Abstract
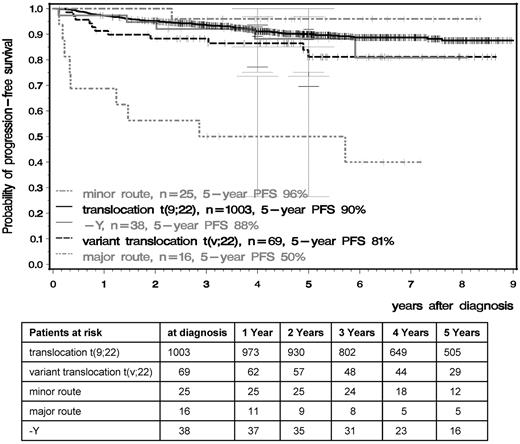
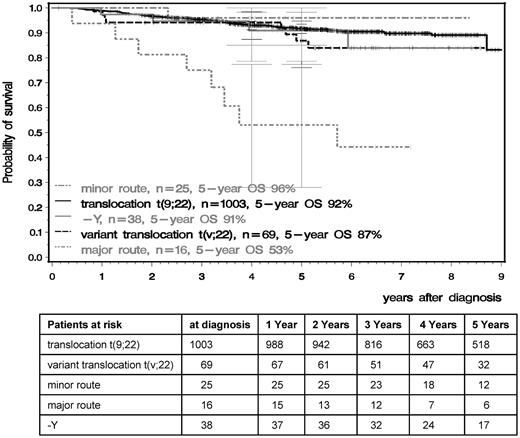
The prognostic relevance of additional cytogenetic findings at diagnosis of chronic myeloid leukemia (CML) is unclear. The impact of additional cytogenetic findings at diagnosis on time to complete cytogenetic (CCR) and major molecular remission (MMR) and progression-free (PFS) and overall survival (OS) was analyzed using data from 1151 Philadelphia chromosome–positive (Ph+) CML patients randomized to the German CML Study IV. At diagnosis, 1003 of 1151 patients (87%) had standard t(9;22)(q34;q11) only, 69 patients (6.0%) had variant t(v;22), and 79 (6.9%) additional cytogenetic aberrations (ACAs). Of these, 38 patients (3.3%) lacked the Y chromosome (−Y) and 41 patients (3.6%) had ACAs except −Y; 16 of these (1.4%) were major route (second Philadelphia [Ph] chromosome, trisomy 8, isochromosome 17q, or trisomy 19) and 25 minor route (all other) ACAs. After a median observation time of 5.3 years for patients with t(9;22), t(v;22), −Y, minor- and major-route ACAs, the 5-year PFS was 90%, 81%, 88%, 96%, and 50%, and the 5-year OS was 92%, 87%, 91%, 96%, and 53%, respectively. In patients with major-route ACAs, the times to CCR and MMR were longer and PFS and OS were shorter (P < .001) than in patients with standard t(9;22). We conclude that major-route ACAs at diagnosis are associated with a negative impact on survival and signify progression to the accelerated phase and blast crisis.
Introduction
Current evidence indicates that acquired genetic instability as a consequence of the Philadelphia (Ph) translocation t(9;22)(q34;q11)1 and the resulting BCR-ABL fusion causes the continuous acquisition of additional chromosomal aberrations (ACAs) and mutations, and thereby progression to the accelerated phase (AP) and blast crisis (BC) of CML.2-4 Approximately 10%-12% of patients in chronic-phase chronic myeloid leukemia (CP-CML) have additional chromosomal findings at diagnosis that include variant translocations, lack of the Y chromosome (−Y), and “true” ACAs, which occur in less than 5% of patients at diagnosis. The proportion of patients with ACAs rises during the course of the disease to approximately 80% in BC.5 It was recognized early that ACAs in CML occurred strictly nonrandomly.5 The most frequent aberrations detected in advanced CML were trisomy 8, a second Ph chromosome, and a partial trisomy of the long arm with partial monosomy of the short arm of chromosome 17 [isochromosome (17)(q10)], which were designated “major-route of karyotypic evolution.”5 ACAs that were rarely observed in AP or BC, such as t(3;12), t(4;6), t(2;16), and t(1;21), were designated minor-route ACAs.6
Chromosomal changes were thought to unequally influence disease progression depending on the type of ACA and its time of appearance. Major-route ACAs were thought to be more correlated with worse prognoses than minor-route ACAs, and as a consequence, patients with major-route ACAs were excluded from the Italian randomized study comparing IFNα with chemotherapy7 and from determination of the Euro-Score.8
The types of chromosome aberrations are not influenced by tyrosine kinase inhibition,9-11 and are similar at diagnosis, during the course of CML, and after treatment.12 Chromosomal abnormalities developing during the course of CML in addition to the Ph chromosome (clonal evolution) are considered a feature of acceleration and indicate a poor prognosis.13,14 Patients with additional chromosomal abnormalities show lower cytogenetic response rates under imatinib.15 The worst outcome has been reported for major-route ACAs and complex abnormalities.11 On the basis of limited evidence, ACAs at diagnosis were defined by the European LeukemiaNet (ELN) recommendations as a warning sign, in contrast to ACAs newly arising under treatment, which define failure.16 Some investigators have reported negative impacts of ACAs at diagnosis,17,18 but these were later thought to be due to inadequate cytogenetic techniques available at the time.
A staging system that included ACAs regardless of subtype as a prognostic factor18,19 was not confirmed when determining the Euro-Score,8 which excluded major-route ACAs from analysis. In a comparison of 29 Ph+ CML patients with ACA at diagnosis with 234 sole Ph+ patients, all treated with IFN, 0 of 4 patients with major-route ACAs had achieved a cytogenetic response.20
Variant translocations [t(v;22)] are characterized by the involvement of one or more chromosomes in addition to chromosomes 9 and 22, and occur in 5%-10% of patients.21,22 In the pre-imatinib era, variant translocations were considered to be indicators of a poor prognosis21 ; however, in the imatinib era, this was no longer found to be the case.22 When treated with imatinib, patients with variant translocations had a similar prognosis to those with standard translocations.22,23 Deletions in derivative chromosome 9 [der(9)], which were a key prognostic factor in the pre-imatinib era, occur with greater frequency in t(v;22) patients.24 The loss of the negative prognostic impact of der(9) deletions with imatinib probably contributes to the loss of negative prognostic impact of variant translocations.25
−Y is observed in approximately 5% of Ph+ patients.6 Although a negative impact on prognosis has been reported in one series,26 −Y is considered a minor-route ACA.12 A physiologic −Y in elderly men has to be considered in evaluating −Y in CML.
To solve the discrepancies between additional chromosomal changes at diagnosis and during progression of the disease, we investigated the effect of clonal chromosomal findings in addition to the standard translocation at diagnosis on the outcome of CML. Using baseline and outcome data of 1151 patients with CP-CML randomized to the German CML-Study IV,27 we report herein a negative prognostic impact of major-route ACAs at diagnosis compared with standard t(9;22), t(v;22), −Y, or minor-route ACAs.
Methods
Patient characteristics and study design
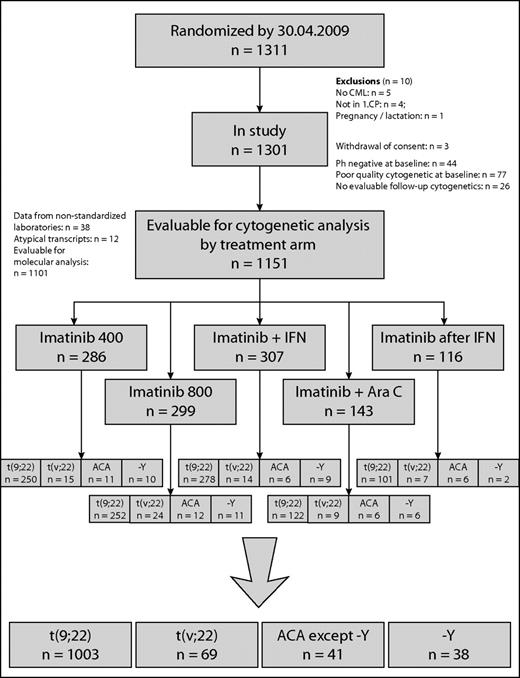
Clinical and cytogenetic data of 1151 of 1311 patients with Ph+ and BCR-ABL+ CP-CML randomized to the German CML-Study IV (imatinib 400 mg vs imatinib 800 mg vs imatinib in combination with IFN vs imatinib in combination with low-dose cytarabine vs imatinib after IFN failure)27 were investigated prospectively. There were 459 female (40%) and 692 male (60%) patients with a median age of 53 years (range, 16-88); median age was lower in ACA patients (48 years) and higher in −Y patients (62 years). The risk profile was similar, except −Y patients had a better (but not significantly different) profile (Table 1). The definitions of CML phases followed the ELN recommendation.16 Patients were randomized into the 5 different treatment arms, as shown in Figure 1.
Patient characteristics
| . | t(9;22) . | t(v;22) . | ACAs (except −Y) . | (−Y) . |
|---|---|---|---|---|
| n | 1003 | 69 | 41 | 38 |
| Median age, y (range) | ||||
| All | 52 (16-87) | 54 (16-88) | 48 (18-72) | 62 (24-79) |
| Male | 51 (16-87) | 54 (16-88) | 45 (23-72) | 62 (24-79) |
| Female | 55 (16-83) | 54.5 (17-70) | 53 (18-69) | - |
| Male, % | 59 | 57 | 59 | 100 |
| EUTOS score, % | ||||
| Low | 90.2 | 80.3 | 87.5 | 94.4 |
| High | 9.8 | 19.7 | 12.5 | 5.6 |
| Euro score, % | ||||
| Low | 35.3 | 27.5 | 43.9 | 39.4 |
| Intermediate | 54.8 | 55.1 | 41.5 | 51.5 |
| High | 9.9 | 17.4 | 14.6 | 9.1 |
| Median follow-up, mo | 64.1 | 63.0 | 72.7 | 58.6 |
| . | t(9;22) . | t(v;22) . | ACAs (except −Y) . | (−Y) . |
|---|---|---|---|---|
| n | 1003 | 69 | 41 | 38 |
| Median age, y (range) | ||||
| All | 52 (16-87) | 54 (16-88) | 48 (18-72) | 62 (24-79) |
| Male | 51 (16-87) | 54 (16-88) | 45 (23-72) | 62 (24-79) |
| Female | 55 (16-83) | 54.5 (17-70) | 53 (18-69) | - |
| Male, % | 59 | 57 | 59 | 100 |
| EUTOS score, % | ||||
| Low | 90.2 | 80.3 | 87.5 | 94.4 |
| High | 9.8 | 19.7 | 12.5 | 5.6 |
| Euro score, % | ||||
| Low | 35.3 | 27.5 | 43.9 | 39.4 |
| Intermediate | 54.8 | 55.1 | 41.5 | 51.5 |
| High | 9.9 | 17.4 | 14.6 | 9.1 |
| Median follow-up, mo | 64.1 | 63.0 | 72.7 | 58.6 |
Cytogenetic analyses were made and interpreted according to ISCN 2009.29
n indicates number; and ACA, additional clonal cytogenetic alterations.
The protocol followed the Declaration of Helsinki and was approved by the ethics committee of the Medizinische Fakultät Mannheim and by local ethics committees of participating centers. Written informed consent was obtained from all patients before they entered the study.
Cytogenetics and FISH analysis
Cytogenetic analyses of 20-25 G- or R-banded bone marrow metaphases (24 or 48 hours of culture) at diagnosis were interpreted according to the International System for Human Cytogenetic Nomenclature (ISCN 2009).28,29 Only patients with cytogenetic alterations in Ph+ clones at diagnosis defined according to ISCN 2009 were evaluated and counted as ACA in this study. In 9 CML patients, constitutional alterations were detected: 5 patients with a pericentric inversion of chromosome 9, inv(9)(p11q13), 2 patients with an additional Y-chromosome, 47,XYY, 1 patient with an additional X chromosome (Klinefelter syndrome), and 1 patient with a constitutional translocation, t(10;19).30,31 Because ACAs in these 9 patients were not disease related, they were assigned to the group with standard t(9;22)(q34;q11). In cases of complex aberrant karyotypes, chromosome-banding analysis was combined with FISH analysis according to the manufacturer's instructions (MetaSystems).32 Cytogenetic remission was defined according to the ELN recommendations.16
RQ-PCR
Measurement of the BCR-ABL fusion transcript was performed in standardized laboratories using the real-time quantitative PCR (RQ-PCR) assay with hybridization probes using ABL for normalization (LightCycler 1.5; Roche Diagnostics).33 Ratios of BCR-ABL/ABL were calculated and expressed according to the international scale.34,35
Statistical analysis
Progression-free survival (PFS) was defined as the time from diagnosis until the beginning of the AP, BC, or death from any cause, whichever came first. For overall survival (OS), death from any cause was the only event. Probabilities of PFS and OS were calculated using the Kaplan-Meier method and compared with the log-rank test. Patients were censored at the date of last follow-up. Cumulative incidences of complete cytogenetic remission (CCR) and major molecular remission (MMR) were estimated by the cumulative incidence function considering competing events.36,37 Death or progression without prior CCR or MMR was seen as a competing event. For the estimation of cumulative incidences of CCR and MMR, patients were censored at the date of stem-cell transplantation (SCT) or at the date of first administration of a second generation tyrosine kinase inhibitor (eg, dasatinib, nilotinib, or bosutinib).
Comparisons of continuous variables (eg, age) were done with the Mann-Whitney-Wilcoxon test. Prognostic scores (eg, Euro and EUTOS) were calculated using published formulas.8,38
P < .05 was considered significant. Because of the explorative character of this work, no adjustment of P values was done, and all P values have to be interpreted descriptively. All analyses were performed with SAS Version 9.1.3 software (SAS Institute).
Results
Patient flow is shown in the CONSORT chart in Figure 1 and patient characteristics are summarized in Table 1. Of 1151 patients, 1003 (87%) had the standard translocation t(9;22)(q34;q11) only, 69 (6.0%) showed a variant translocation t(v;22), and 79 (6.9%) had ACA. Thirty-eight patients (3.3%) had −Y.
In 60 of 69 patients with t(v;22), only 1 additional chromosome was involved in the translocation; in 7 patients, 2 other chromosomes were involved, and in 2 patients, 3 other chromosomes (Table 2).
Cytogenetic data from CP-CML patients with a variant translocation t(v;22) at diagnosis
| No. . | Sex . | Age at diagnosis, y . | Karyotype with variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 61 | 46,XY,t(9;14;22)(q34;p13:q11),inv(9)(p11q13)c[11] |
| 2 | M | 78 | 46,XY,t(3;9;22)(q25;q34;q11)[20] |
| 3 | M | 65 | 46,XY,t(6;9;22;9)(p23;q34;q11;q22)[17] |
| 4 | F | 60 | 46,XX,t(7;9;22)(q36;q34;q11)[25] |
| 5 | M | 66 | 46,XY,t(5;9;22)(q11.2;q34;q11)[25] |
| 6 | F | 17 | 46,XX,t(4;9;22)(q28;q34;q11)[16] |
| 7 | F | 70 | 46,XX,t(9;22;12)(q34;q11;q24)[20] |
| 8 | F | 54 | 46,XX,t(1;9;22)(p36;q34;q11)[20] |
| 9 | F | 55 | 46,XX,t(3;9;22;14;17)(p22;q34;q11;q13;q12)[25] |
| 10 | F | 45 | 46,XX,t(1;9;22;19)(q32;q34;q11;p13)[20] |
| 11 | F | 53 | 46,XX,t(7;22)(q22;q11)[20] |
| 12 | F | 23 | 46,XX,t(5;6;9;22)(q35;p12;q34;q11)[25] |
| 13 | M | 30 | 46,XY,t(9;22;12)(q34;q11;q13)[20] |
| 14 | F | 67 | 46,XX,t(9;22;17)(q34;q11;q12)[25] |
| 15 | F | 51 | 46,XX,t(9;22;19)(q34;q11;q13.3)[25] |
| 16 | F | 50 | 46,XX,t(3;9;22)(p21;q34;q11)[20] |
| 17 | F | 65 | 46,XX,t(9;22;15)(q34;q11;q22)[20] |
| 18 | F | 45 | 46,XX,t(9;22;16)(q34;q11;q22)[21] |
| 19 | M | 56 | 46,XY,t(5;9;22)(p14;q34;q11)[25] |
| 20 | F | 61 | 46,XX,t(9;22;19)(q34;q11;p13)[19]/46,XX[1] |
| 21 | M | 62 | 46,XY,t(1;9;22)(p22;q34;q11)[24]/46,XY[1] |
| 22 | M | 16 | 46,XY,t(2;9;22)(q21;q34;q11)[10] |
| 23 | M | 49 | 46,XY,t(9;22;14)(q34;q11;q32)[25] |
| 24 | F | 65 | 46,XX,t(3;9;22)(q27;q34;q11)[20] |
| 25 | F | 58 | 46,XX,t(3;9;22)(q29;q34;q11)[20] |
| 26 | F | 58 | 46,XX,t(11;22)(q13;q11)[13] |
| 27 | F | 48 | 46,XX,der(1)t(1;22)(p36;q11),der(2)t(2;13)(p16;?)dup(2)(q11q31),del(2)(p13),der(22)t(2;22)(p13;q11)[15] |
| 28 | F | 61 | 46,XX,t(9;22;13)(q34;q11;q13)[20]/46,XX[1] |
| 29 | M | 52 | 46,XY,t(11;22)(q13;q11)[15] |
| 30 | F | 66 | 46,XX,t(1;9;22)(q32;q34;q11)[20] |
| 31 | M | 53 | 46,XY,t(9;22;12;17;17;22)(q34;q11;q15;q11;q23)[10] |
| 32 | M | 71 | 46,XY,t(8;9;22)(q21;q34;q11)[15] |
| 33 | M | 59 | 46,XY,t(9;15;22)(q34;q15;q11)[24]/46,XY[1] |
| 34 | M | 37 | 46,XY,t(9;22;15)(q34;q11;p11)[37] |
| 35 | M | 56 | 46,XY,t(9;22;11;15)(q34;q11;p15;q24) [20] |
| 36 | M | 57 | 46,XY,t(8;9;22)(q22;q34;q11)[15] |
| 37 | M | 47 | 46,XY,t(9;22;16)(q34;q11;q23)[15] |
| 38 | F | 47 | 46,XX,t(3;9;22)(q21;q34;q11)[15] |
| 39 | F | 65 | 46,XX,t(7;9;22)(q32;q34;q11)[20] |
| 40 | M | 18 | 46,XY,t(1;9;19;22)(q21;q34;p13;q11)[15] |
| 41 | M | 27 | 46,XY,t(9;22;17;17)(q34;q11;q25;q12)[15] |
| 42 | F | 54 | 46,XX,t(5;9;22)(p15;q34;q11)[15]/46,XX[6] |
| 43 | F | 35 | 46,XX,t(5;22)(q35;q11)[20] |
| 44 | M | 67 | 45,X,−Y,t(2;9;22)(p13;q34;q11)[20] |
| 45 | F | 41 | 46,XX,t(8;9;22)(q24;q34;q11)[10] |
| 46 | M | 74 | 46,XY.t(6;9;22)(p2?5;34;q11,2)[11] |
| 47 | M | 47 | 46,XY,t(9;22;17)(q34;q11;p12)[25] |
| 48 | M | 88 | 46,XY,t(2;9;22)(p13;q34;q11) [20] |
| 49 | M | 67 | 46,XY,t(9;22;13)(q34;q11;p11)[20] |
| 50 | M | 47 | 46,XY,t(6;9;22)(p12–21.1;q34;q11)[15] |
| 51 | F | 65 | 46,XX,t(1;9;22)(p36;q34;q11)[25] |
| 52 | M | 68 | 46,XY,t(22;2;9)(q11;p1?3;q34)[12]/46,XY[2] |
| 53 | F | 56 | 46,XX,t(22;2;9)(q11;p13;q34)[15] |
| 54 | F | 43 | 46,XX,t(7;7)(p22;q22),t(9;22;9)(q34;q11;p24)[16]/46,XX[4] |
| 55 | M | 60 | 46,XY,t(2;9;22)(p11;q34;q11)[30] |
| 56 | M | 76 | 46,XY,t(3;9;22;12)(p13;q34;q11;p13)[22] |
| 57 | M | 63 | 46,XY,t(9;22;15)(q34;q11;q12)[19]/46,XY[2] |
| 58 | M | 54 | 46,XY,t(9;22;9)(q34;q11;21)[15] |
| 59 | F | 47 | 46,XX,t(9;15;22) (q23;q23;q11)[25] |
| 60 | M | 58 | 46,XY,t(4;9;22)(p15;q34;q11)[14]/46,XY[1] |
| 61 | M | 40 | 46,XY,t(9;22;16)(q34;q11;q24) [20] |
| 62 | F | 56 | 46,XX,t(1;22;9;15;1)(q21;q11;q34;q22;q21)[10] |
| 63 | F | 18 | 46,XX,t(8;9)(q24;p22),der(9)del(9)(q13;q22)t(8;9)(q24;p22),der(20)t(9;20)(q13q22;q13);t(?;9;22)(?;q34;q11)[16]/47,XX,idem,+10[2] |
| 64 | M | 32 | 46,XY,t(9;22;17)(q34;q11;q22)[20] |
| 65 | M | 31 | 46,XY,t(9;22;10)(q34;q11;p15)[13];55,XY,+3,+8,t(9;22;10)(q34;q11;p15),+12,+13,+14,+18,+19,+21,+der(22)t(9;22;10)(q34;q11;p15)[7] |
| 66 | M | 67 | 46,XY,t(9;12;22)(q34;q24;q11.2)[20] |
| 67 | M | 42 | 46,XY,t(9;15;22)(q34;q22;q11)[25] |
| 68 | M | 55 | 46,XY,t(1;12)(p34;q24),t(1;9;22)(p36;q34;q11)[21]/48,idem,+8,+9[2] |
| 69 | M | 42 | 46,XY,t(9;22;10)(q34;q11;q24)[20] |
| No. . | Sex . | Age at diagnosis, y . | Karyotype with variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 61 | 46,XY,t(9;14;22)(q34;p13:q11),inv(9)(p11q13)c[11] |
| 2 | M | 78 | 46,XY,t(3;9;22)(q25;q34;q11)[20] |
| 3 | M | 65 | 46,XY,t(6;9;22;9)(p23;q34;q11;q22)[17] |
| 4 | F | 60 | 46,XX,t(7;9;22)(q36;q34;q11)[25] |
| 5 | M | 66 | 46,XY,t(5;9;22)(q11.2;q34;q11)[25] |
| 6 | F | 17 | 46,XX,t(4;9;22)(q28;q34;q11)[16] |
| 7 | F | 70 | 46,XX,t(9;22;12)(q34;q11;q24)[20] |
| 8 | F | 54 | 46,XX,t(1;9;22)(p36;q34;q11)[20] |
| 9 | F | 55 | 46,XX,t(3;9;22;14;17)(p22;q34;q11;q13;q12)[25] |
| 10 | F | 45 | 46,XX,t(1;9;22;19)(q32;q34;q11;p13)[20] |
| 11 | F | 53 | 46,XX,t(7;22)(q22;q11)[20] |
| 12 | F | 23 | 46,XX,t(5;6;9;22)(q35;p12;q34;q11)[25] |
| 13 | M | 30 | 46,XY,t(9;22;12)(q34;q11;q13)[20] |
| 14 | F | 67 | 46,XX,t(9;22;17)(q34;q11;q12)[25] |
| 15 | F | 51 | 46,XX,t(9;22;19)(q34;q11;q13.3)[25] |
| 16 | F | 50 | 46,XX,t(3;9;22)(p21;q34;q11)[20] |
| 17 | F | 65 | 46,XX,t(9;22;15)(q34;q11;q22)[20] |
| 18 | F | 45 | 46,XX,t(9;22;16)(q34;q11;q22)[21] |
| 19 | M | 56 | 46,XY,t(5;9;22)(p14;q34;q11)[25] |
| 20 | F | 61 | 46,XX,t(9;22;19)(q34;q11;p13)[19]/46,XX[1] |
| 21 | M | 62 | 46,XY,t(1;9;22)(p22;q34;q11)[24]/46,XY[1] |
| 22 | M | 16 | 46,XY,t(2;9;22)(q21;q34;q11)[10] |
| 23 | M | 49 | 46,XY,t(9;22;14)(q34;q11;q32)[25] |
| 24 | F | 65 | 46,XX,t(3;9;22)(q27;q34;q11)[20] |
| 25 | F | 58 | 46,XX,t(3;9;22)(q29;q34;q11)[20] |
| 26 | F | 58 | 46,XX,t(11;22)(q13;q11)[13] |
| 27 | F | 48 | 46,XX,der(1)t(1;22)(p36;q11),der(2)t(2;13)(p16;?)dup(2)(q11q31),del(2)(p13),der(22)t(2;22)(p13;q11)[15] |
| 28 | F | 61 | 46,XX,t(9;22;13)(q34;q11;q13)[20]/46,XX[1] |
| 29 | M | 52 | 46,XY,t(11;22)(q13;q11)[15] |
| 30 | F | 66 | 46,XX,t(1;9;22)(q32;q34;q11)[20] |
| 31 | M | 53 | 46,XY,t(9;22;12;17;17;22)(q34;q11;q15;q11;q23)[10] |
| 32 | M | 71 | 46,XY,t(8;9;22)(q21;q34;q11)[15] |
| 33 | M | 59 | 46,XY,t(9;15;22)(q34;q15;q11)[24]/46,XY[1] |
| 34 | M | 37 | 46,XY,t(9;22;15)(q34;q11;p11)[37] |
| 35 | M | 56 | 46,XY,t(9;22;11;15)(q34;q11;p15;q24) [20] |
| 36 | M | 57 | 46,XY,t(8;9;22)(q22;q34;q11)[15] |
| 37 | M | 47 | 46,XY,t(9;22;16)(q34;q11;q23)[15] |
| 38 | F | 47 | 46,XX,t(3;9;22)(q21;q34;q11)[15] |
| 39 | F | 65 | 46,XX,t(7;9;22)(q32;q34;q11)[20] |
| 40 | M | 18 | 46,XY,t(1;9;19;22)(q21;q34;p13;q11)[15] |
| 41 | M | 27 | 46,XY,t(9;22;17;17)(q34;q11;q25;q12)[15] |
| 42 | F | 54 | 46,XX,t(5;9;22)(p15;q34;q11)[15]/46,XX[6] |
| 43 | F | 35 | 46,XX,t(5;22)(q35;q11)[20] |
| 44 | M | 67 | 45,X,−Y,t(2;9;22)(p13;q34;q11)[20] |
| 45 | F | 41 | 46,XX,t(8;9;22)(q24;q34;q11)[10] |
| 46 | M | 74 | 46,XY.t(6;9;22)(p2?5;34;q11,2)[11] |
| 47 | M | 47 | 46,XY,t(9;22;17)(q34;q11;p12)[25] |
| 48 | M | 88 | 46,XY,t(2;9;22)(p13;q34;q11) [20] |
| 49 | M | 67 | 46,XY,t(9;22;13)(q34;q11;p11)[20] |
| 50 | M | 47 | 46,XY,t(6;9;22)(p12–21.1;q34;q11)[15] |
| 51 | F | 65 | 46,XX,t(1;9;22)(p36;q34;q11)[25] |
| 52 | M | 68 | 46,XY,t(22;2;9)(q11;p1?3;q34)[12]/46,XY[2] |
| 53 | F | 56 | 46,XX,t(22;2;9)(q11;p13;q34)[15] |
| 54 | F | 43 | 46,XX,t(7;7)(p22;q22),t(9;22;9)(q34;q11;p24)[16]/46,XX[4] |
| 55 | M | 60 | 46,XY,t(2;9;22)(p11;q34;q11)[30] |
| 56 | M | 76 | 46,XY,t(3;9;22;12)(p13;q34;q11;p13)[22] |
| 57 | M | 63 | 46,XY,t(9;22;15)(q34;q11;q12)[19]/46,XY[2] |
| 58 | M | 54 | 46,XY,t(9;22;9)(q34;q11;21)[15] |
| 59 | F | 47 | 46,XX,t(9;15;22) (q23;q23;q11)[25] |
| 60 | M | 58 | 46,XY,t(4;9;22)(p15;q34;q11)[14]/46,XY[1] |
| 61 | M | 40 | 46,XY,t(9;22;16)(q34;q11;q24) [20] |
| 62 | F | 56 | 46,XX,t(1;22;9;15;1)(q21;q11;q34;q22;q21)[10] |
| 63 | F | 18 | 46,XX,t(8;9)(q24;p22),der(9)del(9)(q13;q22)t(8;9)(q24;p22),der(20)t(9;20)(q13q22;q13);t(?;9;22)(?;q34;q11)[16]/47,XX,idem,+10[2] |
| 64 | M | 32 | 46,XY,t(9;22;17)(q34;q11;q22)[20] |
| 65 | M | 31 | 46,XY,t(9;22;10)(q34;q11;p15)[13];55,XY,+3,+8,t(9;22;10)(q34;q11;p15),+12,+13,+14,+18,+19,+21,+der(22)t(9;22;10)(q34;q11;p15)[7] |
| 66 | M | 67 | 46,XY,t(9;12;22)(q34;q24;q11.2)[20] |
| 67 | M | 42 | 46,XY,t(9;15;22)(q34;q22;q11)[25] |
| 68 | M | 55 | 46,XY,t(1;12)(p34;q24),t(1;9;22)(p36;q34;q11)[21]/48,idem,+8,+9[2] |
| 69 | M | 42 | 46,XY,t(9;22;10)(q34;q11;q24)[20] |
Cytogenetic analyses were made and interpreted according to ISCN 2009.29
No. indicates number of patients; F, female; M, male; and CP, chronic phase.
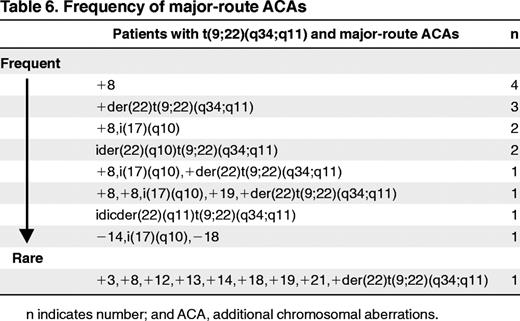
Seventy-three of 79 ACA patients had ACA in addition to t(9;22) and 6 in addition to t(v;22) (Tables 3 and 4). Twenty-five of the 41 patients (2.2% of total) with ACAs except −Y (Table 3 patients 1-25) showed minor-route alterations such as t(3;12), t(4;6), t(2;16), or t(1;21). Sixteen patients (1.4%) had major-route aberrations such as +8, +der(22)t(9;22)(q34;q11), ider(22)(q10) t(9;22)(q34,q11), and isochromosome(17)(q10) (Table 3 patients 26-41). In patients with major-route ACAs, trisomy 8 was the most frequent additional alteration (n = 9) (Tables 5 and 6). +der(22)t(9;22)(q34;q11) was observed in 6 patients, isochromosome i(17) (q10) in 5 patients, and ider(22)(q10)t(9;22)(q34;11) in 3 patients. Trisomy 19 was observed in 2 patients. The median percentage of metaphases with major-route ACAs was 59% (range 8%-100%; Table 5) and with minor-route ACAs, 85% (range 15%-100%). The frequency of major-route ACAs is summarized in Table 6. Twenty-seven of the 41 patients showed only the aberrant clone, whereas in 14 patients, normal metaphases (46,XX or 46,XY) existed in parallel to the aberrant clone. In 6 of 16 patients with major-route ACAs and in 5 of 25 patients with minor-route ACAs, complex aberrant karyotypes with 3 or more alterations were detected.
Cytogenetic data of CP-CML patients with ACAs in Ph+ cells at diagnosis
| No. . | Sex . | Age at diagnosis, y . | Cytogenetic aberrations in addition to the t(9;22)(q34;q11) or the variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 60 | 46,XY,t(9;22)(q34;q11),del(15)(q22),add(17)(p11)[8] |
| 2 | F | 53 | 46,XX,t(3;12)(p14;q23),t(9;22)(q34;q11)[16]/46,XX[4] |
| 3 | M | 48 | 46,XY,t(4;6)(q21;p23),t(9;22)(q34;q11)[20] |
| 4 | M | 47 | 46,XY,t(9;22)(q34;q11),t(14;17)(p11;p11)[20] |
| 5 | M | 46 | 46,XY,der(9)t(9;22)(q34;q11)del(9)(q33q34),del(22)(q11q12), der(22)t(9;22)(q34;q11)[20] |
| 6 | M | 27 | 45,XY,t(9;22)(q34;q11),-21[4]/46,XY[16] |
| 7 | F | 52 | 46,XX,t(2;16)(p2?3;p1?3),t(9;22)(q34;q11)[24] |
| 8 | M | 30 | 46,XY,der(9)inv(9)(p22q34)t(9;22)(q34;q11),der(22)t(9;22)(q34;q11)[15] |
| 9 | F | 56 | 46,XX,t(1;21)(q21;q22),t(9;22)(q34;q11)[20] |
| 10 | F | 60 | 46,XX,t(1;9)(q24;q31)t(9;22)(q34;q11)[15]/46,XX[5] |
| 11 | F | 46 | 46,XX,der(2)t(2;4)(q37;q21),del(4)(q21),t(9;22)(q34;q11)[20] |
| 12 | F | 56 | 46,XX,der(9)add(9)(p23)t(9;22)(q34;q11)inv(9)(p23q34),der(22)t(9;22)(q34;q11)[15] |
| 13 | F | 70 | 46,XX,t(9;22)(q34;q11),t(10;22)(q25;q13)[17] |
| 14 | M | 49 | 92,XXYY,i(7)(q10)x2,t(9;22)(q34;q11)x2[5]/46,XY[14] |
| 15 | F | 48 | 46,XX,der(1)t(1;22)(p36;q11),der(2)t(2;13)(p16;?)dup(2)(q11q31),del(2)(p13),der(22)t(2;22)(p13;q11)[15] |
| 16 | F | 43 | 46,XX,t(7;7)(p22;q22),t(9;22;9)(q34;q11;p24)[16]/46,XX[20] |
| 17 | F | 38 | 46,XX,del(1)(q21),t(9;22;1)(q34;q11;q21)[20] |
| 18 | F | 18 | 46,XX,t(8;9)(q24;p22),der(9)del(9)(q13q22)t(8;9)(q24;p22),der(20)t(9;20)(q13q22;q13);t(?;9;22)(?;q34;q11)[16]/47,XX,idem,+10[2] |
| 19 | F | 61 | 46,XX,t(9;22)(q34;q11)[21]/46,XX,del(6)(q15q23),t(9;22)(q34;q11)[4] |
| 20 | F | 68 | 46,XX,del(5)(q13q22),t(9;22)(q34;q11)[24] |
| 21 | M | 42 | 46,XY,inv(3)(p13q25),t(9;22)(q34;q11)[17] |
| 22 | M | 36 | 46,XY,t(9;22)(q34;q11),t(11;19)(q14.1;q13)[16] |
| 23 | F | 49 | 46,XX,del(3)(p11p21),t(9,22)(q34;q11)[13] |
| 24 | M | 44 | 46,XY,t(9;22)(q34;q11)[17]/46,XY,dup(1)(q31q21),t(9;22)(q34;q11)[3] |
| 25 | F | 47 | 46,XX,t(9;22)(q34;q11)[2]/46,XX,der(7;11)ins(7;11)(p14;p11q25)t(7;11)(p22;p11),t(9;22)(q34;q11)[13] |
| 26 | M | 59 | 46,XY,der(9)t(9;22)(q34;q11),ider(22)(q10)t(9;22)(q34;q11)[11]/46,XY[14] |
| 27 | M | 39 | 48,XY,+8,t(9;22)(q34;q11),i(17)(q10),+der(22)t(9;22)(q34;q11)[10]/46,XY[4] |
| 28 | M | 72 | 47,XY,+8,t(9;22)(q34;q11)[20] |
| 29 | M | 28 | 50,XY,+8,+8,t(9;22)(q34;q11),i(17)(q10),+19,+der(22)t(9;22)(q34;q11)[6]/46,XX[22] |
| 30 | M | 53 | 46,XY,der(9)t(9;22)(q34;q11),idicder(22)(q11)t(9;22)(q34;q11)[22]/46,XY[3] |
| 31 | M | 40 | 47,XY,+8,t(9;22)(q34;q11)[19]/46,XY[6] |
| 32 | M | 41 | 44,XY,t(9;22)(q34;q11),-14,i(17)(q10),-18[15]/46,XY[11] |
| 33 | M | 23 | 47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[2]/46,XY[18] |
| 34 | F | 59 | 47,XX,+8,t(9;22)(q34;q11)[2]/46,XX[23] |
| 35 | M | 48 | 46,XY,t(9;22)(q34;q11)[6]/47,XY,+8,t(9;22)(q34;q11)[12]/46,XY[2] |
| 36 | M | 31 | 46,XY,t(9;22;10)(q34;q11;p15)[13];55,XY,+3,+8,t(9;22;10)(q34;q11;p15),+12,+13,+14,+18,+19,+21,+der(22)t(9;22;10)(q34;q11;p15)[7] |
| 37 | M | 37 | 46,XY,t(9;22)(q34;q11)[10]/47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[10] |
| 38 | M | 28 | 47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[20] |
| 39 | M | 51 | 46,XY,t(9;22)(q34;q11)[8]/47,XY,+8,t(9;22)(q34;q11),i(17)(q10)[18] |
| 40 | F | 52 | 47,XX,+8,t(9;22)(q34,q11),i(17)(q10)[10] |
| 41 | F | 69 | 46,XX,t(9;22)(q34;q11)[9]/46,XX,der(9)t(9;22)(q34;q11),ider(22)(q10)t(9;22)(q34;q11)[13] |
| No. . | Sex . | Age at diagnosis, y . | Cytogenetic aberrations in addition to the t(9;22)(q34;q11) or the variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 60 | 46,XY,t(9;22)(q34;q11),del(15)(q22),add(17)(p11)[8] |
| 2 | F | 53 | 46,XX,t(3;12)(p14;q23),t(9;22)(q34;q11)[16]/46,XX[4] |
| 3 | M | 48 | 46,XY,t(4;6)(q21;p23),t(9;22)(q34;q11)[20] |
| 4 | M | 47 | 46,XY,t(9;22)(q34;q11),t(14;17)(p11;p11)[20] |
| 5 | M | 46 | 46,XY,der(9)t(9;22)(q34;q11)del(9)(q33q34),del(22)(q11q12), der(22)t(9;22)(q34;q11)[20] |
| 6 | M | 27 | 45,XY,t(9;22)(q34;q11),-21[4]/46,XY[16] |
| 7 | F | 52 | 46,XX,t(2;16)(p2?3;p1?3),t(9;22)(q34;q11)[24] |
| 8 | M | 30 | 46,XY,der(9)inv(9)(p22q34)t(9;22)(q34;q11),der(22)t(9;22)(q34;q11)[15] |
| 9 | F | 56 | 46,XX,t(1;21)(q21;q22),t(9;22)(q34;q11)[20] |
| 10 | F | 60 | 46,XX,t(1;9)(q24;q31)t(9;22)(q34;q11)[15]/46,XX[5] |
| 11 | F | 46 | 46,XX,der(2)t(2;4)(q37;q21),del(4)(q21),t(9;22)(q34;q11)[20] |
| 12 | F | 56 | 46,XX,der(9)add(9)(p23)t(9;22)(q34;q11)inv(9)(p23q34),der(22)t(9;22)(q34;q11)[15] |
| 13 | F | 70 | 46,XX,t(9;22)(q34;q11),t(10;22)(q25;q13)[17] |
| 14 | M | 49 | 92,XXYY,i(7)(q10)x2,t(9;22)(q34;q11)x2[5]/46,XY[14] |
| 15 | F | 48 | 46,XX,der(1)t(1;22)(p36;q11),der(2)t(2;13)(p16;?)dup(2)(q11q31),del(2)(p13),der(22)t(2;22)(p13;q11)[15] |
| 16 | F | 43 | 46,XX,t(7;7)(p22;q22),t(9;22;9)(q34;q11;p24)[16]/46,XX[20] |
| 17 | F | 38 | 46,XX,del(1)(q21),t(9;22;1)(q34;q11;q21)[20] |
| 18 | F | 18 | 46,XX,t(8;9)(q24;p22),der(9)del(9)(q13q22)t(8;9)(q24;p22),der(20)t(9;20)(q13q22;q13);t(?;9;22)(?;q34;q11)[16]/47,XX,idem,+10[2] |
| 19 | F | 61 | 46,XX,t(9;22)(q34;q11)[21]/46,XX,del(6)(q15q23),t(9;22)(q34;q11)[4] |
| 20 | F | 68 | 46,XX,del(5)(q13q22),t(9;22)(q34;q11)[24] |
| 21 | M | 42 | 46,XY,inv(3)(p13q25),t(9;22)(q34;q11)[17] |
| 22 | M | 36 | 46,XY,t(9;22)(q34;q11),t(11;19)(q14.1;q13)[16] |
| 23 | F | 49 | 46,XX,del(3)(p11p21),t(9,22)(q34;q11)[13] |
| 24 | M | 44 | 46,XY,t(9;22)(q34;q11)[17]/46,XY,dup(1)(q31q21),t(9;22)(q34;q11)[3] |
| 25 | F | 47 | 46,XX,t(9;22)(q34;q11)[2]/46,XX,der(7;11)ins(7;11)(p14;p11q25)t(7;11)(p22;p11),t(9;22)(q34;q11)[13] |
| 26 | M | 59 | 46,XY,der(9)t(9;22)(q34;q11),ider(22)(q10)t(9;22)(q34;q11)[11]/46,XY[14] |
| 27 | M | 39 | 48,XY,+8,t(9;22)(q34;q11),i(17)(q10),+der(22)t(9;22)(q34;q11)[10]/46,XY[4] |
| 28 | M | 72 | 47,XY,+8,t(9;22)(q34;q11)[20] |
| 29 | M | 28 | 50,XY,+8,+8,t(9;22)(q34;q11),i(17)(q10),+19,+der(22)t(9;22)(q34;q11)[6]/46,XX[22] |
| 30 | M | 53 | 46,XY,der(9)t(9;22)(q34;q11),idicder(22)(q11)t(9;22)(q34;q11)[22]/46,XY[3] |
| 31 | M | 40 | 47,XY,+8,t(9;22)(q34;q11)[19]/46,XY[6] |
| 32 | M | 41 | 44,XY,t(9;22)(q34;q11),-14,i(17)(q10),-18[15]/46,XY[11] |
| 33 | M | 23 | 47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[2]/46,XY[18] |
| 34 | F | 59 | 47,XX,+8,t(9;22)(q34;q11)[2]/46,XX[23] |
| 35 | M | 48 | 46,XY,t(9;22)(q34;q11)[6]/47,XY,+8,t(9;22)(q34;q11)[12]/46,XY[2] |
| 36 | M | 31 | 46,XY,t(9;22;10)(q34;q11;p15)[13];55,XY,+3,+8,t(9;22;10)(q34;q11;p15),+12,+13,+14,+18,+19,+21,+der(22)t(9;22;10)(q34;q11;p15)[7] |
| 37 | M | 37 | 46,XY,t(9;22)(q34;q11)[10]/47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[10] |
| 38 | M | 28 | 47,XY,t(9;22)(q34;q11),+der(22)t(9;22)(q34;q11)[20] |
| 39 | M | 51 | 46,XY,t(9;22)(q34;q11)[8]/47,XY,+8,t(9;22)(q34;q11),i(17)(q10)[18] |
| 40 | F | 52 | 47,XX,+8,t(9;22)(q34,q11),i(17)(q10)[10] |
| 41 | F | 69 | 46,XX,t(9;22)(q34;q11)[9]/46,XX,der(9)t(9;22)(q34;q11),ider(22)(q10)t(9;22)(q34;q11)[13] |
ACAs except −Y. Patients 1-25 show minor-route ACAs in addition to the standard or variant translocation; patients 26-41 show major-route ACAs. Cytogenetic analyses were made and interpreted according to ISCN 2009.29
No. indicates number of patients; F, female; M, male; and CP, chronic phase.
Cytogenetic data from CP-CML patients with −Y in Ph+ cells at diagnosis
| No. . | Sex . | Age at diagnosis, y . | −Y in addition to the t(9;22)(q34;q11) or the variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 48 | 45,X,−Y,t(9;22)(q34;q11)[13]/46,XY[12] |
| 2 | M | 50 | 45,X,−Y,t(9;22)(q34;q11)[23]/46,XY[2] |
| 3 | M | 44 | 45,X,−Y,t(9;22)(q34;q11)[5]/46,XY[6] |
| 4 | M | 40 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 5 | M | 56 | 45,X,−Y,t(9;22)(q34;q11)[3]/46,XY[22] |
| 6 | M | 65 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 7 | M | 42 | 45,X,−Y,t(9;22)(q34;q11)[13]/46,XY[7] |
| 8 | M | 70 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 9 | M | 59 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 10 | M | 58 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 11 | M | 74 | 45,X,−Y,t(9;22)(q34;q11)[14]/46,XY[11] |
| 12 | M | 70 | 45,X,−Y,t(9;22)(q34;q11)[12]/46,XY[5] |
| 13 | M | 75 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 14 | M | 81 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 15 | M | 32 | 45,X,−Y,t(9;22)(q34;q11)[17]/46,XY[8] |
| 16 | M | 68 | 45,X,−Y,t(9;22)(q34;q11)[18]/46,XY[2] |
| 17 | M | 60 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 18 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[18] |
| 19 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[3]/46,XY[22] |
| 20 | M | 63 | 45,X,−Y,t(9;22)(q34;q11)[4]/46,XY[16] |
| 21 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[6]/46,XY[14] |
| 22 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[6]/46,XY[19] |
| 23 | M | 30 | 45,X,−Y,t(9;22)(q34;q11)[7]/46,XY[15] |
| 24 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[19]/46,XY[5] |
| 25 | M | 79 | 45,X,−Y,t(9;22)(q34;q11)[17]/46,XY[3] |
| 26 | M | 67 | 45,X,−Y,t(9;22)(q34;q11)[16] |
| 27 | M | 67 | 45,X,−Y,t(2;9;22)(p13;q34;q11)[20] |
| 28 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 29 | M | 56 | 45,X,−Y,t(9;22)(q34;q11)[30] |
| 30 | M | 52 | 45,X,−Y,t(9;22)(q34;q11)[7] |
| 31 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 32 | M | 72 | 45,X,−Y,t(9;22)(q34;q11)[14] |
| 33 | M | 68 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 34 | M | 67 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 35 | M | 73 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 36 | M | 78 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 37 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 38 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| No. . | Sex . | Age at diagnosis, y . | −Y in addition to the t(9;22)(q34;q11) or the variant t(v;22) at diagnosis [no. of analyzed metaphases] . |
|---|---|---|---|
| 1 | M | 48 | 45,X,−Y,t(9;22)(q34;q11)[13]/46,XY[12] |
| 2 | M | 50 | 45,X,−Y,t(9;22)(q34;q11)[23]/46,XY[2] |
| 3 | M | 44 | 45,X,−Y,t(9;22)(q34;q11)[5]/46,XY[6] |
| 4 | M | 40 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 5 | M | 56 | 45,X,−Y,t(9;22)(q34;q11)[3]/46,XY[22] |
| 6 | M | 65 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 7 | M | 42 | 45,X,−Y,t(9;22)(q34;q11)[13]/46,XY[7] |
| 8 | M | 70 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 9 | M | 59 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 10 | M | 58 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 11 | M | 74 | 45,X,−Y,t(9;22)(q34;q11)[14]/46,XY[11] |
| 12 | M | 70 | 45,X,−Y,t(9;22)(q34;q11)[12]/46,XY[5] |
| 13 | M | 75 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 14 | M | 81 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 15 | M | 32 | 45,X,−Y,t(9;22)(q34;q11)[17]/46,XY[8] |
| 16 | M | 68 | 45,X,−Y,t(9;22)(q34;q11)[18]/46,XY[2] |
| 17 | M | 60 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 18 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[18] |
| 19 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[3]/46,XY[22] |
| 20 | M | 63 | 45,X,−Y,t(9;22)(q34;q11)[4]/46,XY[16] |
| 21 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[6]/46,XY[14] |
| 22 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[6]/46,XY[19] |
| 23 | M | 30 | 45,X,−Y,t(9;22)(q34;q11)[7]/46,XY[15] |
| 24 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[19]/46,XY[5] |
| 25 | M | 79 | 45,X,−Y,t(9;22)(q34;q11)[17]/46,XY[3] |
| 26 | M | 67 | 45,X,−Y,t(9;22)(q34;q11)[16] |
| 27 | M | 67 | 45,X,−Y,t(2;9;22)(p13;q34;q11)[20] |
| 28 | M | 62 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 29 | M | 56 | 45,X,−Y,t(9;22)(q34;q11)[30] |
| 30 | M | 52 | 45,X,−Y,t(9;22)(q34;q11)[7] |
| 31 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 32 | M | 72 | 45,X,−Y,t(9;22)(q34;q11)[14] |
| 33 | M | 68 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 34 | M | 67 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 35 | M | 73 | 45,X,−Y,t(9;22)(q34;q11)[25] |
| 36 | M | 78 | 45,X,−Y,t(9;22)(q34;q11)[15] |
| 37 | M | 61 | 45,X,−Y,t(9;22)(q34;q11)[20] |
| 38 | M | 66 | 45,X,−Y,t(9;22)(q34;q11)[25] |
All patients show −Y in addition to the translocation t(9;22)(q34;q11) or to a variant translocation (patient 27); patients 1-28 show no metaphases with −Y under CR (karyotypes 46,XY), whereas patients 29-38 show cells without t(9;22)(q34;q11) in CR but with −Y (45,X,−Y). Cytogenetic analyses were made and interpreted according to ISCN 2009.29
No. indicates number of patients; M, male; and CP, chronic phase.
Characteristics of CP-CML patients with major-route ACAs
| No acc. to Table 3 . | Trisomy 8 . | Karyotype (short) in addition to t(9;22)(q34;q11) . | Metaphases with ACAs, % . | Complex . | Best response . | Follow-up, y . | Course . | Comments . |
|---|---|---|---|---|---|---|---|---|
| 26 | ider(22) | 44 | CMR4 | 6,9 | No progress | Alive in CMR4 | ||
| 27 | 1 | +8,i(17)(q10),+der(22) | 71 | 1 | none | 1,7 | BC* | SCT after BC |
| 28 | 1 | +8 | 100 | CMR4 | 5,7 | * | Died in remission, no follow-up | |
| 29 | 1 | +8,+8,i(17)(q10),+19,+der(22) | 21 | 1 | none | 6,9 | BC | Alive after SCT |
| 30 | idicder(22) | 88 | CCR | 6,5 | Imatinib failure | Alive after SCT | ||
| 31 | 1 | +8 | 76 | CHR | 7,2 | Alive after SCT | ||
| 32 | -14,i(17)(q10),-18 | 58 | 1 | none | 0,4 | BC* | ||
| 33 | +der(22) | 10 | CHR | 3,2 | Imatinib failure* | SCT | ||
| 34 | 1 | +8 | 8 | CMR4 | 6,1 | No progress | Alive in CMR4 | |
| 35 | 1 | +8 | 60 | none | 2,7 | BC* | ||
| 36 | 1 | +3,+8,+12,+13,+14,+18,+19, +21,+der(22) | 35 | 1 | none | 4,9 | AP | Alive after SCT |
| 37 | +der(22) | 50 | none | 1,3 | Sepsis* | Imatinib failure | ||
| 38 | +der(22) | 100 | CMR4 | 3,0 | CP | Alive in MMR | ||
| 39 | 1 | +8,i(17)(q10) | 69 | 1 | MMR | 3,3 | No progress | Alive in MMR |
| 40 | 1 | +8,i(17)(q10) | 100 | 1 | CMR4 | 3,5 | BC* | SCT after BC |
| 41 | ider(22) | 59 | CHR | 3,8 | * | Aplasia after imatinib failure | ||
| Total | 9 | 6 | 8 patients died |
| No acc. to Table 3 . | Trisomy 8 . | Karyotype (short) in addition to t(9;22)(q34;q11) . | Metaphases with ACAs, % . | Complex . | Best response . | Follow-up, y . | Course . | Comments . |
|---|---|---|---|---|---|---|---|---|
| 26 | ider(22) | 44 | CMR4 | 6,9 | No progress | Alive in CMR4 | ||
| 27 | 1 | +8,i(17)(q10),+der(22) | 71 | 1 | none | 1,7 | BC* | SCT after BC |
| 28 | 1 | +8 | 100 | CMR4 | 5,7 | * | Died in remission, no follow-up | |
| 29 | 1 | +8,+8,i(17)(q10),+19,+der(22) | 21 | 1 | none | 6,9 | BC | Alive after SCT |
| 30 | idicder(22) | 88 | CCR | 6,5 | Imatinib failure | Alive after SCT | ||
| 31 | 1 | +8 | 76 | CHR | 7,2 | Alive after SCT | ||
| 32 | -14,i(17)(q10),-18 | 58 | 1 | none | 0,4 | BC* | ||
| 33 | +der(22) | 10 | CHR | 3,2 | Imatinib failure* | SCT | ||
| 34 | 1 | +8 | 8 | CMR4 | 6,1 | No progress | Alive in CMR4 | |
| 35 | 1 | +8 | 60 | none | 2,7 | BC* | ||
| 36 | 1 | +3,+8,+12,+13,+14,+18,+19, +21,+der(22) | 35 | 1 | none | 4,9 | AP | Alive after SCT |
| 37 | +der(22) | 50 | none | 1,3 | Sepsis* | Imatinib failure | ||
| 38 | +der(22) | 100 | CMR4 | 3,0 | CP | Alive in MMR | ||
| 39 | 1 | +8,i(17)(q10) | 69 | 1 | MMR | 3,3 | No progress | Alive in MMR |
| 40 | 1 | +8,i(17)(q10) | 100 | 1 | CMR4 | 3,5 | BC* | SCT after BC |
| 41 | ider(22) | 59 | CHR | 3,8 | * | Aplasia after imatinib failure | ||
| Total | 9 | 6 | 8 patients died |
ACA indicates additional chromosomal aberrations; SCT, stem cell transplantation; BC, blast crisis; CMR4; complete molecular response; MMR, major molecular remission; AP, accelerated phase; CCR, complete cytogenetic remission; CP, chromic phase; and CHR, complete haematological remission.
Patient died.
Time to CCR and MMR
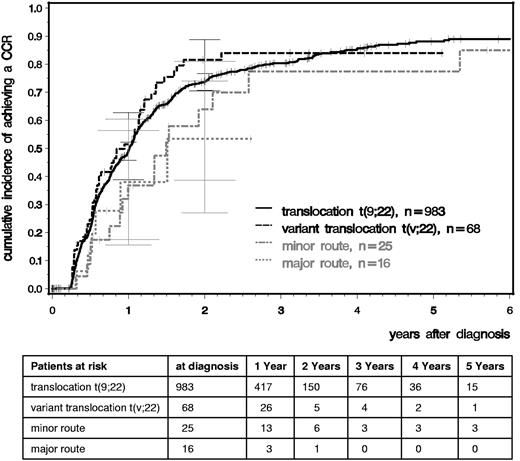
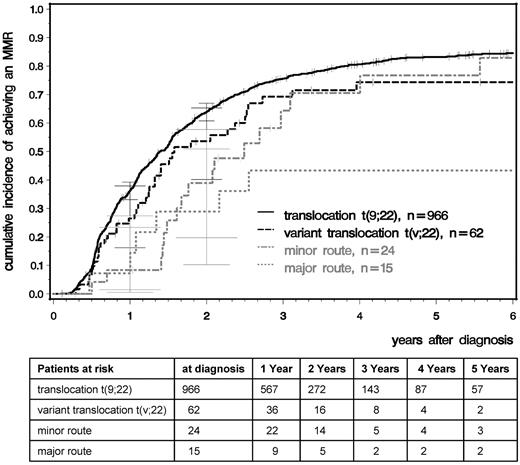
We analyzed time to CCR and MMR comparing patients with standard t(9;22), variant t(v;22), −Y, and major- and minor-route ACAs (Figures 2 and 3). After a median observation time of 5.3 years, the median times to CCR were 1.01 years for t(9,22), 0.95 for t(v,22), 0.98 for −Y, and 1.49 for minor-route ACAs (Figure 2). The median times to MMR were 1.40, 1.58, 1.65, and 2.49 years, respectively (Figure 3). No difference in the cumulative incidence of CCR or MMR was seen among patients with standard t(9;22), variant translocation, −Y (not shown), and minor-route ACAs (Figures 2 and 3).
Cumulative incidences of CCR for patients with t(9;22), t(v;22), and minor- and major-route ACAs estimated by the cumulative incidence function considering competing risks.
Cumulative incidences of CCR for patients with t(9;22), t(v;22), and minor- and major-route ACAs estimated by the cumulative incidence function considering competing risks.
Cumulative incidences of MMR for patients with t(9;22), t(v;22), and minor- and major-route ACAs estimated by the cumulative incidence function considering competing risks.
Cumulative incidences of MMR for patients with t(9;22), t(v;22), and minor- and major-route ACAs estimated by the cumulative incidence function considering competing risks.
For the major-route ACA group, the number of patients with MMR and CCR was lower and the time to remission was deferred. However, patient numbers were too small to obtain reliable probability estimations because the observations of MMR and CCR depend very much on the numbers and times of evaluation.
Survival
OS and PFS were significantly inferior in patients with major-route ACAs compared with patients with standard t(9;22), t(v;22), −Y, or minor-route ACAs (Figures 4 and 5). The 5-year PFS of standard t(9;22), t(v;22), −Y, minor-route ACAs, and major-route ACAs was 90%, 81%, 88%, 96%, and 50% (P < .001) and the 5-year OS was 92%, 87%, 91%, 96%, and 53% (P < .001), respectively. Variant t(v;22) and −Y had no influence on PFS or OS.
PFS in the t(9;22), t(v;22), −Y, and minor- and major-route ACA groups calculated by the Kaplan-Meier method and compared by the log-rank test. Patients were censored at the date of last follow-up. The difference between the standard and the major-route groups was significant at P < .001.
PFS in the t(9;22), t(v;22), −Y, and minor- and major-route ACA groups calculated by the Kaplan-Meier method and compared by the log-rank test. Patients were censored at the date of last follow-up. The difference between the standard and the major-route groups was significant at P < .001.
OS in the t(9;22), t(v;22), and minor- and major-route ACA groups calculated by the Kaplan-Meier method and compared by the log-rank test. Patients were censored at the last follow-up. The difference between the standard and the major-route groups was significant at P < .001.
OS in the t(9;22), t(v;22), and minor- and major-route ACA groups calculated by the Kaplan-Meier method and compared by the log-rank test. Patients were censored at the last follow-up. The difference between the standard and the major-route groups was significant at P < .001.
Comparing the groups with major- and minor-route ACAs, 24 of the 25 patients with minor-route ACAs were still alive, whereas 8 of 16 patients with major-route ACAs died (P < .01). Six of these 8 had trisomy 8, 1 an additional Ph chromosome, and 1 ider(22) (Table 3 patients 33 and 41 and Table 5). Eight patients with major-route ACAs were alive with a median observation time of 6.5 years, 5 with trisomy 8, 1 with a double Ph chromosome (Table 3 patient 38), and 2 with ider(22) (patients 26 and 30). Of the 5 living patients with trisomy 8, 3 received an allogeneic stem cell transplantation (SCT), 2 after progression to AP and BC. Two patients were alive under imatinib in complete molecular remission (CMR). Patients 26 and 38 (Table 3) were in MMR and CMR under imatinib treatment and patient 30 received a SCT after imatinib failure.
−Y
In 28 patients with −Y (Table 4 patients 1-28), the aberrant Ph+ clone, including −Y, disappeared during therapy. In CCR, all analyzed cells showed the normal male karyotype 46,XY. In 10 patients (Table 4 patients 29-38), the clone with t(9;22) disappeared in CCR, but a clone with −Y was still detectable.
Discussion
Our data from 1151 uniformly diagnosed and treated patients from a large, randomized study show that the prognostic impact of ACAs at diagnosis, as determined by time to CCR and MMR and PFS and OS, depends on the type of ACA. Probably the most relevant finding is the highly significant negative impact of major-route ACAs at diagnosis on time to CCR and MMR and PFS and OS. In contrast to MMR and CCR, the observation of which is very much dependent on the number and times of evaluation, the date of death is known exactly; therefore, because of its concomitant effects, AP and BC cannot be missed and will be noticed quite promptly. Together with a 3-year minimum observation time of living patients for PFS and for OS (median observation times 6.1 and 6.5 years, respectively) in the major-route ACA group, the significantly worse results of this group are therefore quite meaningful, even though the number of cases was small at only 16 patients. All other additional cytogenetic findings at diagnosis (ie, variant translocation, −Y, and minor-route ACAs) had no recognizable impact on prognosis. Trisomy 8 was the most common observation, mostly in combination with other aberrations, followed by a second Ph chromosome and isochromosome(17)(q10).
Our findings are in agreement with earlier observations on smaller numbers of patients20 and reconcile the discrepancies in prognostic impact between ACAs at diagnosis and the appearance of new ACAs under treatment.16 This correlation likely required the large patient number with a sufficiently long follow-up period provided by the CML Study IV. It is of interest that the median age of patients with ACAs as a group was lower than that of the other groups, which underlines the strength of their prognostic impact.
Only major-route, not minor-route ACAs, had the observed prognostic impact; therefore, the evaluation of these groups together may obscure the impact.39 Major-route ACAs involving complex cytogenetic aberrations at diagnosis appear to have a particularly poor prognosis, because 5 of 6 patients with complex karyotypes have progressed (3 of 6 have died and 2 are alive after SCT), whereas complex cytogenetic aberrations in minor-route ACAs had no recognizable impact (5 of 5 patients are alive in CCR after a median of 56 months; range, 36-100 months). We could not detect an impact of the percentage of metaphases affected by ACAs that has been reported by others.40 In comparing the percentages of metaphases affected by major-route ACAs (median of 59%) and minor-route ACAs (median of 85%), the impact of cytogenetic aberrations depends on the type of ACA independently of the percentage of metaphases.
Our patients with variant translocations treated with imatinib show the same prognosis as patients with standard t(9;22) reported by others.22,23 In the imatinib era, the prognostic impact of derivative 9 deletions, which are frequently associated with variant translocations, is similarly overruled by therapy25,41-43 as that of minor-route ACAs.
Minor-route ACAs are observed mostly sporadically, with the possible exception of t(3;21) or inv(3), and typically disappear under treatment. No clear correlation with disease progression is recognizable. Failure to separate the prognostically different groups might explain the observations on ACAs at diagnosis in the past, because no impact on prognosis can be detected if all ACAs, including major-route ACAs, minor-route ACAs, and −Y, are evaluated together.
Furthermore, patients with −Y have a prognosis not significantly different from those with standard t(9,22). Because the −Y frequently occurs in healthy elderly men, −Y after therapy in complete remission of the Ph+ clone indicates that it was not disease related. The fact that the majority (60%) of patients with −Y (Table 4 patients 1-28) showed a mosaic of aberrant and normal cells led us to the conclusion that −Y was acquired and therefore disease related. No patient in the group that lost −Y under therapy (Table 4 patients 29-38) showed a mosaic, supporting the hypothesis that −Y could be a consequence of age. Current analysis showed no differences in outcome between patients 1-28 and patients 29-38.
The nonrandom association of chromosomal aberrations with disease progression and survival is a feature also observed with other cancers and in animal tumor models, is probably required for maintenance of the malignant state, and appears to be a general principle of carcinogenesis.44,45
Although nonrandom, more than one abnormal chromosomal constellation is associated with an unfavorable prognosis and disease progression. Trisomy 8, isochromosome(17)(q10), and a second Ph chromosome are regularly found in CML progression, but in variable combinations. This chromosomal individuality of each clonal evolution and cancer has been compared with speciation in evolution.46
Taking all cytogenetic findings at diagnosis together, the proportion of patients with additional cytogenetic findings at diagnosis is 12.9%, which is still in the range of published data. The proportions of patients with variant translocations (6.0%), ACAs (3.6%), and −Y (3.3%) are also within the published range. The proportion of patients with major-route ACAs at diagnosis is small at 1.4%, but the data on their impact on prognosis are clear.
We conclude that the prognostic impact of ACAs at diagnosis of CML is heterogeneous and that consideration of their type is important. Major-route ACAs at diagnosis identify a small group of patients with significantly poorer prognosis compared with all other patients, requiring close observation and early and intensive intervention such as early SCT.47
The online version of this article contains a data supplement.
Presented in part at the 52nd Annual Meeting of the American Society of Hematology (ASH) in Orlando, FL, in December 2010; the Annual Meeting of the American Society of Clinical Oncology (ASCO) in Chicago, IL, in June 2011; and the 16th Congress of the European Hematology Association (EHA) in London, United Kingdom, in June 2011.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Angelika Adler, Gabriele Bartsch, Sabine Dean, Christine Folz, Michaela Hausmann, Ute Kossak, Elke Matzat, Barbara Müller, Regina Pleil-Lösch, Nicole Schomber, Inge Stalljann, Cornelia Willersinn, the Steering Group (Rüdiger Hehlmann, Gerhard Ehninger, Joerg Hasford, Andreas Hochhaus, Dieter Hossfeld, Hans-Jochem Kolb, Stefan Krause, Christoph Nerl, Hans Pralle, Dominik Heim, Gabriela M. Baerlocher, Hermann Heimpel), and all CML trial participants.
Authorship
Contribution: A.F., S.S., and R.H. had primary responsibility for the manuscript; A.F., A.L., A.H., M.C.M., B.H., C.H., G.G., B.S., M.J., A.R., S.J-M., U.P., J.S., W.-K.H., J.S., H.E., A.D.H., C.F., L.K., A.N., M.K., F.S., M. Pfreundschuh, C.F.W., K.S., G.M.B., M.L., M. Pfirrmann, J.H., S.S., and R.H. contributed to the design of the study, to the statistical analysis and to the interpretation of the results; and all authors checked and approved the final version of the manuscript.
Conflict-of-interest disclosure: The CML Study IV is supported by the Deutsche Krebshilfe (number 106642), Novartis (Nürnberg, Germany), Kompetenznetz für Akute and Chronische Leukämien (BMBF 01GI0270), José-Carreras Leukämiestiftung (DJCLS H09/01f, H06/04v, H03/01), and the European LeukemiaNet (LSHC-CT-2004-503216). C.H. declares employment and equity ownership to Müchner Leukämie Labor (MLL). The remaining authors declare no competing financial interests.
For a complete list of German CML Study Group participants, see the supplemental Appendix (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Correspondence: Dr Susanne Saussele, III. Medizinische Klinik, Medizinische Fakultät Mannheim, der Universität Heidelberg, Pettenkoferstrasse 22, 68169 Mannheim, Germany; e-mail: susanne.saussele@medma.uni-heidelberg.de.
References
Author notes
A.F and A.L contributed equally to this work.
S.S. and R.H. are co–senior authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal