mismatch) | 0.376 | 0.731 | −1.156 | 1.663 | .719 |
| LEL match (vs LEL mismatch) | 0.467 | 0.698 | −0.768 | 1.922 | .758 |
| AHA+, DSA+ (vs DSA−) | −3.070 | 0.809 | −4.647 | −1.302 | < .001 |
| Combining HEL and LEL mismatch | |||||
| Intercept | 3.506 | 0.264 | 3.027 | 4.050 | |
| HEL and LEL match (vs any mismatch in HEL or LEL) | 0.876 | 0.860 | −0.539 | 2.769 | .857 |
| AHA+, DSA+ (vs DSA−) | −2.923 | 0.849 | −4.507 | −1.144 | .001 |
AHA indicates anti-HLA Abs.
Univariate Bayesian logistic regression models of probability of engraftment in 592 matched unrelated donor transplantation patients
| Variable . | Mean . | SD . | Probability (P) of graft failure . | OR . |
|---|---|---|---|---|
| Male (vs female) | 0.59 | 0.47 | .893 | 1.80 |
| White race (vs others) | −0.45 | 0.84 | .302 | 0.64 |
| AHA yes (vs no) | −0.87 | 0.50 | .048 | 0.42 |
| AHA yes and DSA yes (vs no) | −3.00 | 0.79 | < .001 | 0.05 |
| HEL 8 (vs 7) | 0.17 | 0.68 | .630 | 1.18 |
| LEL 6 (vs < 6) | 0.65 | 0.69 | .825 | 1.92 |
| Log (CD34 cell numbers infused) | 0.38 | 0.28 | .916 | 1.46 |
| CMV mismatch yes (vs no) | −0.24 | 0.48 | .296 | 0.79 |
| Sex mismatch yes (vs no) | −0.57 | 0.47 | .121 | 0.57 |
| ABO mismatch yes (vs no) | −1.51 | 0.69 | .015 | 0.22 |
| Variable . | Mean . | SD . | Probability (P) of graft failure . | OR . |
|---|---|---|---|---|
| Male (vs female) | 0.59 | 0.47 | .893 | 1.80 |
| White race (vs others) | −0.45 | 0.84 | .302 | 0.64 |
| AHA yes (vs no) | −0.87 | 0.50 | .048 | 0.42 |
| AHA yes and DSA yes (vs no) | −3.00 | 0.79 | < .001 | 0.05 |
| HEL 8 (vs 7) | 0.17 | 0.68 | .630 | 1.18 |
| LEL 6 (vs < 6) | 0.65 | 0.69 | .825 | 1.92 |
| Log (CD34 cell numbers infused) | 0.38 | 0.28 | .916 | 1.46 |
| CMV mismatch yes (vs no) | −0.24 | 0.48 | .296 | 0.79 |
| Sex mismatch yes (vs no) | −0.57 | 0.47 | .121 | 0.57 |
| ABO mismatch yes (vs no) | −1.51 | 0.69 | .015 | 0.22 |
AHA indicates anti-HLA Abs.
No differences between DSA patients who did and did not engraft were found in terms of median age (52 vs 56 years), degree of matching (all DSA occurred in patients with 8 of 8 allele match), median number of CD34+ cells infused (6.7 × 106/kg vs 11.2 × 106/kg), conditioning regimen (3 of 5 vs 2 of 3 had nonmyeloablative conditioning), intensity of Ab levels (strong in both groups, median 3696 vs 5059 FI; Table 2).
Risk factors for the development of anti-HLA Abs
We have previously observed a striking association between the sex of patients who experience graft failure and the development of allosensitization; all patients with DSAs were multiparous young women with a median of 3 pregnancies.10 This study confirms a significant association among female sex, the development of allosensitization, and the presence of DSAs; 30.8% of women versus 12.1% of men had anti-HLA Abs (P < .0001) and 7 of 8 patients with DSAs were women, all of whom except 1 had at least 2 prior pregnancies. When the presence of anti-HLA Abs was evaluated in women with no pregnancies compared with the male recipients, no significant association was identified (P = .24, OR = 2.1, 95% confidence interval [95% CI] = 0.62-6.94). However, when allosensitization was compared between women with 1 prior pregnancy and men, a higher proportion of female patients had anti-HLA Abs (P = .008, OR = 6.3, 95% CI = 1.62-24.95). Moreover, a stronger association was found between women with 2 or more pregnancies and men (P = .0003, OR = 9.5, 95% CI = 2.83-32.02). These results suggest that pregnancy confers a significant risk for allosensitization, and that this risk is further increased with a higher number of pregnancies. Although the majority of allosensitized individuals in this study were women, ∼ 12% of patients with anti-HLA Abs were men (and 1 patient had DSAs), suggesting that other factors are associated with the development of anti-HLA Abs in these patients. The likely cause of allosensitization is transfusion of blood products, a factor not evaluated in this study because of incomplete transfusion history data (many patients were initially treated or received blood products intermittently at outside institutions).
Expression of HLA-DP Ags on normal progenitor cell surface
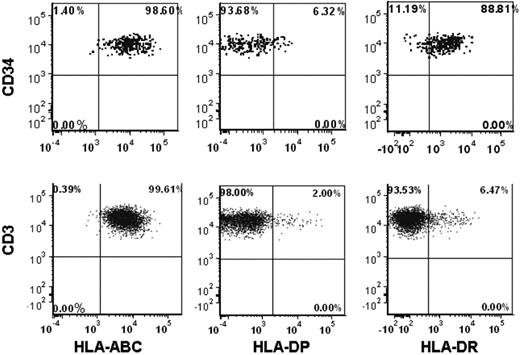
Compared with our previous experience in T cell–depleted haploidentical stem cell transplantation, the risk of graft failure in the presence of DSAs appears to be less in T cell–replete unrelated donor transplantations.10 We hypothesized that a lower expression of HLA-DP Ags on the progenitor cell surface could explain this difference, at least in part, and evaluated 8 normal donor BM and peripheral blood samples for the presence of anti-HLA Ags on the CD34+ progenitor cell surface (Figure 2). HLA-DP was found to be significantly less expressed on normal CD34+ cells compared with HLA class I Ags (HLA-A, HLA-B, and HLA-C; median 36.1% vs 99.9%, respectively; P < .001). Significant differences in HLA-DP expression were seen also between CD34+ cells derived from peripheral blood and BM samples (15%, range 10.5%-29.9% vs median 59.3%, range 42.2%-71.5%, respectively; P = .004). A significantly lower expression of HLA-DP compared with class I HLA Ags (HLA-A, HLA-B, and HLA-C) was also seen on the surface of CD3+ cells (median 1.9% vs 99.9%, respectively). HLA-DR was expressed in a high proportion of CD34+ cells (median 94.1%, range 85.6-99.5%), whereas CD3+ cells expressed only low levels of HLA-DR (median 4.6%, range 1.8-13.0) (Figure 2).
Representative flow cytometric analysis of class I and class II HLA Ag expression on CD34+ and CD3+ cell surface for a peripheral blood sample from a normal donor.
Representative flow cytometric analysis of class I and class II HLA Ag expression on CD34+ and CD3+ cell surface for a peripheral blood sample from a normal donor.
Discussion
Allosensitization remains an important problem in transplantation. Anti-HLA Abs have been shown to be associated with graft rejection in solid-organ transplantation.18-20 More recently, an association between anti-HLA Abs and graft failure has been found in hematopoietic stem cell transplantation.10,21,22 All of these studies confirmed a higher risk of graft failure in the presence of DSAs in different types of transplantations with mismatches.
In the present study, we provide evidence that DSAs against low-expression HLA Ag DPB1 are also deleterious to engraftment using a homogenous cohort of MUD transplantation patients. In multivariate analysis, DSAs were the single most important factor associated with graft failure in these patients; none of the other factors except ABO mismatch was associated with an increased risk of graft failure, including mismatching in HEL or LEL. Interestingly, having an HLA mismatch by itself was not associated with a significantly higher risk of graft failure unless it was associated with DSAs. Moreover, alloimmunization per se did not cause a significant increase risk of graft failure unless Abs were directed against the donor HLA Ags, suggesting that DSAs are the key to the development of graft failure. These results, combined with our previous experience in haploidentical transplantation patients, strongly suggest that anti-HLA Abs against the mismatched HLA Ags have an important role in the development of graft failure in hematopoietic stem cell transplantation.
The lower risk of graft rejection in T cell–replete MUD transplantations in this study, compared with our previous experience with T cell–depleted haploidentical transplantations10 could be explained by a lower expression of HLA-DP Ags on the CD34 cell surface or by the retention of other cells in the graft such as T cells (CD3+), B cells, hematogones, monocytes, and dendritic cells. The presence of these other hematopoietic cells that uniformly expressed class I HLA Ags and had variable expression of HLA-DR and HLA-DP Ags (data not shown) could be associated with a protective effect from the development of graft failure in these patients. Dendritic cells, especially plasmacytoid dendritic cells, play a key role in concert with other collaborative cell types to allow optimal hematopoietic cell engraftment.23 The plasmacytoid dendritic cells have been shown to express a high level of HLA-DP Ags.24 In the 8 patients that we studied, although the dendritic cells were not specifically studied, the monocytes—including dendritic cells as identified by CD45/side scatter—showed variable HLA-DP expression.
The results of the present study also show that one cause of allosensitization is the exposure of fetal HLA Ags of paternal origin to the maternal immune system during early pregnancy. Moreover, a higher risk of allosensitization occurs with a higher number of pregnancies, strongly suggesting that pregnancy is the main cause of the development of anti-HLA Abs in these patients. A second role, not evaluated in the present study, is attributed to transfusions of blood products, because men were also allosensitized, albeit to a significantly lower proportion than women. This observation was more difficult to evaluate because of incomplete data on transfusions of blood products before referral for hematopoietic transplantation.
Anti-HLA Abs were detected in ∼ 20% of MUD transplantation recipients, making it a common occurrence in stem cell transplantation. Anti-DP Abs were found in 20 of 592 (3.4%) patients. Of these 20 patients, 4 received transplantation with a matched donor in HLA-DPB1. Sixteen patients with anti-DP Abs were transplanted with an HLA-DPB1–mismatched donor, and DSAs were found in 8 of 16 patients (50%). Three of these 8 patients had graft failure. Virtually all anti-DP reactive sera showed reactivity with multiple alleles of HLA-DPB1 (broadly reactive) recognizing public (supertypic) determinants on the HLA-DPB1 molecules. In other words, given the cross-reactive nature of anti-DP Abs, when they are present, they are likely to react with many donors.
Whereas the alleles of DQA1 were not typed in this study, we queried for the possible impact of Abs directed against DQA1. The alleles of DQA1 showed tight linkage disequilibrium with alleles of DQB1 and DRB112 ; in the studies by Lee et al6 and Hurley et al,25 > 99.8% of the transplantations matching in DRB1-DQB1 loci also match in DQA1. In the present study, > 93% of the transplantations were matched in those loci; therefore, this group of patients most likely includes a few or no mismatches in DQA1. The serum of only 2 patients in the 40 transplantations that presented a mismatch in DQB1 or DRB1 and could be mismatched in DQA1 showed anti-DQ reactivity; neither of these sera presented specificity that could be ascribed to the recognition of epitopes present in DQA1 alleles. Furthermore, we analyzed and made predictions about the possible occurrence of mismatching in DQA1 in the pairs mismatched in either DRB1 or DQB1, and identified only 1 patient who presented with a mismatch in DRB1 in the HVG direction and was likely to present an additional mismatch in DQA1; in the latter case, the serum of the patient did not show any anti-HLA reactivity. We estimated that there were very few or no cases in which anti-DQA1 Abs might have resulted in undetected DSAs in the present study. Despite this evaluation, we propose that prospective typing of DQA1 alleles and screening for anti-DQA1 Abs may be applicable in allogeneic hematopoietic stem cell transplantation, in particular in transplantations presenting mismatches in DRB1 and/or DQB1 alleles.
The results of the present study indicate that the presence of DSAs in the patient's pretransplantation serum reactive with the donor HLA-DPB1–mismatched Ags is associated with increased risk for graft failure. The immunologic basis for this observation may result from a direct deleterious effect of Abs that leads to the destruction of the graft cells via complement fixation or by Ab-dependent cell-mediated cytotoxicity. Alternatively, the presence of DSAs in these patients may just reflect immunization and expansion of alloreactive T lymphocytes that may mediate graft failure. The fact that the DSAs are of the IgG subclass suggests the potential recognition of the donor's mismatched Ags by the patient's T lymphocytes. It should be noted that the T-cell and B-cell epitopes may be distinct in nature. The sera of all patients with anti-DP DSAs reacted with multiple-DP Ag preparations, indicating the recognition of supertypic or public epitopes; therefore, the T-cell epitopes may not fully overlap or actually may be restricted only to a few alleles. It is possible that some of the DSAs may not match specifically the anti-DP Ags involved in the sensitization. If T cells were the effectors in graft failure, one could hypothesize that in the 3 patients with DSAs and graft failure, both the humoral and T-cell epitopes (TCEs) coincide; in contrast, there could be a dissociation between T-cell and B-cell epitopes in the 5 patients with engraftment, in which the DSA results from sharing humoral epitopes between the mismatched anti-DP Ags and different anti-DP Ags present in the initial immunizing event. These mechanisms deserve further evaluation, including the investigation of donor-specific alloreactive T lymphocytes.
In the 8 patients with preformed DSAs discussed here, we also evaluated the types of T-cell epitope mismatches in the HVG direction in patients with and without engraftment according to the epitope-matching scoring defined by Fleischhauer et al for DP-immunogenic mismatches.26 In the 5 transplantations in which there was donor engraftment, 2 patients presented with nonpermissive TCE mismatches (DPB1*1001 and DPB1*1401), whereas in the 3 patients without engraftment, 2 presented with nonpermissive TCE mismatches (DPB1*0301 in both of them). The analysis of the TCE mismatches and the anti-DP serologic epitopes defined by our group27 in previous studies, and additional possible epitopes listed in Table 2, did not identify any specific epitope present in the nonengraftment group that was absent in the group of transplantations with engraftment. Similarly, neither the nature of the cross-reactivity nor the ranges of MFIs identified distinctive characteristics between the patients with and without engraftment. These analyses performed in our dataset did not allow us to favor either of the hypotheses, including humoral or cellular mechanisms for graft failure in the allosensitized patient groups against donor Ags.
Preclinical studies on animal models demonstrated that passive transfer of anti-HLA Abs can reject the transplanted donor hematopoietic stem cells, and that preformed anti-HLA Abs are sufficient to cause graft failure.28,29 Consequently, attempts to remove anti-HLA Abs directed against the donor cells before transplantation make intuitive sense. In the present study, only 2 of the 8 patients with preformed DSAs could be treated prospectively using rituximab and plasma exchange.10 Although there was not complete Ab removal by these steps, the FI against the anti-DP mismatched Ags decreased significantly in both of them from very high levels (MFI 9497 and 7393) to moderate or low values after treatment (MFI 3733 and 2759, respectively). One patient showed engraftment, whereas the other 1 failed to engraft.
In conclusion, anti-DPB1 DSAs are associated with graft failure in MUD hematopoietic stem cell transplantation. Screening for anti-HLA Abs is warranted when considering donors with mismatches, and attempts to reduce the Ab levels before transplantation deserve further investigation in hematopoietic stem cell transplantation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by an institutional research grant from the M. D. Anderson Cancer Center to S.O.C.
Authorship
Contribution: S.O.C. contributed to the study design, collected and analyzed the data, interpreted the results, and wrote the manuscript; X.W. and P.F.T. performed the statistical analysis and reviewed, edited, and approved the manuscript; S.A.W. and Y.H. performed the HLA Ab staining and flow cytometry studies and reviewed and approved the manuscript; G.R. and F.A. contributed to data collection and reviewed and approved the manuscript; P.C., J.J.M., M.K., and E.J.S. critically reviewed, edited, and approved the manuscript; M.d.L. and R.E.C. contributed to data interpretation and critically reviewed, edited, and approved the manuscript; and M.F.-V. performed the Ab testing, contributed to the study design, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.F.-V. is Department of Pathology, Stanford Medical School, Stanford University, Stanford, CA.
Correspondence: Marcelo Fernandez-Vina, PhD, 3373 Hillview Ave, Palo Alto, CA 94304; e-mail: marcelof@standford.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal